



生物多样性 ›› 2023, Vol. 31 ›› Issue (3): 22346. DOI: 10.17520/biods.2022346 cstr: 32101.14.biods.2022346
所属专题: 生物入侵
蒲佳佳1, 杨平俊2, 戴洋3, 陶可欣1, 高磊4, 杜予州5, 曹俊3, 俞晓平1, 杨倩倩1,*( )
)
收稿日期:2022-06-24
接受日期:2022-09-05
出版日期:2023-03-20
发布日期:2022-12-30
通讯作者:
杨倩倩
作者简介:* E-mail: yqq@cjlu.edu.cn基金资助:
Jiajia Pu1, Pingjun Yang2, Yang Dai3, Kexin Tao1, Lei Gao4, Yuzhou Du5, Jun Cao3, Xiaoping Yu1, Qianqian Yang1,*( )
)
Received:2022-06-24
Accepted:2022-09-05
Online:2023-03-20
Published:2022-12-30
Contact:
Qianqian Yang
摘要: 福寿螺(Pomacea spp.)已广泛分布在我国长江以南各省, 且逐年向北扩散。本研究采集了长江下游上海及江苏分布区11个种群的福寿螺样品, 测序获得270条线粒体COI基因序列, 生成10个单倍型(Hap1–10)。基于遗传距离及系统发育分析将Hap1–9鉴定为小管福寿螺(Pomacea canaliculata), Hap10为斑点福寿螺(P. maculata)。其中, 小管福寿螺在所有采样点均有分布; AMOVA层次分析将小管福寿螺种群分成跨长江分布的3个组群, 且分子变异主要来源于组群间。进一步结合已发表的我国其他地区(大陆和香港), 以及日本和原产地阿根廷、巴西种群的福寿螺序列, 形成包含972条COI序列的数据集进行种群遗传学分析。单倍型网络分析中, 所采集的小管福寿螺种群分布于3个包含阿根廷单倍型的子网络, 其中包含Hap5和Hap7的子网络在我国首次被发现, 表明长江下游地区小管福寿螺从阿根廷多次入侵, 并发现一个新的入侵历史事件。斑点福寿螺仅在长江以北江苏地区检测到, 单倍型Hap10也是我国大陆其他地区的主要单倍型, 表明长江以北江苏地区的斑点福寿螺可能由国内已有分布区扩散而来, 均起源于巴西。我国不同地区种群的遗传多样性比较发现, 长江以南的小管福寿螺遗传多样性最高(Hd = 0.627), 而香港种群斑点福寿螺的遗传多样性最高(Hd = 0.356)。基于核EF1α基因分型分析检测表明, 所采集福寿螺的杂交种比例为52.6%, 高于原产地种群, 表明种间渐渗杂交在入侵过程中持续发生。本研究对于福寿螺监测预警及有效防控具有重要意义。
蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩 (2023) 长江下游外来生物福寿螺的种类及其种群遗传结构. 生物多样性, 31, 22346. DOI: 10.17520/biods.2022346.
Jiajia Pu, Pingjun Yang, Yang Dai, Kexin Tao, Lei Gao, Yuzhou Du, Jun Cao, Xiaoping Yu, Qianqian Yang (2023) Species identification and population genetic structure of non-native apple snails (Ampullariidea: Pomacea) in the lower reaches of the Yangtze River. Biodiversity Science, 31, 22346. DOI: 10.17520/biods.2022346.
| 序号 Code | 地点 Locality | 编号 Code | 经度 Longitude (E) | 纬度 Latitude (N) | 生境 Habitat | 序列数量 No. of sequences |
|---|---|---|---|---|---|---|
| 1 | 上海市宝山区三星村 Sanxing Village, Baoshan District, Shanghai | SHBS | 121.36° | 31.31° | 河道 River | 20 |
| 2 | 苏州市吴中区甪直镇 Luzhi Town, Wuzhong District, Suzhou City | WZLZ | 120.48° | 31.14° | 水生蔬菜田 Aquatic vegetable field | 30 |
| 3 | 苏州市吴中区马家浜村 Majiabang Village, Wuzhong District, Suzhou City | WZMJB | 120.46° | 31.14° | 水生蔬菜田 Aquatic vegetable field | 30 |
| 4 | 昆山市周庄镇 Zhouzhuang Town, Kunshan City | KSZZ | 120.56° | 31.21° | 稻田 Paddy field | 30 |
| 5 | 常熟市尚湖镇 Shanghu Town, Changshu City | CSSH | 120.42° | 31.36° | 稻田 Paddy field | 30 |
| 6 | 常熟市古里镇 Guli Town, Changshu City | CSGL | 120.85° | 31.65° | 稻田 Paddy field | 15 |
| 7 | 张家港市凤凰镇鸷山 Fenghuang Town, Zhangjiagang City | ZJGFH | 120.62° | 31.76° | 沟渠 Ditch | 17 |
| 8 | 张家港市凤凰镇杏市村 Xingshi Village, Fenghuang Town, Zhangjiagang City | ZJGXSC | 120.68° | 31.78° | 稻田 Paddy field | 28 |
| 9 | 扬州市广陵区沙头镇 Shatou Town, Guangling District, Yangzhou City | JSYZ | 119.50° | 32.34° | 河道 River | 30 |
| 10 | 泰州市兴化市大邹镇顾马村 Guma Village, Dazou Town, Xinghua City, Taizhou City | JSTZ | 119.92° | 33.15° | 河道、蟹田 River, crab field | 30 |
| 11 | 宿迁市泗洪县 Sihong County, Suqian City | JSSQ | 118.25° | 33.47° | 湖泊 Lake | 10 |
表1 长江下游分布区福寿螺样品采集信息
Table 1 Sampling information of the apple snails collected from the lower reaches of the Yangtze River
| 序号 Code | 地点 Locality | 编号 Code | 经度 Longitude (E) | 纬度 Latitude (N) | 生境 Habitat | 序列数量 No. of sequences |
|---|---|---|---|---|---|---|
| 1 | 上海市宝山区三星村 Sanxing Village, Baoshan District, Shanghai | SHBS | 121.36° | 31.31° | 河道 River | 20 |
| 2 | 苏州市吴中区甪直镇 Luzhi Town, Wuzhong District, Suzhou City | WZLZ | 120.48° | 31.14° | 水生蔬菜田 Aquatic vegetable field | 30 |
| 3 | 苏州市吴中区马家浜村 Majiabang Village, Wuzhong District, Suzhou City | WZMJB | 120.46° | 31.14° | 水生蔬菜田 Aquatic vegetable field | 30 |
| 4 | 昆山市周庄镇 Zhouzhuang Town, Kunshan City | KSZZ | 120.56° | 31.21° | 稻田 Paddy field | 30 |
| 5 | 常熟市尚湖镇 Shanghu Town, Changshu City | CSSH | 120.42° | 31.36° | 稻田 Paddy field | 30 |
| 6 | 常熟市古里镇 Guli Town, Changshu City | CSGL | 120.85° | 31.65° | 稻田 Paddy field | 15 |
| 7 | 张家港市凤凰镇鸷山 Fenghuang Town, Zhangjiagang City | ZJGFH | 120.62° | 31.76° | 沟渠 Ditch | 17 |
| 8 | 张家港市凤凰镇杏市村 Xingshi Village, Fenghuang Town, Zhangjiagang City | ZJGXSC | 120.68° | 31.78° | 稻田 Paddy field | 28 |
| 9 | 扬州市广陵区沙头镇 Shatou Town, Guangling District, Yangzhou City | JSYZ | 119.50° | 32.34° | 河道 River | 30 |
| 10 | 泰州市兴化市大邹镇顾马村 Guma Village, Dazou Town, Xinghua City, Taizhou City | JSTZ | 119.92° | 33.15° | 河道、蟹田 River, crab field | 30 |
| 11 | 宿迁市泗洪县 Sihong County, Suqian City | JSSQ | 118.25° | 33.47° | 湖泊 Lake | 10 |
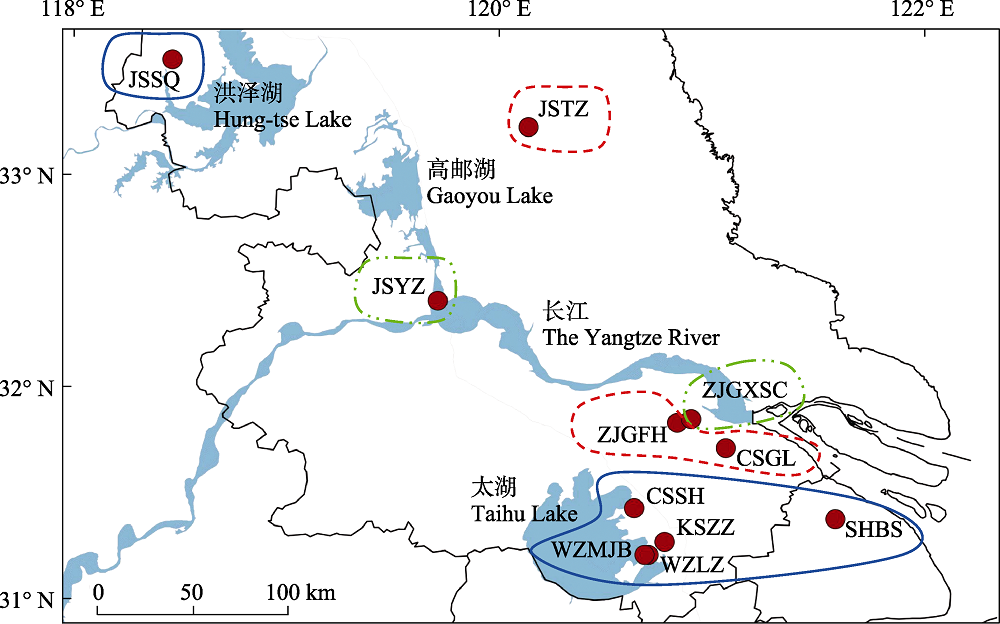
图1 本研究福寿螺采样点及基于AMOVA层次分析的分组。实心圆表示采样点, 同一线型框表示同一组群, 图中缩写含义见表1。
Fig. 1 Sampling locations of the apple snails and grouping based on AMOVA hierarchical analysis. Solid circles represent sampling points, the frames in same linetype indicates a population group, the meaning of the abbreviation in the figure is shown in Table 1.
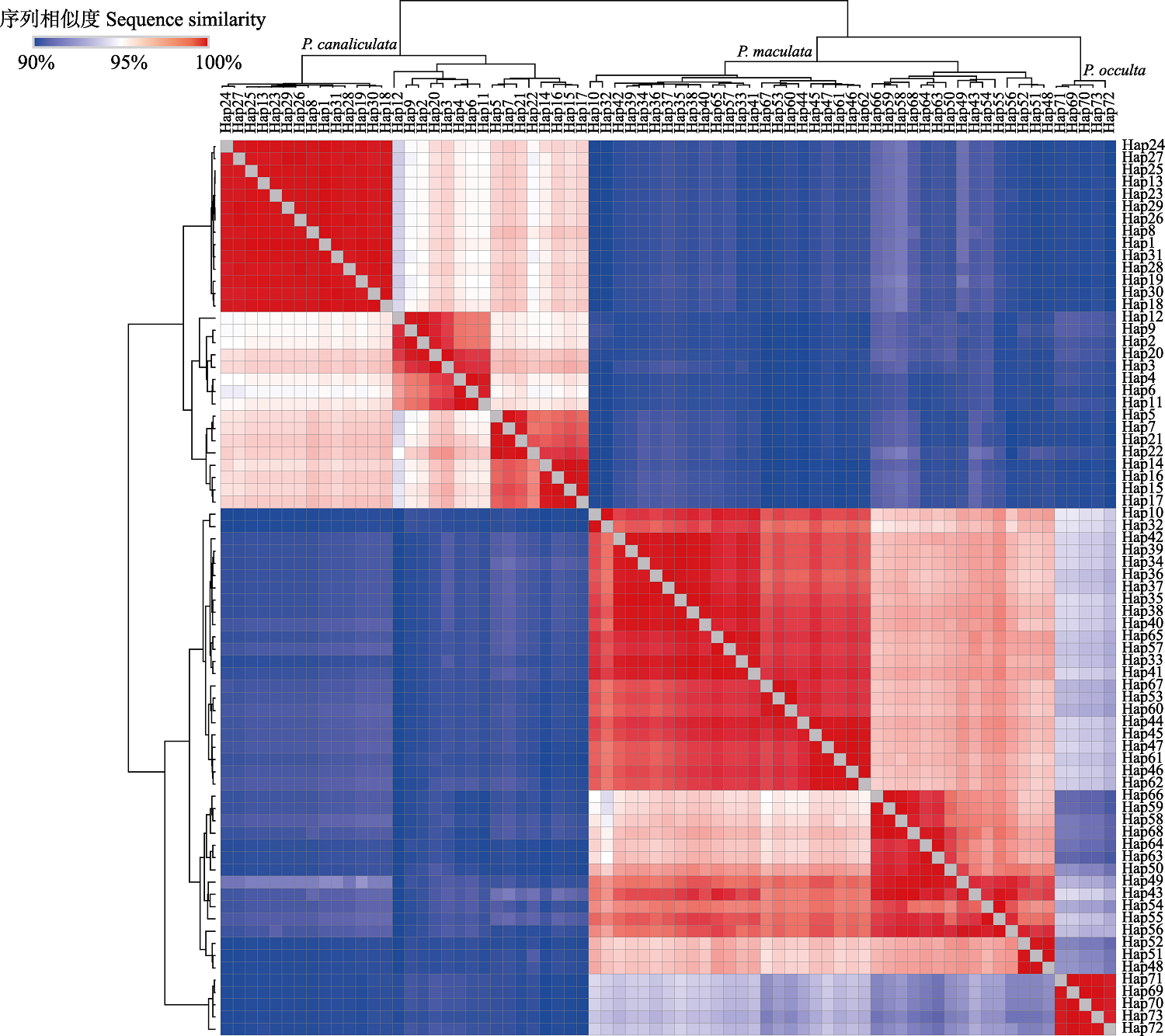
图2 基于K2P遗传距离的单倍型序列相似性热图。P. canaliculata: 小管福寿螺; P. maculata: 斑点福寿螺; P. occulta: 隐秘福寿螺。
Fig. 2 Heatmap of the sequence similarities based on K2P genetic distance of the haplotypes

图3 长江下游福寿螺单倍型构建的系统发育树。系统发育分支节点为相邻连接法/贝叶斯法(NJ/BI)系统发育树的置信值, 仅显示> 60%的数值; 其中加粗显示的Hap1?10为本研究所测COI序列生成的单倍型。
Fig. 3 The phylogenetic tree constructed by the haplotypes of apple snails from the lower reaches of the Yangtze River. The branch nodes of phylogenetic trees denote neighbor-joining/ Bayesian inference bootstrap supports. Only the values > 60% are displayed; Hap1-10 shown in bold are haplotypes generated from COI sequences from this study.
| 单倍型 Haplotype | 序列数量 No. of sequences (%) | SHBS | WZLZ | WZMJB | KSZZ | CSSH | CSGL | ZJGFH | ZJGXSC | JSYZ | JSTZ | JSSQ | 物种 Species |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap1 | 126 (46.67) | 19 | 23 | 16 | 30 | 27 | - | - | - | - | 2 | 9 | 小管福寿螺Pomacea canaliculata |
| Hap2 | 15 (5.56) | - | - | - | - | - | - | 10 | - | - | 5 | - | 小管福寿螺 P. canaliculata |
| Hap3 | 21 (7.78) | - | - | - | - | - | 15 | 6 | - | - | - | - | 小管福寿螺 P. canaliculata |
| Hap4 | 28 (10.37) | - | - | - | - | - | - | - | 28 | - | - | - | 小管福寿螺 P. canaliculata |
| Hap5 | 22 (8.15) | - | 7 | 14 | - | 1 | - | - | - | - | - | - | 小管福寿螺 P. canaliculata |
| Hap6 | 18 (6.67) | - | - | - | - | - | - | - | - | 18 | - | - | 小管福寿螺 P. canaliculata |
| Hap7 | 1 (0.37) | 1 | - | - | - | - | - | - | - | - | 小管福寿螺 P. canaliculata | ||
| Hap8 | 2 (0.74) | - | - | - | - | 2 | - | - | - | - | 小管福寿螺 P. canaliculata | ||
| Hap9 | 1 (0.37) | - | - | - | - | - | - | 1 | - | - | - | - | 小管福寿螺 P. canaliculata |
| Hap10 | 36 (13.33) | - | - | - | - | - | - | - | - | 12 | 23 | 1 | 斑点福寿螺 P. maculata |
表2 本研究福寿螺种群的COI单倍型分布。地点编号见表1。
Table 2 Distribution of COI haplotypes of the populations from this study. Location code see Table 1.
| 单倍型 Haplotype | 序列数量 No. of sequences (%) | SHBS | WZLZ | WZMJB | KSZZ | CSSH | CSGL | ZJGFH | ZJGXSC | JSYZ | JSTZ | JSSQ | 物种 Species |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap1 | 126 (46.67) | 19 | 23 | 16 | 30 | 27 | - | - | - | - | 2 | 9 | 小管福寿螺Pomacea canaliculata |
| Hap2 | 15 (5.56) | - | - | - | - | - | - | 10 | - | - | 5 | - | 小管福寿螺 P. canaliculata |
| Hap3 | 21 (7.78) | - | - | - | - | - | 15 | 6 | - | - | - | - | 小管福寿螺 P. canaliculata |
| Hap4 | 28 (10.37) | - | - | - | - | - | - | - | 28 | - | - | - | 小管福寿螺 P. canaliculata |
| Hap5 | 22 (8.15) | - | 7 | 14 | - | 1 | - | - | - | - | - | - | 小管福寿螺 P. canaliculata |
| Hap6 | 18 (6.67) | - | - | - | - | - | - | - | - | 18 | - | - | 小管福寿螺 P. canaliculata |
| Hap7 | 1 (0.37) | 1 | - | - | - | - | - | - | - | - | 小管福寿螺 P. canaliculata | ||
| Hap8 | 2 (0.74) | - | - | - | - | 2 | - | - | - | - | 小管福寿螺 P. canaliculata | ||
| Hap9 | 1 (0.37) | - | - | - | - | - | - | 1 | - | - | - | - | 小管福寿螺 P. canaliculata |
| Hap10 | 36 (13.33) | - | - | - | - | - | - | - | - | 12 | 23 | 1 | 斑点福寿螺 P. maculata |
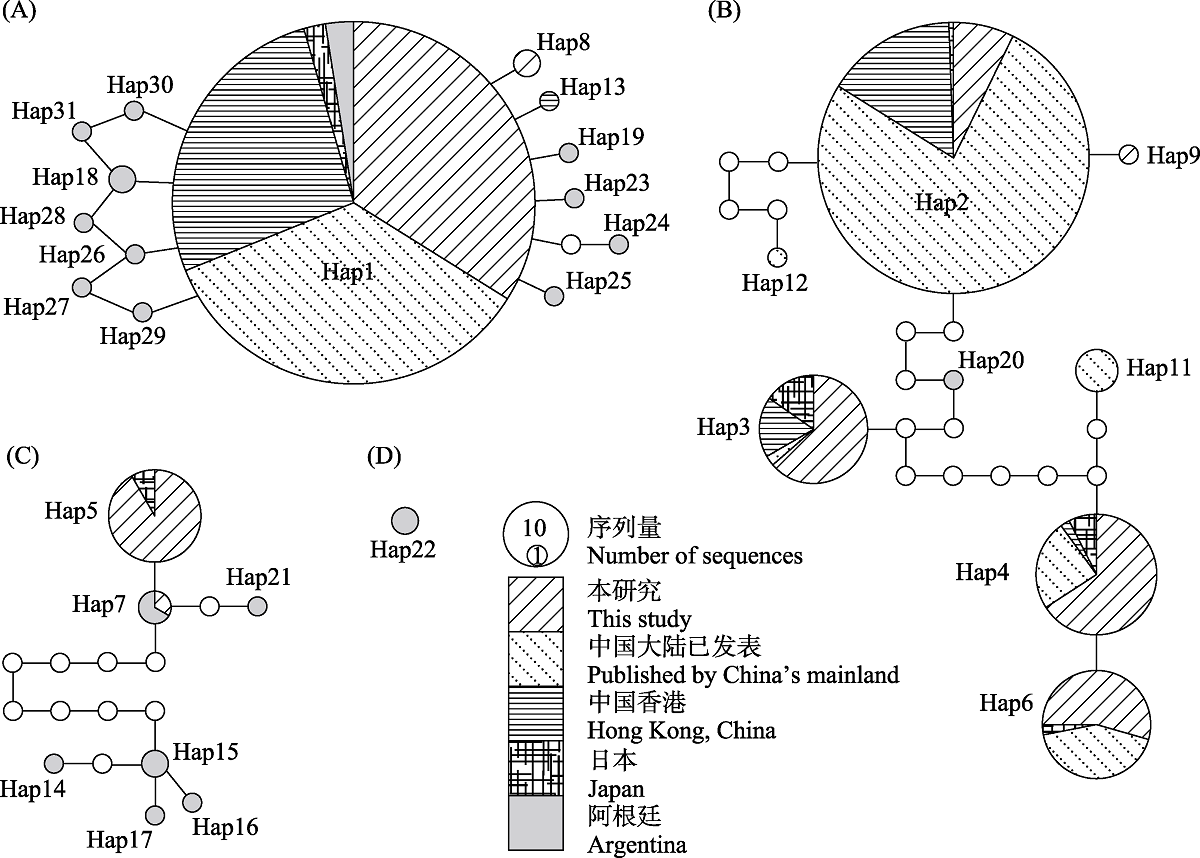
图4 基于TCS模型建立的小管福寿螺单倍型网络。95%简约连接界限下分成4个子网络(A?D); 圆的大小与序列量成正比, 不同图案代表不同地理种群, 白色圆表示缺失单倍型。
Fig. 4 Haplotype network of Pomacea canaliculata based on TCS model. The haplotypes network of P. canaliculata splits into four sub-networks (A?D) under 95% parsimony limit. The size of the circles represents the number of sequences. Pattern types represent different geographical populations, with the white circles indicating missing haplotypes.
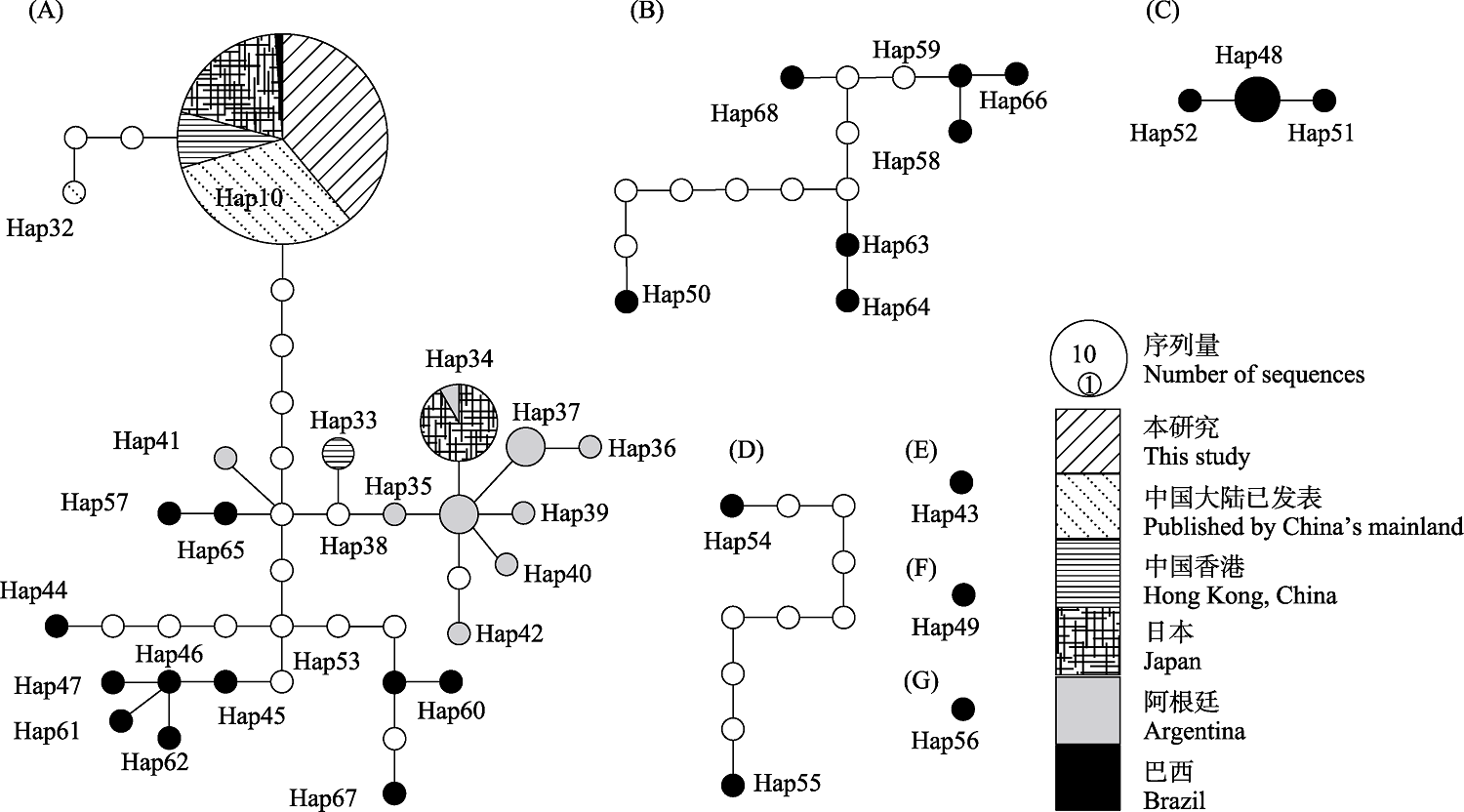
图5 基于TCS模型建立的斑点福寿螺单倍型网络。95%简约连接界限下分成7个子网络(A?G); 圆的大小与序列量成正比, 不同图案代表不同地理种群, 白色圆表示缺失单倍型。
Fig. 5 Haplotype network of Pomacea maculata based on TCS model. The haplotypes network of P. maculata splits into seven sub-networks (A?G) under 95% parsimony limit. The size of the circles represents the number of sequences. Pattern types represent different geographical populations, with the white circles indicating missing haplotypes.
| 物种 Species | 地理种群 Region | 序列数 No. of sequences | 单倍型数 No. of haplotypes | 单倍型多样性 Haplotype diversity (Hd) | 核苷酸多样性 Nucleotide diversity (π) | 核苷酸平均差异数 Average number of nucleotide difference (k) |
|---|---|---|---|---|---|---|
| 小管福寿螺 Pomacea canaliculata | 长江以南 South of the Yangtze River | 200 | 8 | 0.627 | 0.02546 | 14.744 |
| 长江以北 North of the Yangtze River | 34 | 3 | 0.611 | 0.02541 | 14.713 | |
| 中国大陆已发表 Published by China’s mainland | 319 | 7 | 0.587 | 0.02543 | 14.722 | |
| 香港 Hong Kong | 139 | 5 | 0.441 | 0.01956 | 11.324 | |
| 斑点福寿螺 P. maculata | 江苏 Jiangsu | 36 | 1 | 0.000 | 0.00000 | 0.000 |
| 中国大陆已发表 Published by China’s mainland | 30 | 2 | 0.067 | 0.00035 | 0.200 | |
| 香港 Hong Kong | 10 | 2 | 0.356 | 0.00430 | 2.489 |
表3 本研究种群、已发表的中国大陆及香港种群的小管福寿螺和斑点福寿螺的遗传多样性
Table 3 Distribution of population genetic diversity of Pomacea canaliculata and P. maculata from this study, published by China’s mainland and Hong Kong, and this study
| 物种 Species | 地理种群 Region | 序列数 No. of sequences | 单倍型数 No. of haplotypes | 单倍型多样性 Haplotype diversity (Hd) | 核苷酸多样性 Nucleotide diversity (π) | 核苷酸平均差异数 Average number of nucleotide difference (k) |
|---|---|---|---|---|---|---|
| 小管福寿螺 Pomacea canaliculata | 长江以南 South of the Yangtze River | 200 | 8 | 0.627 | 0.02546 | 14.744 |
| 长江以北 North of the Yangtze River | 34 | 3 | 0.611 | 0.02541 | 14.713 | |
| 中国大陆已发表 Published by China’s mainland | 319 | 7 | 0.587 | 0.02543 | 14.722 | |
| 香港 Hong Kong | 139 | 5 | 0.441 | 0.01956 | 11.324 | |
| 斑点福寿螺 P. maculata | 江苏 Jiangsu | 36 | 1 | 0.000 | 0.00000 | 0.000 |
| 中国大陆已发表 Published by China’s mainland | 30 | 2 | 0.067 | 0.00035 | 0.200 | |
| 香港 Hong Kong | 10 | 2 | 0.356 | 0.00430 | 2.489 |
| 变异来源 Source of variation | 自由度 Degree of freedom | 平方和 Sum of squares | 方差组分 Variance components | 变异百分率 Percentage of variation (%) |
|---|---|---|---|---|
| 组群间 Among groups | 2 | 122.265 | 9.56034 | 7.37 |
| 组群内种群间 Among populations within groups | 8 | 165.622 | 0.89486 | 7.24 |
| 种群内个体间 Among individuals within populations | 223 | 423.865 | 1.90074 | 15.38 |
| 总变异 Total variation | 233 | 1,811.752 | 12.35594 | 100.00 |
表4 长江下游分布区小管福寿螺种群COI分子变异方差分析
Table 4 Variance analysis of COI molecular variation among populations of Pomacea canaliculata in the lower reaches of the Yangtze River
| 变异来源 Source of variation | 自由度 Degree of freedom | 平方和 Sum of squares | 方差组分 Variance components | 变异百分率 Percentage of variation (%) |
|---|---|---|---|---|
| 组群间 Among groups | 2 | 122.265 | 9.56034 | 7.37 |
| 组群内种群间 Among populations within groups | 8 | 165.622 | 0.89486 | 7.24 |
| 种群内个体间 Among individuals within populations | 223 | 423.865 | 1.90074 | 15.38 |
| 总变异 Total variation | 233 | 1,811.752 | 12.35594 | 100.00 |
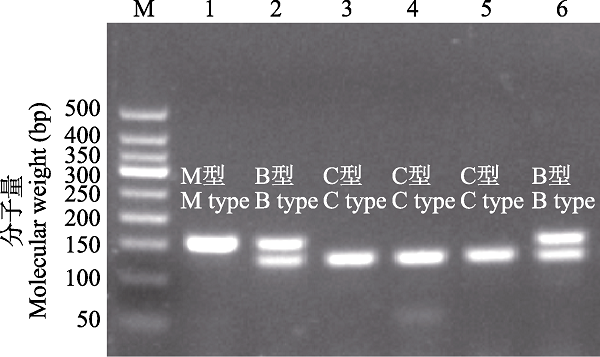
图6 基于核EF1α基因多重PCR扩增的长江下游福寿螺杂交渐渗型检测。M为50 bp DNA梯带; 1?6为长江下游福寿螺样品, 分别为M型、B型、C型、C型、C型、B型。
Fig. 6 Introgressive hybridization of the apple snails in the lower reaches of the Yangtze River based on EF1α gene multiplex PCR method. M, 50 bp DNA ladder; 1?6 represent for M type, B type, C type, C type, C type, and B type of samples of apple snails collected in the lower reaches of the Yangtze River.
| [1] |
Brito FC, Joshi RC (2016) The golden apple snail Pomacea canaliculata: A review on invasion, dispersion and control. Outlooks on Pest Management, 27, 157-163.
DOI URL |
| [2] |
Clement M, Posada D, Crandall KA (2000) TCS: A computer program to estimate gene genealogies. Molecular Ecology, 9, 1657-1659.
DOI PMID |
| [3] | Cowie RH, Hayes KA, Thiengo SC (2006) What are apple snails? Confused taxonomy and some preliminary resolution. In: Global Advances in Ecology and Management of Golden Apple Snails (eds Joshi RC, Sebastian LS), pp. 3-23. Philippine Rice Research Institute, Munñoz. |
| [4] |
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131, 479-491.
DOI PMID |
| [5] |
Eyer PA, Blumenfeld AJ, Johnson LNL, Perdereau E, Shults P, Wang SC, Dedeine F, Dupont S, Bagnères AG, Vargo EL (2021) Extensive human-mediated jump dispersal within and across the native and introduced ranges of the invasive termite Reticulitermes flavipes. Molecular Ecology, 30, 3948-3964.
DOI URL |
| [6] |
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294-299.
PMID |
| [7] |
Freeland JR, Boag PT (1999) The mitochondrial and nuclear genetic homogeneity of the phenotypically diverse Darwin’s ground finches. Evolution, 53, 1553-1563.
DOI PMID |
| [8] |
Glasheen PM, Burks RL, Campos SR, Hayes KA (2020) First evidence of introgressive hybridization of apple snails (Pomacea spp.) in their native range. Journal of Molluscan Studies, 86, 96-103.
DOI PMID |
| [9] |
Hayes KA, Burks RL, Castro-Vazquez A, Darby PC, Heras H, Martín PR, Qiu JW, Thiengo SC, Vega IA, Wada T, Yusa Y, Burela S, Pilar Cadierno M, Cueto JA, Dellagnola FA, Dreon MS, Victoria Frassa M, Giraud-Billoud M, Godoy MS, Ituarte S, Koch E, Matsukura K, Yanina Pasquevich M, Rodriguez C, Saveanu L, Seuffert ME, Strong EE, Sun J, Tamburi NE, Tiecher MJ, Turner RL, Valentine-Darby PL, Cowie RH (2015) Insights from an integrated view of the biology of apple snails (Caenogastropoda: Ampullariidae). Malacologia, 58, 245-302.
DOI URL |
| [10] |
Hayes KA, Cowie RH, Thiengo SC (2009) A global phylogeny of apple snails: Gondwanan origin, generic relationships, and the influence of outgroup choice (Caenogastropoda: Ampullariidae). Biological Journal of the Linnean Society, 98, 61-76.
DOI URL |
| [11] |
Hayes KA, Cowie RH, Thiengo SC, Strong EE (2012) Comparing apples with apples: Clarifying the identities of two highly invasive Neotropical Ampullariidae (Caenogas- tropoda). Zoological Journal of the Linnean Society, 166, 723-753.
DOI URL |
| [12] |
Hayes KA, Joshi RC, Thiengo SC, Cowie RH (2008) Out of South America: Multiple origins of non-native apple snails in Asia. Diversity and Distributions, 14, 701-712.
DOI URL |
| [13] | Ji XM, Wang AX, Fang LC, Li CL, Xu L, Yang SK, Liu XB, Zhong L, Liu YM (2020) Investigation on the distribution of apple snail (Pomacea canaliculata) in the lower reaches of Yangtze River in China. Hubei Agricultural Sciences, 59, 111-116, 124. (in Chinese with English abstract) |
| [戢小梅, 王爱新, 方林川, 李长林, 许林, 杨守坤, 刘宪葆, 钟兰, 刘义满 (2020) 中国长江下游地区福寿螺分布现状考察. 湖北农业科学, 59, 111-116, 124.] | |
| [14] |
Kagawa K, Takimoto G (2018) Hybridization can promote adaptive radiation by means of transgressive segregation. Ecology Letters, 21, 264-274.
DOI PMID |
| [15] |
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647-1649.
DOI PMID |
| [16] |
Lei JC, Chen L, Li H (2017) Using ensemble forecasting to examine how climate change promotes worldwide invasion of the golden apple snail (Pomacea canaliculata). Environmental Monitoring and Assessment, 189, 404.
DOI PMID |
| [17] |
Liu QQ, Dong ZJ (2018) Population genetic structure of Gonionemus vertens based on the mitochondrial COI sequence. Biodiversity Science, 26, 1204-1211. (in Chinese with English abstract)
DOI URL |
|
[刘青青, 董志军 (2018) 基于线粒体COI基因分析钩手水母的群体遗传结构. 生物多样性, 26, 1204-1211.]
DOI |
|
| [18] | Liu YM, Li CL, Jin L, Pei X, Chen XB, Wang AX, Ji XM, Zhong L (2020) Investigation on northernmost distribution areas of apple snail (Pomacea canaliculata) in upper reaches of Yangtze River. Journal of Changjiang Vegetables, (12), 53-57. (in Chinese with English abstract) |
| [刘义满, 李长林, 金莉, 裴忺, 陈绪柏, 王爱新, 戢小梅, 钟兰 (2020) 长江上游地区福寿螺北缘分布地区调查. 长江蔬菜, (12), 53-57.] | |
| [19] | Luo D, Wang XJ, Xu M, Gu DE, Mu XD, Wei H, Yang YX, Demayo CG, Hu YC (2018) Correlation between shell-body mass ratio and hydrostatic settling characteristics of mollusc species. Acta Ecologica Sinica, 38, 6778-6785. (in Chinese with English abstract) |
| [罗渡, 汪学杰, 徐猛, 顾党恩, 牟希东, 韦慧, 杨叶欣, Cesar G. Demayo, 胡隐昌 (2018) 贝类壳-体质量比和静水沉降特性的相关性. 生态学报, 38, 6778-6785.] | |
| [20] |
Lv S, Zhang Y, Steinmann P, Yang GJ, Yang K, Zhou XN, Utzinger J (2011) The emergence of angiostrongyliasis in the People’s Republic of China: The interplay between invasive snails, climate change and transmission dynamics. Freshwater Biology, 56, 717-734.
DOI URL |
| [21] |
Matsukura K, Izumi Y, Yoshida K, Wada T (2016) Cold tolerance of invasive freshwater snails, Pomacea canaliculata, P. maculata, and their hybrids helps explain their different distributions. Freshwater Biology, 61, 80-87.
DOI URL |
| [22] |
Matsukura K, Okuda M, Cazzaniga NJ, Wada T (2013) Genetic exchange between two freshwater apple snails, Pomacea canaliculata and Pomacea maculata invading East and Southeast Asia. Biological Invasions, 15, 2039-2048.
DOI URL |
| [23] |
Matsukura K, Okuda M, Kubota K, Wada T (2008) Genetic divergence of the genus Pomacea (Gastropoda: Ampullariidae) distributed in Japan, and a simple molecular method to distinguish P. canaliculata and P. insularum. Applied Entomology and Zoology, 43, 535-540.
DOI URL |
| [24] |
Morrison WE, Hay ME (2011) Feeding and growth of native, invasive and non-invasive alien apple snails (Ampullariidae) in the United States: Invasives eat more and grow more. Biological Invasions, 13, 945-955.
DOI URL |
| [25] |
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution, 29, 1-10.
DOI PMID |
| [26] | Qian ZJ, Lin YF, Yang YT, Chen SH, Hu YP, Zhou X, Li H, Ding H, Chen L (2021) Genetic diversity of invasive Pomacea snails in Suzhou City. Chinese Journal of Zoology, 56, 929-938. (in Chinese with English abstract) |
| [钱子衿, 林友福, 杨雨婷, 陈书涵, 胡亚萍, 周旭, 李宏, 丁晖, 陈炼 (2021) 苏州市入侵福寿螺的遗传多样性. 动物学杂志, 56, 929-938.] | |
| [27] |
Rawlings TA, Hayes KA, Cowie RH, Collins TM (2007) The identity, distribution, and impacts of non-native apple snails in the continental United States. BMC Evolutionary Biology, 7, 97.
PMID |
| [28] |
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572-1574.
DOI PMID |
| [29] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34, 3299-3302.
DOI PMID |
| [30] |
VanWallendael A, Alvarez M, Franks SJ (2021) Patterns of population genomic diversity in the invasive Japanese knotweed species complex. American Journal of Botany, 108, 857-868.
DOI PMID |
| [31] | Wang L, Mo KL, Chen QW, Zhang JY, Xia J, Lin YQ (2019) Estimating ecological flows for fish overwintering in plain rivers using a method based on water temperature and critical water depth. Ecohydrology, 12, e2098. |
| [32] | Yang HF, Yang SP, Wang P, Zhou WC (2018) Prediction of potential geographical distribution area of golden apple snail (Pomacea canaliculata) in China. Acta Agriculturae Jiangxi, 30(3), 70-73. (in Chinese with English abstract) |
| [杨海芳, 杨姗萍, 王沛, 周卫川 (2018) 福寿螺在中国的潜在地理分布区预测. 江西农业学报, 30(3), 70-73.] | |
| [33] |
Yang QQ, He C, Liu GF, Yin CL, Xu YP, Liu SW, Qiu JW, Yu XP (2020) Introgressive hybridization between two non-native apple snails in China: Widespread hybridization and homogenization in egg morphology. Pest Management Science, 76, 4231-4239.
DOI URL |
| [34] |
Yang QQ, Ip J C-H, Zhao XX, Li JN, Jin YJ, Yu XP, Qiu JW (2022) Molecular analyses revealed three morphologically similar species of non-native apple snails and their patterns of distribution in freshwater wetlands of Hong Kong. Diversity and Distributions, 28, 97-111.
DOI URL |
| [35] |
Yang QQ, Liu SW, He C, Cowie RH, Yu XP, Hayes KA (2019) Invisible apple snail invasions: Importance of continued vigilance and rigorous taxonomic assessments. Pest Management Science, 75, 1277-1286.
DOI PMID |
| [36] |
Yang QQ, Liu SW, He C, Yu XP (2018) Distribution and the origin of invasive apple snails, Pomacea canaliculata and P. maculata (Gastropoda: Ampullariidae) in China. Scientific Reports, 8, 1185.
DOI |
| [37] |
Yang QQ, Liu SW, Ru WD, Liu GF, Yu XP (2016) Molecular identification of invasive golden apple snails in Zhejiang Province based on DNA barcoding. Biodiversity Science, 24, 341-350. (in Chinese with English abstract)
DOI |
|
[杨倩倩, 刘苏汶, 茹炜岽, 刘光富, 俞晓平 (2016) 基于DNA条形码技术对浙江省外来入侵福寿螺进行分子鉴定. 生物多样性, 24, 341-350.]
DOI |
|
| [38] | Yang QQ, Yu XP (2019) A new species of apple snail in the genus Pomacea (Gastropoda: Caenogastropoda: Ampullarii- dae). Zoological Studies, 58, e13. |
| [39] | Yang YX, Hu YC, Li XH, Wang XJ, Mu XD, Song HM, Wang PX, Liu C, Luo JR (2010) Historical invasion, expansion process and harm investigation of Pomacea canaliculata in China. Chinese Agricultural Science Bulletin, 26, 245-250. (in Chinese with English abstract) |
| [杨叶欣, 胡隐昌, 李小慧, 汪学杰, 牟希东, 宋红梅, 王培欣, 刘超, 罗建仁 (2010) 福寿螺在中国的入侵历史、扩散规律和危害的调查分析. 中国农学通报, 26, 245-250.] | |
| [40] |
Yin YX, He Q, Pan XW, Liu QY, Wu YJ, Li XR (2022) Predicting current potential distribution and the range dynamics of Pomacea canaliculata in China under global climate change. Biology, 11, 110.
DOI URL |
| [41] |
Yoshida K, Matsukura K, Cazzaniga NJ, Wada T (2014) Tolerance to low temperature and desiccation in two invasive apple snails, Pomacea canaliculata and P. maculata (Caenogastropoda: Ampullariidae), collected in their original distribution area (northern and central Argentina). Journal of Molluscan Studies, 80, 62-66.
DOI URL |
| [1] | 施国杉, 刘峰, 曹光宏, 陈典, 夏尚文, 邓云, 王彬, 杨效东, 林露湘. 西双版纳热带季节雨林木本植物的beta多样性: 空间、环境与林分结构的作用[J]. 生物多样性, 2024, 32(12): 24285-. |
| [2] | 杜聪聪, 冯学宇, 陈志林. 桥头堡效应中气候生态位差异的缩小促进了红火蚁的入侵[J]. 生物多样性, 2024, 32(11): 24276-. |
| [3] | 原雪姣, 张渊媛, 张衍亮, 胡璐祎, 桑卫国, 杨峥, 陈颀. 基于飞机草历史分布数据拟合的物种分布模型及其预测能力[J]. 生物多样性, 2024, 32(11): 24288-. |
| [4] | 韩丽霞, 王永健, 刘宣. 外来物种入侵与本土物种分布区扩张的异同[J]. 生物多样性, 2024, 32(1): 23396-. |
| [5] | 王明慧, 陈昭铨, 李帅锋, 黄小波, 郎学东, 胡子涵, 尚瑞广, 刘万德. 云南普洱季风常绿阔叶林不同种子扩散方式的优势种空间点格局分析[J]. 生物多样性, 2023, 31(9): 23147-. |
| [6] | 魏博, 刘林山, 谷昌军, 于海彬, 张镱锂, 张炳华, 崔伯豪, 宫殿清, 土艳丽. 紫茎泽兰在中国的气候生态位稳定且其分布范围仍有进一步扩展的趋势[J]. 生物多样性, 2022, 30(8): 21443-. |
| [7] | 曲梦君, 努尔依拉·阿巴拜克, 邹旭阁, 赵航, 朱威霖, 王健铭, 李景文. 地理距离和环境因子对阿拉善戈壁植物群落β多样性的影响[J]. 生物多样性, 2022, 30(11): 22029-. |
| [8] | 米湘成, 王绪高, 沈国春, 刘徐兵, 宋晓阳, 乔秀娟, 冯刚, 杨洁, 毛子昆, 徐学红, 马克平. 中国森林生物多样性监测网络: 二十年群落构建机制探索的回顾与展望[J]. 生物多样性, 2022, 30(10): 22504-. |
| [9] | 刘艳杰, 黄伟, 杨强, 郑玉龙, 黎绍鹏, 吴昊, 鞠瑞亭, 孙燕, 丁建清. 近十年植物入侵生态学重要研究进展[J]. 生物多样性, 2022, 30(10): 22438-. |
| [10] | 张丹, 马松梅, 魏博, 王春成, 张林, 闫涵. 中国梭梭属植物历史分布格局及其驱动机制[J]. 生物多样性, 2022, 30(1): 21192-. |
| [11] | 严靖, 闫小玲, 李惠茹, 杜诚, 马金双. 华东地区归化植物的组成特征、引入时间及时空分布[J]. 生物多样性, 2021, 29(4): 428-438. |
| [12] | 武鹏峰, 崔淑艳, Abid Ali, 郑国. 蜘蛛飞航研究进展[J]. 生物多样性, 2021, 29(4): 517-530. |
| [13] | 杨锡福, 张洪茂, 张知彬. 植物大年结实及其与动物贮食行为之间的关系[J]. 生物多样性, 2020, 28(7): 821-832. |
| [14] | 姚志良,温韩东,邓云,曹敏,林露湘. 哀牢山亚热带中山湿性常绿阔叶林树种beta多样性格局形成的驱动力[J]. 生物多样性, 2020, 28(4): 445-454. |
| [15] | 何维明. 生物入侵的影响是否准确可知?[J]. 生物多样性, 2020, 28(2): 253-255. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn