



Biodiv Sci ›› 2018, Vol. 26 ›› Issue (11): 1204-1211. DOI: 10.17520/biods.2018044 cstr: 32101.14.biods.2018044
• Original Papers: Animal Diversity • Previous Articles Next Articles
Qingqing Liu1,2, Zhijun Dong1,3,*( )
)
Received:2018-03-08
Accepted:2018-07-04
Online:2018-11-20
Published:2019-01-08
Contact:
Dong Zhijun
About author:# Co-first authors
Qingqing Liu, Zhijun Dong. Population genetic structure of Gonionemus vertens based on the mitochondrial COI sequence[J]. Biodiv Sci, 2018, 26(11): 1204-1211.
| 采样点 Site | 样本量 N | 单倍型 H | 单倍型组成(样本数) Haplotype composition (No. of inds.) | 单倍型多样性 Hd | 核苷酸多样性 π (%) | GenBank序列号 GenBank accession no. |
|---|---|---|---|---|---|---|
| 西北大西洋 Northwest Atlantic (NWA) | ||||||
| 美国新罕布什尔州大海湾 Great Bay, New Hampshire, America (GB) | 7 | 2 | Hap1(3); Hap14(4) | 0.571 ± 0.119 | 0.912 ± 0.191 | KY437905-911 |
| 美国雅茅斯巴斯河 Bass River, Yarmouth, America (BR) | 17 | 3 | Hap1(2); Hap12(12); Hap14(3) | 0.485 ± 0.126 | 1.004 ± 0.270 | KY437888-904 |
| 美国马什皮汉布林池塘 Hamblin Pond, Mashpee, America (HP) | 18 | 1 | Hap12(18) | 0 | 0 | KY437834-851 |
| 美国奥克布拉夫斯农场池塘 Farm Pond, Oak Bluffs, America (FP) | 17 | 2 | Hap1(1); Hap12(16) | 0.118 ± 0.101 | 0.164 ± 0.141 | KY437814-830 |
| 美国埃德加敦森格肯塔克池塘 Sengekontacket Pond, Edgartown, America (SG) | 3 | 2 | Hap1(2); Hap12(1) | 0.667 ± 0.314 | 0.931 ± 0.439 | KY437831-833 |
| 美国北金斯敦波特池塘 Potter Pond, North Kingston, America (PP) | 22 | 2 | Hap1(2); Hap12(20) | 0.173 ± 0.101 | 0.242 ± 0.141 | KY437912-933 |
| 美国格罗顿芒福德湾 Mumford Cove, Groton, America (MC) | 14 | 2 | Hap1(13); Hap12(1) | 0.143 ± 0.119 | 0.200 ± 0.166 | KY437934-947 |
| 美国格罗顿松岛 Pine Island, Groton, America (PI) | 24 | 2 | Hap1(23); Hap12(1) | 0.083 ± 0.075 | 0.116 ± 0.105 | KY437864-887 |
| 西北太平洋 Northwest Pacific (NWP) | ||||||
| 俄罗斯阿穆尔湾 Amur Bay, Peter the Great Gulf, Russia (AB) | 3 | 1 | Hap1(3) | 0 | 0 | KY437948-950 |
| 俄罗斯沃斯托克湾 Vostok Bay, Peter the Great Gulf, Russia (VB) | 30 | 2 | Hap1(26); Hap11(4) | 0.239 ± 0.092 | 0.048 ± 0.018 | KY437951-980 |
| 日本越喜来湾 Okirai Bay, Japan (JP) | 12 | 2 | Hap1(1); Hap2(11) | 0.167 ± 0.134 | 0.100 ± 0.080 | KY437852-863 |
| 中国厦门 Xiamen, China (XM) | 10 | 1 | Hap10(10) | 0 | 0 | KF962130-139 |
| 中国烟台 Yantai, China (YT) | 27 | 5 | Hap2(23); Hap3(1); Hap4(1); Hap8(1); Hap9(1) | 0.279 ± 0.112 | 0.148 ± 0.095 | MH020717-743 |
| 中国大连 Dalian, China (DL) | 30 | 4 | Hap1(6); Hap2(19); Hap5(4); Hap6(1) | 0.559 ± 0.086 | 0.307 ± 0.058 | MH020640-669 |
| 中国东营 Dongying, China (DY) | 18 | 1 | Hap2(18) | 0 | 0 | MH020670-687 |
| 中国乐亭 Laoting, China (LT) | 29 | 3 | Hap1(2); Hap2(25); Hap7(2) | 0.256 ± 0.102 | 0.106 ± 0.050 | MH020688-716 |
| 东北太平洋 Northeast Pacific (NEP) | ||||||
| 美国圣胡安岛 San Juan Island, America (FH) | 4 | 1 | Hap13(4) | 0 | 0 | KY437982-985 |
| 东北大西洋 Northeast Atlantic (NEA) | ||||||
| 冰岛阿夫塔内斯 Álftanes, Iceland (IC) | 1 | 1 | Hap13(1) | 0 | 0 | KY437981 |
| 总群体 Total | - | 286 | 14 | - | 0.743 ± 0.012 | 1.046 ± 0.097 |
Table 1 Locations, number of individuals and diversity parameters for the population of Gonionemus vertens
| 采样点 Site | 样本量 N | 单倍型 H | 单倍型组成(样本数) Haplotype composition (No. of inds.) | 单倍型多样性 Hd | 核苷酸多样性 π (%) | GenBank序列号 GenBank accession no. |
|---|---|---|---|---|---|---|
| 西北大西洋 Northwest Atlantic (NWA) | ||||||
| 美国新罕布什尔州大海湾 Great Bay, New Hampshire, America (GB) | 7 | 2 | Hap1(3); Hap14(4) | 0.571 ± 0.119 | 0.912 ± 0.191 | KY437905-911 |
| 美国雅茅斯巴斯河 Bass River, Yarmouth, America (BR) | 17 | 3 | Hap1(2); Hap12(12); Hap14(3) | 0.485 ± 0.126 | 1.004 ± 0.270 | KY437888-904 |
| 美国马什皮汉布林池塘 Hamblin Pond, Mashpee, America (HP) | 18 | 1 | Hap12(18) | 0 | 0 | KY437834-851 |
| 美国奥克布拉夫斯农场池塘 Farm Pond, Oak Bluffs, America (FP) | 17 | 2 | Hap1(1); Hap12(16) | 0.118 ± 0.101 | 0.164 ± 0.141 | KY437814-830 |
| 美国埃德加敦森格肯塔克池塘 Sengekontacket Pond, Edgartown, America (SG) | 3 | 2 | Hap1(2); Hap12(1) | 0.667 ± 0.314 | 0.931 ± 0.439 | KY437831-833 |
| 美国北金斯敦波特池塘 Potter Pond, North Kingston, America (PP) | 22 | 2 | Hap1(2); Hap12(20) | 0.173 ± 0.101 | 0.242 ± 0.141 | KY437912-933 |
| 美国格罗顿芒福德湾 Mumford Cove, Groton, America (MC) | 14 | 2 | Hap1(13); Hap12(1) | 0.143 ± 0.119 | 0.200 ± 0.166 | KY437934-947 |
| 美国格罗顿松岛 Pine Island, Groton, America (PI) | 24 | 2 | Hap1(23); Hap12(1) | 0.083 ± 0.075 | 0.116 ± 0.105 | KY437864-887 |
| 西北太平洋 Northwest Pacific (NWP) | ||||||
| 俄罗斯阿穆尔湾 Amur Bay, Peter the Great Gulf, Russia (AB) | 3 | 1 | Hap1(3) | 0 | 0 | KY437948-950 |
| 俄罗斯沃斯托克湾 Vostok Bay, Peter the Great Gulf, Russia (VB) | 30 | 2 | Hap1(26); Hap11(4) | 0.239 ± 0.092 | 0.048 ± 0.018 | KY437951-980 |
| 日本越喜来湾 Okirai Bay, Japan (JP) | 12 | 2 | Hap1(1); Hap2(11) | 0.167 ± 0.134 | 0.100 ± 0.080 | KY437852-863 |
| 中国厦门 Xiamen, China (XM) | 10 | 1 | Hap10(10) | 0 | 0 | KF962130-139 |
| 中国烟台 Yantai, China (YT) | 27 | 5 | Hap2(23); Hap3(1); Hap4(1); Hap8(1); Hap9(1) | 0.279 ± 0.112 | 0.148 ± 0.095 | MH020717-743 |
| 中国大连 Dalian, China (DL) | 30 | 4 | Hap1(6); Hap2(19); Hap5(4); Hap6(1) | 0.559 ± 0.086 | 0.307 ± 0.058 | MH020640-669 |
| 中国东营 Dongying, China (DY) | 18 | 1 | Hap2(18) | 0 | 0 | MH020670-687 |
| 中国乐亭 Laoting, China (LT) | 29 | 3 | Hap1(2); Hap2(25); Hap7(2) | 0.256 ± 0.102 | 0.106 ± 0.050 | MH020688-716 |
| 东北太平洋 Northeast Pacific (NEP) | ||||||
| 美国圣胡安岛 San Juan Island, America (FH) | 4 | 1 | Hap13(4) | 0 | 0 | KY437982-985 |
| 东北大西洋 Northeast Atlantic (NEA) | ||||||
| 冰岛阿夫塔内斯 Álftanes, Iceland (IC) | 1 | 1 | Hap13(1) | 0 | 0 | KY437981 |
| 总群体 Total | - | 286 | 14 | - | 0.743 ± 0.012 | 1.046 ± 0.097 |
| 变异来源 Source of variation | 自由度 d.f. | 平方和 Sum of squares | 方差比例 % of variation | F | P |
|---|---|---|---|---|---|
| 群组间 Among groups | 3 | 362.274 | 60.17 | Fsc = 0.66436 | 0.0000 |
| 组内群体间 Among populations within groups | 14 | 247.509 | 26.46 | Fst = 0.86631 | 0.0000 |
| 群体内 Within populations | 268 | 136.735 | 13.37 | Fct = 0.60168 | 0.0002 |
| 总计 Total | 285 | 746.517 | 100 |
Table 2 Spatial analysis of molecular variance among population of Gonionemus vertens
| 变异来源 Source of variation | 自由度 d.f. | 平方和 Sum of squares | 方差比例 % of variation | F | P |
|---|---|---|---|---|---|
| 群组间 Among groups | 3 | 362.274 | 60.17 | Fsc = 0.66436 | 0.0000 |
| 组内群体间 Among populations within groups | 14 | 247.509 | 26.46 | Fst = 0.86631 | 0.0000 |
| 群体内 Within populations | 268 | 136.735 | 13.37 | Fct = 0.60168 | 0.0002 |
| 总计 Total | 285 | 746.517 | 100 |
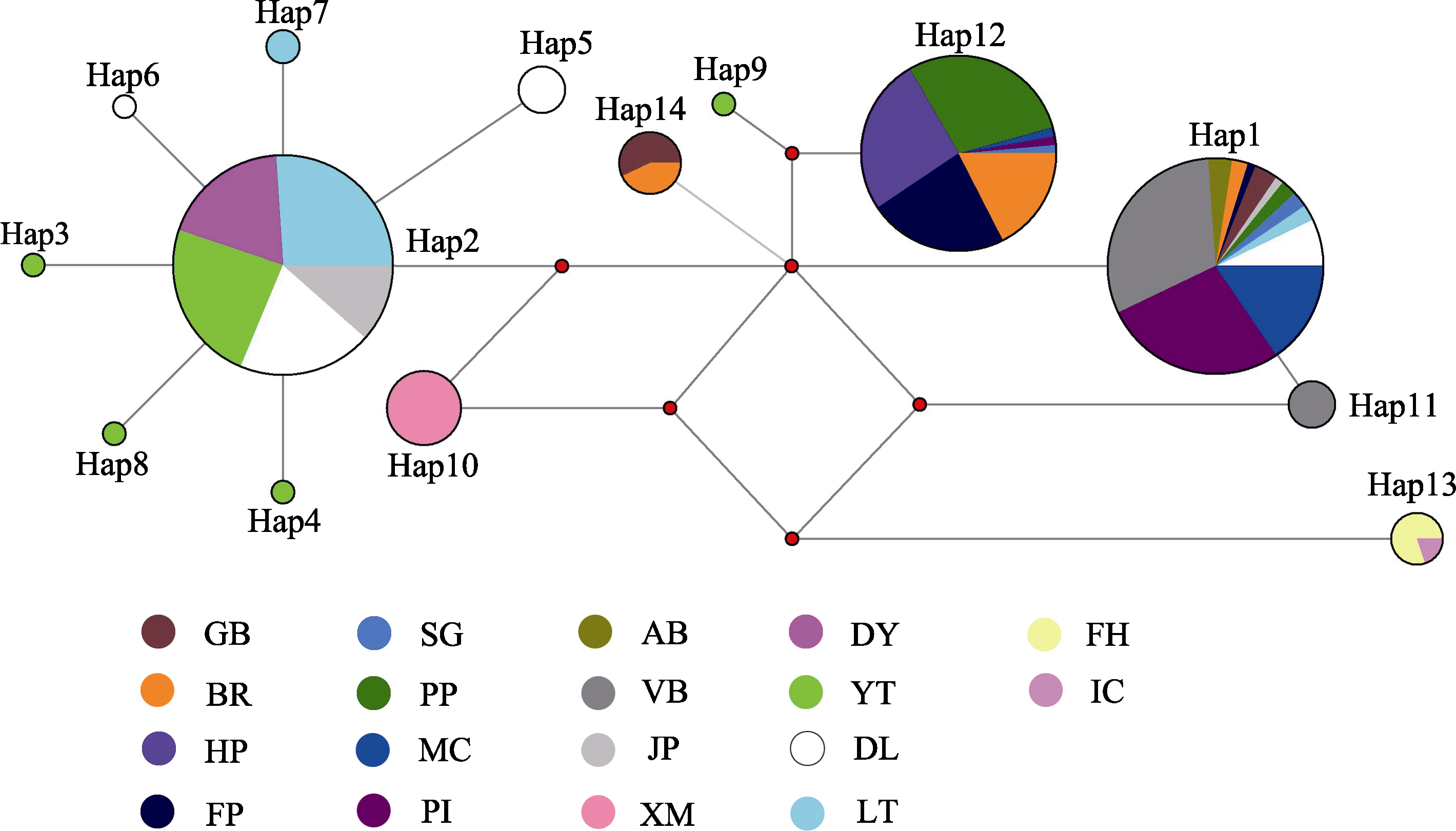
Fig. 3 Median-joining haplotype network of Gonionemus vertens based on the combined sequence of COI. The color fan area indicates the proportion of each sample population in the same haplotype; the circle area indicates the frequency of haplotype occurrence; the red dot represents the intermediate mutation node. The abbreviation of the sampling points in the figure is consistent with Table 1.
| [5] | Dawson MN, Gupta AS, England MH (2005) Coupled biophysical global ocean model and molecular genetic analyses identify multiple introductions of cryptogenic species. Proceedings of the National Academy of Sciences, USA, 102, 11968-11973. |
| [6] | Dong ZJ, Liu ZY, Liu DY (2015) Genetic characterization of the scyphozoan jellyfish Aurelia spp. in Chinese coastal waters using mitochondrial markers. Biochemical Systematics and Ecology, 60, 15-23. |
| [7] | Dong ZJ, Liu ZY, Liu DY, Liu QQ, Sun TT (2016) Low genetic diversity and lack of genetic structure in the giant jellyfish Nemopilema nomurai in Chinese coastal waters. Marine Biology Research, 12, 769-775. |
| [8] | Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567. |
| [9] | Govindarajan AF, Boero F, Halanych KM (2006) Phylogenetic analysis with multiple markers indicates repeated loss of the adult medusa stage in Campanulariidae (Hydrozoa, Cnidaria). Molecular Phylogenetics and Evolution, 38, 820-834. |
| [10] | Govindarajan AF, Carman MR, Khaidarov MR, Semenchenko A, Wares JP (2017) Mitochondrial diversity in Gonionemus (Trachylina: Hydrozoa) and its implications for understanding the origins of clinging jellyfish in the Northwest Atlantic Ocean. PeerJ, 5, e3205. |
| [11] | Hall BG (2004) Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Molecular Biology and Evolution, 22, 792-802. |
| [12] | Hall TA (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98. |
| [13] | Harrison JS (2004) Evolution, biogeography, and the utility of mitochondrial 16S and COI genes in phylogenetic analysis of the crab genus Austinixa (Decapoda: Pinnotheridae). Molecular Phylogenetics and Evolution, 30, 743-754. |
| [14] | Hebert PDN, Cywinska A, Ball SL (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences, 270, 313-321. |
| [15] | Jiang HC, Liu N, Gao JQ, Su B, Li JH, He JL, Liu AY (2017) Zooplankton community structure in Sishili Bay and its relationship with environmental factors. Acta Ecologica Sinica, 37, 1318-1327. (in Chinese with English abstract) |
| [姜会超, 刘宁, 高继庆, 苏博, 李佳蕙, 何健龙, 刘爱英 (2017) 烟台四十里湾浮游动物群落特征及与环境因子的关系. 生态学报, 37, 1318-1327.] | |
| [16] | Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870-1874. |
| [17] | Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452. |
| [18] | Li YL, Dong J, Wang B, Li YP, Yu XG, Fu J, Wang WB (2016) Genetic characterization of different populations of Rhopilema esculentum based on the mitochondrial COI sequence. Chinese Journal of Applied Ecology, 27, 2340-2347. (in Chinese with English abstract) |
| [李玉龙, 董婧, 王彬, 李轶平, 于旭光, 付杰, 王文波 (2016) 基于线粒体COI基因的海蜇不同地理群体遗传特征. 应用生态学报, 27, 2340-2347.] | |
| [19] | Li YL, Kong XY, Yu ZN, Kong J, Ma S, Chen LM (2009) Genetic diversity and historical demography of Chinese shrimp Feneropenaeus chinensis in Yellow Sea and Bohai Sea based on mitochondrial DNA analysis. African Journal of Biotechnology, 8, 1193-1202. |
| [20] | Neigel JE, Avise JC (1993) Application of a random walk model to geographic distributions of animal mitochondrial DNA variation. Genetics, 135, 1209-1220. |
| [21] | Nylander JAA, Olsson U, Alström P, Sanmartín I (2008) Accounting for phylogenetic uncertainty in biogeography: A Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus). Systematic Biology, 57, 257-268. |
| [22] | Palumbi SR (1994) Genetic divergence, reproductive isolation, and marine speciation. Annual Review of Ecology and Systematics, 25, 547-572. |
| [23] | Pigulevsky SV, Michaleff PV (1969) Poisoning by the medusa Gonionemus vertens in the sea of Japan. Toxicon, 7, 145-149. |
| [24] | Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539-542. |
| [25] | Stabile J, Waldman JR, Parauka F, Wirgin I (1996) Stock structure and homing fidelity in Gulf of Mexico sturgeon (Acipenser oxyrinchus desotoi) based on restriction fragment length polymorphism and sequence analyses of mitochondrial DNA. Genetics, 144, 767-775. |
| [26] | Stopar K, Ramšak A, Trontelj P, Malej A (2010) Lack of genetic structure in the jellyfish Pelagia noctiluca (Cnidaria: Scyphozoa: Semaeostomeae) across European seas. Molecular Phylogenetics and Evolution, 57, 417-428. |
| [27] | Tian JL (1987) Preliminary exploration of Gonionemus vertens in the sea of Jiaozhou Bay. Journal of Biology, 15, 17-18. (in Chinese) |
| [田金良 (1987) 胶州湾钩手水母初探. 生物学杂志, 15, 17-18.] | |
| [28] | Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294-299. |
| [29] | Ward RD, Woodwark M, Skibinskid OF (1994) A comparison of genetic diversity levels in marine, freshwater, and anadromous fishes. Journal of Fish Biology, 44, 213-232. |
| [30] | Wright S (1943) Isolation by distance. Genetics, 28, 114. |
| [31] | Wright S (1949) The genetical structure of populations. Annals of Human Genetics, 15, 323-354. |
| [32] | Yakovlev YM, Vaskovsky VE (1993) The toxic krestovik medusa Gonionemus vertens. Russian Journal of Marine Biology, 19, 287-294. |
| [33] | Zhang DN, Zheng LM, He JR, Zhang WJ, Lin YS, Li Y (2015) DNA barcoding of hydromedusae in northern Beibu Gulf for species identification. Biodiversity Science, 23, 50-60. (in Chinese with English abstract) |
| [张珰妮, 郑连明, 何劲儒, 张文静, 林元烧, 李阳 (2015) 基于线粒体COI和16S片段序列的北部湾北部水螅水母 DNA 条形码分析. 生物多样性, 23, 50-60.] | |
| [34] | Zhao M, Song W, Ma CY, Zhang FY, Jiang KJ, Song ZM, Ma LB (2015) Population genetic structure of Collichthys lucidus based on the mitochondrial cytochrome oxidase subunit I sequence. Journal of Fishery Sciences of China, 22, 233-242. (in Chinese with English abstract) |
| [1] | Avise JC, Neigel JE, Arnold J (1984) Demographic influences on mitochondrial DNA lineage survivorship in animal populations. Journal of Molecular Evolution, 20, 99-105. |
| [2] | Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48. |
| [3] | Brown WM, George M, Wilson AC (1979) Rapid evolution of animal mitochondrial DNA. Proceedings of the National Academy of Sciences, USA, 76, 1967-1971. |
| [4] | Cheng FP, Wang MX, Wang YT, Zhang F, Li CL, Sun S (2012) DNA barcoding of common medusozoa in northern China based on mtCOI sequence. Oceanologia et Limnologia Sinica, 43, 451-459. (in Chinese with English abstract) |
| [34] | [赵明, 宋炜, 马春艳, 张凤英, 蒋科技, 宋志明, 马凌波 (2015) 基于线粒体COI基因序列的棘头梅童鱼7个野生群体遗传结构分析. 中国水产科学, 22, 233-242.] |
| [4] | [程方平, 王敏晓, 王彦涛, 张芳, 李超伦, 孙松 (2012) 中国北方习见水母类的DNA条形码分析. 海洋与湖沼, 43, 451-459.] |
| [1] | Xiaoyan Luo, Qiang Li, Xiaolei Huang. DNA barcode reference dataset for flower-visiting insects in Daiyun Mountain National Nature Reserve [J]. Biodiv Sci, 2023, 31(8): 23236-. |
| [2] | Fan Wu, Shenyun Liu, Huqiang Jiang, Qian Wang, Kaiwei Chen, Hongliang Li. Pollination difference between Apis cerana cerana and Apis mellifera ligustica during the late autumn and winter [J]. Biodiv Sci, 2023, 31(5): 22528-. |
| [3] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [4] | Xiaofeng Niu, Xiaomei Wang, Yan Zhang, Zhipeng Zhao, Enyuan Fan. Integration and application of sturgeon identification methods [J]. Biodiv Sci, 2022, 30(6): 22034-. |
| [5] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [6] | Zhengsen Yu, Na Song, Hiroyuki Motomura, Tianxiang Gao. Taxonomic revision of the cardinalfish genus Jaydia in China [J]. Biodiv Sci, 2021, 29(7): 971-979. |
| [7] | Rui Hu, Ruxiao Wang, Shiyu Du, Meng Li, Yuhui Xing, Da Pan, Haigen Xu, Hongying Sun. Biodiversity and spatiotemporal variations of benthic macroinvertebrates in the Baoying Lake, Yangzhou, Jiangsu [J]. Biodiv Sci, 2020, 28(12): 1558-1569. |
| [8] | Xingtong Wu, Lu Chen, Minqiu Wang, Yuan Zhang, Xueying Lin, Xinyu Li, Hong Zhou, Yafeng Wen. Population structure and genetic divergence in Firmiana danxiaensis [J]. Biodiv Sci, 2018, 26(11): 1168-1179. |
| [9] | Ruoyu Liu, Zhongmin Sun, Jianting Yao, Zimin Hu, Delin Duan. Genetic diversity of the habitat-forming red alga Gracilaria vermiculophylla along Chinese coasts [J]. Biodiv Sci, 2016, 24(7): 781-790. |
| [10] | Gangbiao Xu,Yan Liang,Yan Jiang,Xiongsheng Liu,Shangli Hu,Yufei Xiao,Bobo Hao. Genetic diversity and population structure of Bretschneidera sinensis, an endangered species [J]. Biodiv Sci, 2013, 21(6): 723-731. |
| [11] | Kai He, Wenzhi Wang, Quan Li, Peipeng Luo, Yuehua Sun, Xuelong Jiang. DNA barcoding in surveys of small mammal community: a case study in Lianhuashan, Gansu Province, China [J]. Biodiv Sci, 2013, 21(2): 197-205. |
| [12] | Rong Zhou, Jiaqi Li, You Li, Naifa Liu, Fengjie Fang, Limin Shi, Ying Wang. Genetic variation in rusty-necklaced partridge (Alectoris magna) detected by mitochondrial DNA [J]. Biodiv Sci, 2012, 20(4): 451-459. |
| [13] | Biyun Chen, Qiong Hu, Christina Dixelius, Guoqing Li, Xiaoming Wu. Genetic diversity in Sclerotinia sclerotiorum assessed with SRAP markers [J]. Biodiv Sci, 2010, 18(5): 509-515. |
| [14] | Junhong Zhang, Huahong Huang, Zaikang Tong, Longjun Cheng, Yuelong Liang, Yiliang Chen. Genetic diversity in six natural populations of Betula luminifera from southern China [J]. Biodiv Sci, 2010, 18(3): 233-240. |
| [15] | Gen Zhang, Yilong Xi, Yinghao Xue, Xin Hu, Xianling Xiang, Xinli Wen. Effects of coal ash pollution on the genetic diversity of Brachionus calyciflorus based on rDNA ITS sequences [J]. Biodiv Sci, 2010, 18(3): 241-250. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()