



生物多样性 ›› 2025, Vol. 33 ›› Issue (1): 24251. DOI: 10.17520/biods.2024251 cstr: 32101.14.biods.2024251
王嘉陈1( ), 徐汤俊1(
), 徐汤俊1( ), 许唯1(
), 许唯1( ), 张高季1(
), 张高季1( ), 尤艺瑾1, 阮宏华2(
), 尤艺瑾1, 阮宏华2( ), 刘宏毅1,2,*(
), 刘宏毅1,2,*( )(
)( )
)
收稿日期:2024-06-22
接受日期:2024-08-06
出版日期:2025-01-20
发布日期:2024-09-13
通讯作者:
* E-mail: 基金资助:
Jiachen Wang1( ), Tangjun Xu1(
), Tangjun Xu1( ), Wei Xu1(
), Wei Xu1( ), Gaoji Zhang1(
), Gaoji Zhang1( ), Yijin You1, Honghua Ruan2(
), Yijin You1, Honghua Ruan2( ), Hongyi Liu1,2,*(
), Hongyi Liu1,2,*( )(
)( )
)
Received:2024-06-22
Accepted:2024-08-06
Online:2025-01-20
Published:2024-09-13
Contact:
* E-mail: Supported by:摘要:
城市化改变了原有的自然景观格局, 导致生物多样性丧失。土壤动物在食物链中扮演着关键角色, 对维持生态系统稳定起着重要作用。研究城市景观格局变化对土壤动物遗传多样性和种群结构的影响, 可以为维持城市生态系统稳定及其生物多样性保护提供理论依据。本研究采集了南京地区7个种群共133个大蚰蜒(Thereuopoda clunifera)样品, 以线粒体Cytb基因和6个微卫星位点为分子标记, 分析了大蚰蜒的种群遗传结构及其影响因素。结果显示: 7个种群中共检测出6个Cytb单倍型和14个突变位点, 各种群核苷酸多样性指数均低于0.005, 遗传多样性处在较低水平。而微卫星标记反映各种群的平均等位基因数在4.167-5.167之间, 观测杂合度在0.470-0.603之间, 各种群微卫星多样性较高。栖息地面积和周边城市化程度与种群遗传多样性间不存在相关性。种群间两两遗传分化系数介于0.020-0.106, 基因流介于2.108-12.266, 表明种群间遗传分化水平较低, 存在较为频繁的基因交流。遗传分化水平与预测的地理阻力呈显著正相关, 大蚰蜒的种群遗传结构可能受到城市化的影响。斑块间良好的连通性保证了种群间的基因流, 散布的城市绿地可作为廊道, 为大蚰蜒提供扩散机会。本研究结果可为城市土壤动物多样性的保护提供理论依据。
王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅 (2025) 城市景观格局对大蚰蜒种群遗传结构的影响. 生物多样性, 33, 24251. DOI: 10.17520/biods.2024251.
Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu (2025) Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China. Biodiversity Science, 33, 24251. DOI: 10.17520/biods.2024251.
| 种群 Populations | 样地名称 Localities | 样地面积 Area of sampling sites (ha) | 样品数量 Sample size | 采样时间 Sampling time | 样地距市中心距离 Distance from the city center (km) |
|---|---|---|---|---|---|
| BJG | 北极阁 Beijige | 26.87 | 16 | 2022.6.3‒6.20 | 2.15 |
| XHS | 仙鹤山 Xianhe Mountain | 222.10 | 20 | 2022.6.25‒7.18 | 14.19 |
| TS | 汤山 Tang Mountain | 559.12 | 18 | 2022.6.21‒7.12 | 23.35 |
| LWS | 龙王山 Longwang Mountain | 210.07 | 13 | 2022.6.7‒6.24 | 18.12 |
| MFS | 幕府山 Mufu Mountain | 564.52 | 21 | 2022.7.19‒8.8 | 9.54 |
| ZJS | 紫金山 Zijin Mountain | 2,828.34 | 23 | 2022.8.9‒8.21 | 7.03 |
| FS | 方山 Fang Mountain | 587.82 | 22 | 2022.7.21‒8.12 | 18.26 |
表1 南京市7个大蚰蜒种群样品采集信息
Table 1 Sample information of seven Thereuopoda clunifera populations in Nanjing
| 种群 Populations | 样地名称 Localities | 样地面积 Area of sampling sites (ha) | 样品数量 Sample size | 采样时间 Sampling time | 样地距市中心距离 Distance from the city center (km) |
|---|---|---|---|---|---|
| BJG | 北极阁 Beijige | 26.87 | 16 | 2022.6.3‒6.20 | 2.15 |
| XHS | 仙鹤山 Xianhe Mountain | 222.10 | 20 | 2022.6.25‒7.18 | 14.19 |
| TS | 汤山 Tang Mountain | 559.12 | 18 | 2022.6.21‒7.12 | 23.35 |
| LWS | 龙王山 Longwang Mountain | 210.07 | 13 | 2022.6.7‒6.24 | 18.12 |
| MFS | 幕府山 Mufu Mountain | 564.52 | 21 | 2022.7.19‒8.8 | 9.54 |
| ZJS | 紫金山 Zijin Mountain | 2,828.34 | 23 | 2022.8.9‒8.21 | 7.03 |
| FS | 方山 Fang Mountain | 587.82 | 22 | 2022.7.21‒8.12 | 18.26 |
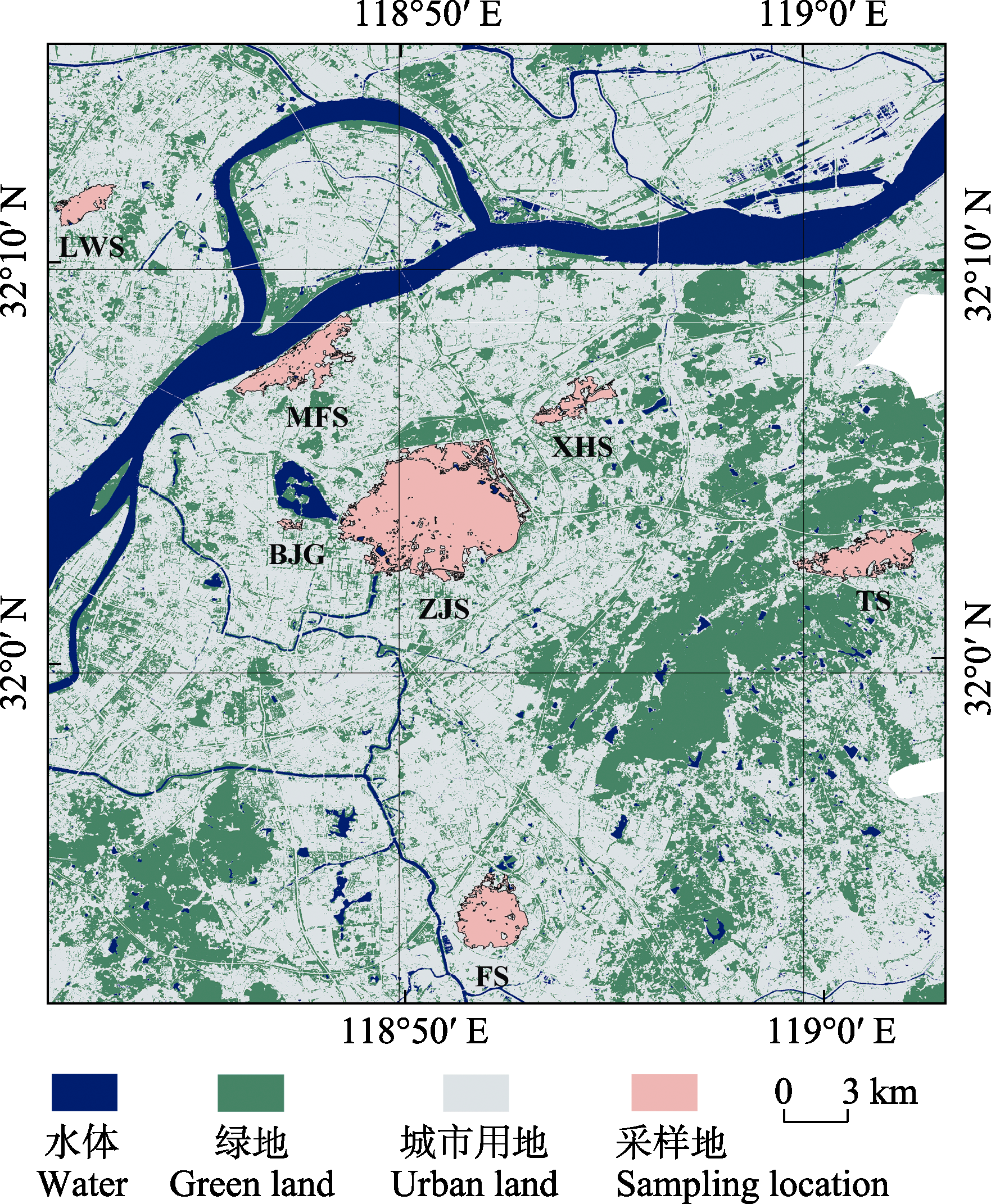
图1 南京7个大蚰蜒种群采样图。种群名称缩写含义同表1。
Fig. 1 Sampling sites of seven Thereuopoda clunifera populations in Nanjing. Population name abbreviations are shown in Table 1.
| 种群 Populations | Cytb基因 Cytb gene | 微卫星位点 Microsatellite locus | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | n | S | Hd | Pi | Na | Ne | Ho | He | |
| BJG | 16 | 1 | 0 | 0 | 0 | 5.167 | 2.849 | 0.479 | 0.604 |
| XHS | 20 | 1 | 0 | 0 | 0 | 5.000 | 2.982 | 0.492 | 0.645 |
| TS | 18 | 3 | 9 | 0.216 | 0.00186 | 5.167 | 2.638 | 0.470 | 0.587 |
| LWS | 13 | 2 | 3 | 0.282 | 0.00143 | 4.167 | 2.867 | 0.536 | 0.624 |
| MFS | 21 | 2 | 1 | 0.095 | 0.00016 | 5.000 | 3.192 | 0.603 | 0.655 |
| ZJS | 23 | 1 | 0 | 0 | 0 | 5.000 | 3.332 | 0.566 | 0.651 |
| FS | 22 | 2 | 1 | 0.247 | 0.00042 | 4.833 | 2.807 | 0.500 | 0.579 |
表2 基于线粒体DNA Cytb基因和微卫星的南京大蚰蜒遗传多样性参数
Table 2 Genetic diversity of Thereuopoda clunifera in Nanjing based on mtDNA Cytb gene and microsatellites
| 种群 Populations | Cytb基因 Cytb gene | 微卫星位点 Microsatellite locus | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | n | S | Hd | Pi | Na | Ne | Ho | He | |
| BJG | 16 | 1 | 0 | 0 | 0 | 5.167 | 2.849 | 0.479 | 0.604 |
| XHS | 20 | 1 | 0 | 0 | 0 | 5.000 | 2.982 | 0.492 | 0.645 |
| TS | 18 | 3 | 9 | 0.216 | 0.00186 | 5.167 | 2.638 | 0.470 | 0.587 |
| LWS | 13 | 2 | 3 | 0.282 | 0.00143 | 4.167 | 2.867 | 0.536 | 0.624 |
| MFS | 21 | 2 | 1 | 0.095 | 0.00016 | 5.000 | 3.192 | 0.603 | 0.655 |
| ZJS | 23 | 1 | 0 | 0 | 0 | 5.000 | 3.332 | 0.566 | 0.651 |
| FS | 22 | 2 | 1 | 0.247 | 0.00042 | 4.833 | 2.807 | 0.500 | 0.579 |
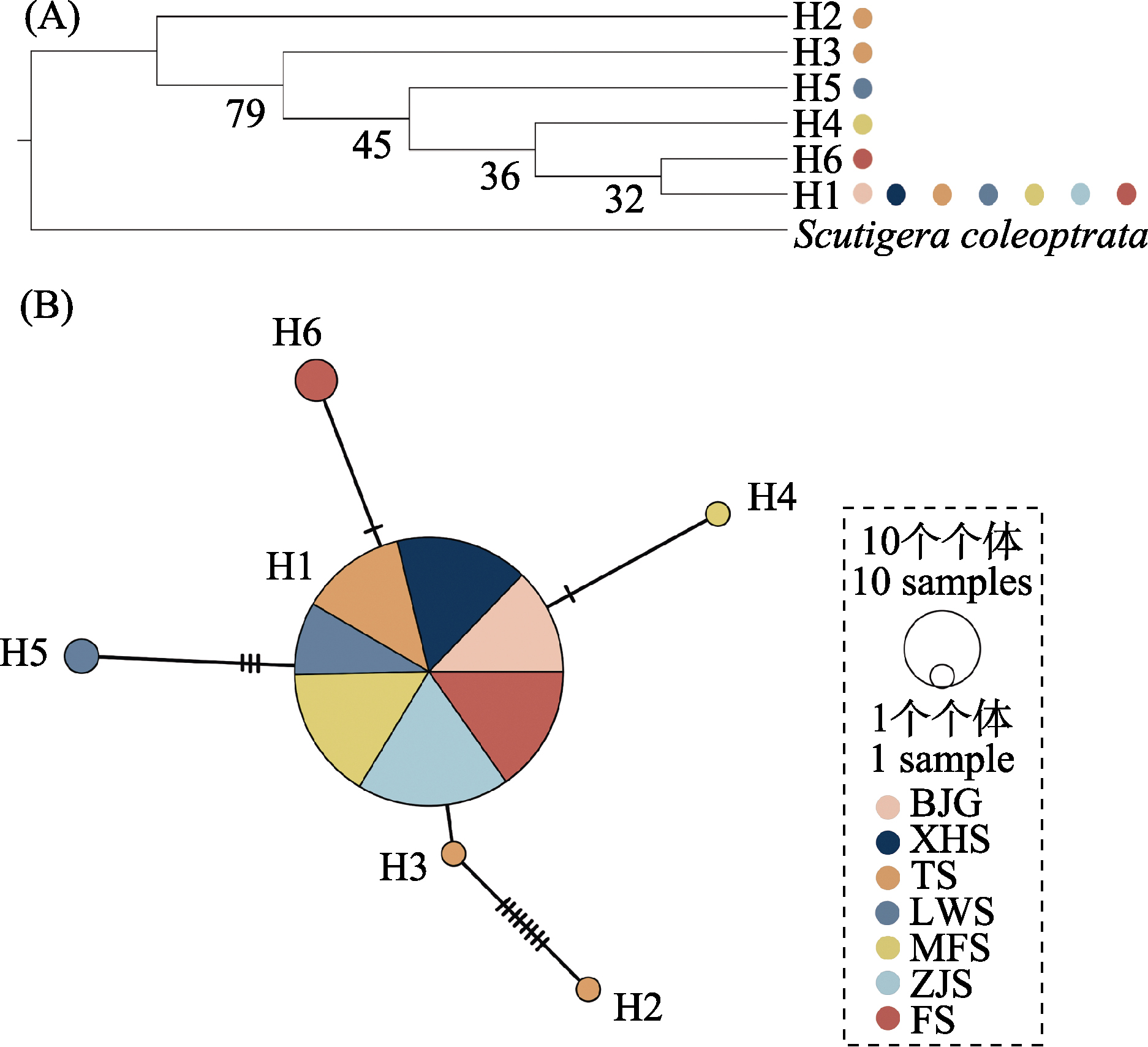
图2 基于南京大蚰蜒种群的线粒体DNA Cytb基因的系统发育树(A)和单倍型网络关系图(B)。其中H1-H6为本研究所测线粒体DNA Cytb序列生成的单倍型, 系统发育树的外类群为地中海蚰蜒(Scutigera coleoptrata), 在单倍型网络关系图中不同颜色代表不同地理种群。种群名称缩写含义同表1。
Fig. 2 The molecular phylogenetic tree (A) and the haplotype network (B) based on mtDNA Cytb gene in Nangjing populations of Thereuopoda clunifera. H1-H6 are haplotypes generated from mtDNA Cytb sequences from this study. The outgroup of the molecular phylogenetic tree is Scutigera coleoptrata. Different colors represent different geographical populations in haplotype network. Population name abbreviations are shown in Table 1.
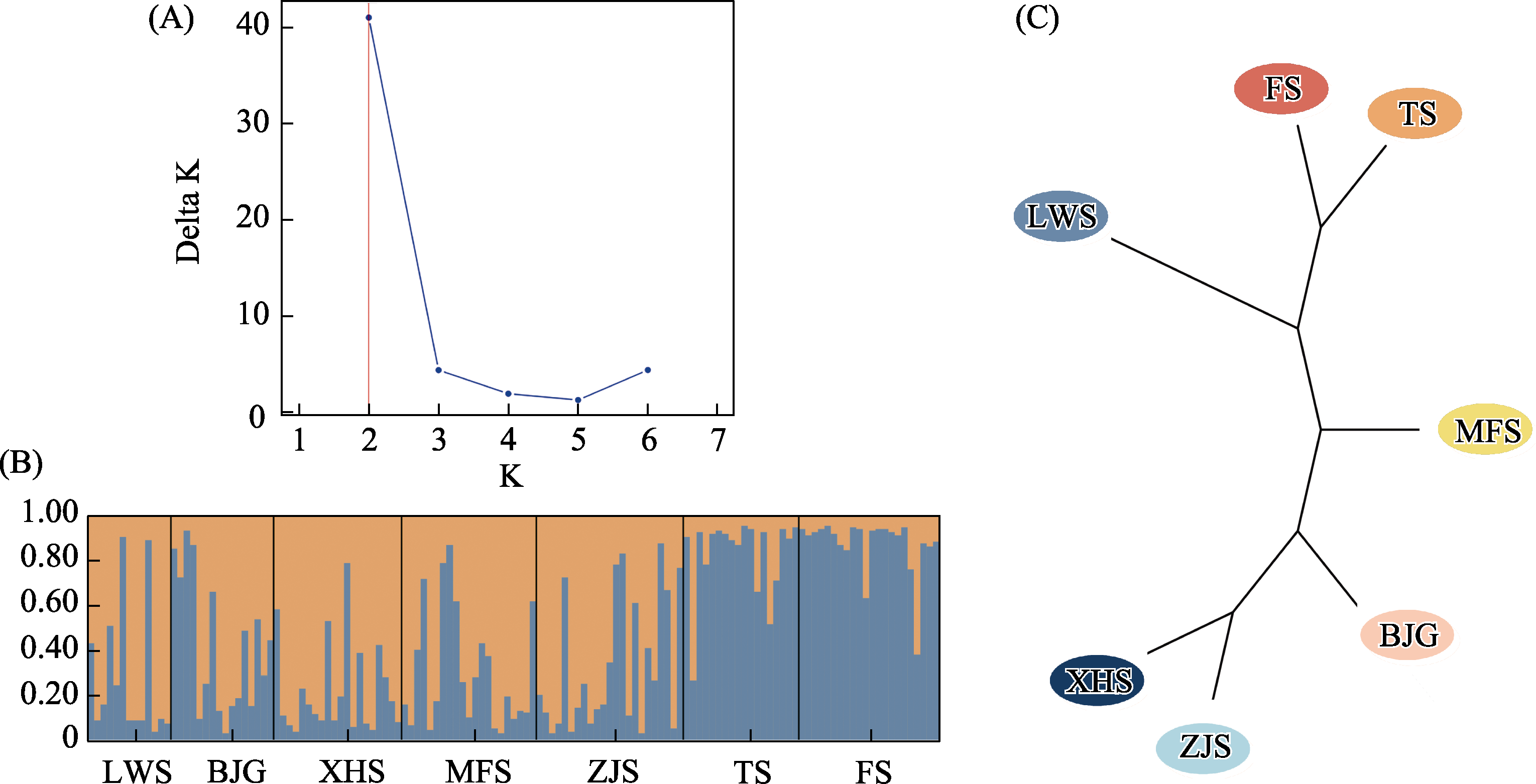
图3 用于确定种群最优数量(K)的Delta K图(A)、南京大蚰蜒种群的遗传结构图(K = 2时) (B)和基于种群间遗传距离的UPGMA树(C)。种群名称缩写含义同表1。
Fig. 3 Delta K plot for determining optimal number of populations (K) (A), structure of Thereuopoda clunifera populations in Nanjing (K = 2) (B), and UPGMA tree based on genetic distances between populations (C). Population name abbreviations are shown in Table 1.
| 变异来源 Source of variation | 自由度 Degree of freedom | 方差平方和 Sum of squares of variance | 变异组分 Variance components | 变异百分比 Percentage of variation (%) | 固定指数 Fixation index |
|---|---|---|---|---|---|
| 种群间 Among populations | 6 | 36.148 | 0.11700 | 6.81 | FST = 0.06811; P < 0.01 |
| 种群内 Within populations | 259 | 414.634 | 1.60090 | 93.19 | |
| 合计 Total | 265 | 450.782 | 1.7179 |
表3 基于微卫星位点的南京大蚰蜒种群的分子方差分析
Table 3 Analysis of molecular variance on microsatellites among Thereuopoda clunifera populations in Nanjing
| 变异来源 Source of variation | 自由度 Degree of freedom | 方差平方和 Sum of squares of variance | 变异组分 Variance components | 变异百分比 Percentage of variation (%) | 固定指数 Fixation index |
|---|---|---|---|---|---|
| 种群间 Among populations | 6 | 36.148 | 0.11700 | 6.81 | FST = 0.06811; P < 0.01 |
| 种群内 Within populations | 259 | 414.634 | 1.60090 | 93.19 | |
| 合计 Total | 265 | 450.782 | 1.7179 |
| 种群 Populations | BJG | XHS | TS | LWS | MFS | ZJS | FS |
|---|---|---|---|---|---|---|---|
| BJG | 0.052 | 0.079 | 0.106 | 0.061 | 0.046 | 0.067 | |
| XHS | 4.520 | 0.090 | 0.064 | 0.051 | 0.020 | 0.090 | |
| TS | 2.912 | 2.542 | 0.094 | 0.062 | 0.072 | 0.039 | |
| LWS | 2.112 | 3.675 | 2.400 | 0.082 | 0.053 | 0.106 | |
| MFS | 3.860 | 4.617 | 3.763 | 2.792 | 0.031 | 0.065 | |
| ZJS | 5.212 | 12.266 | 3.225 | 4.507 | 7.747 | 0.065 | |
| FS | 3.473 | 2.538 | 6.205 | 2.108 | 3.579 | 3.574 |
表4 基于微卫星位点的南京大蚰蜒种群的遗传分化系数(对角线上)和基因流(对角线下)
Table 4 Fixation index (FST) (above the diagonal) and gene flow (Nm) (below the diagonal) of Thereuopoda clunifera populations in Nanjing based on microsatellites. Population name abbreviations are shown in Table 1.
| 种群 Populations | BJG | XHS | TS | LWS | MFS | ZJS | FS |
|---|---|---|---|---|---|---|---|
| BJG | 0.052 | 0.079 | 0.106 | 0.061 | 0.046 | 0.067 | |
| XHS | 4.520 | 0.090 | 0.064 | 0.051 | 0.020 | 0.090 | |
| TS | 2.912 | 2.542 | 0.094 | 0.062 | 0.072 | 0.039 | |
| LWS | 2.112 | 3.675 | 2.400 | 0.082 | 0.053 | 0.106 | |
| MFS | 3.860 | 4.617 | 3.763 | 2.792 | 0.031 | 0.065 | |
| ZJS | 5.212 | 12.266 | 3.225 | 4.507 | 7.747 | 0.065 | |
| FS | 3.473 | 2.538 | 6.205 | 2.108 | 3.579 | 3.574 |
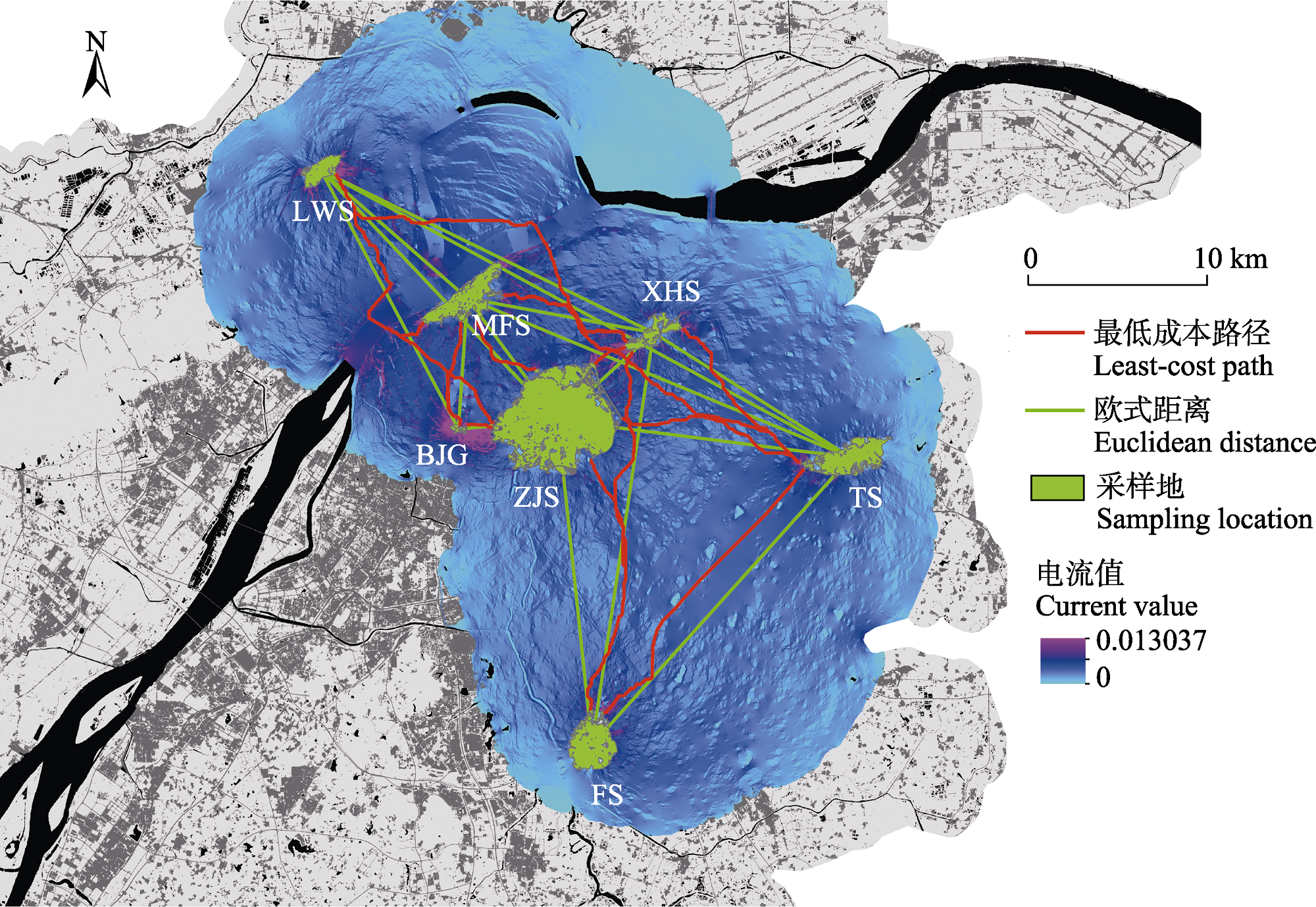
图4 南京大蚰蜒种群间欧式距离及最低成本路径示意图和扩散路径电流图。种群名称缩写含义同表1。
Fig. 4 The Euclidean distance, least-cost path and disperse path of Thereuopoda clunifera populations in Nanjing. The abbreviation of the population name has the same meaning as in Table 1.
| [1] | Adriaensen F, Chardon JP, De Blust G, Swinnen E, Villalba S, Gulinck H, Matthysen E (2003) The application of ‘least- cost’ modelling as a functional landscape model. Landscape and Urban Planning, 64, 233-247. |
| [2] | Björklund M, Ruiz I, Senar JC (2010) Genetic differentiation in the urban habitat: The great tits (Parus major) of the parks of Barcelona City. Biological Journal of the Linnean Society, 99, 9-19. |
| [3] | Bohonak AJ (1999) Dispersal, gene flow, and population structure. The Quarterly Review of Biology, 74, 21-45. |
| [4] | Cao LZ, Wu KM (2019) Genetic diversity and demographic history of globe skimmers (Odonata: Libellulidae) in China based on microsatellite and mitochondrial DNA markers. Scientific Reports, 9, 8619. |
| [5] | Chen CD, Wu SJ, Douglas MC, Lü MQ, Wen ZF, Jiang Y, Chen JL (2015) Effects of changing cost values on landscape connectivity simulation. Acta Ecologica Sinica, 35, 7367-7376. (in Chinese with English abstract) |
| [陈春娣, 吴胜军, Meurk Colin Douglas, 吕明权, 温兆飞, 姜毅, 陈吉龙 (2015) 阻力赋值对景观连接模拟的影响. 生态学报, 35, 7367-7376.] | |
| [6] | Chen Y (1959) Chinese Animal Atlas:Annelids (attached Myriapods). Science Press, Beijing. (in Chinese) |
| [陈义 (1959) 中国动物图谱: 环节动物(附多足类). 科学出版社, 北京.] | |
| [7] |
Clobert J, Le Galliard JF, Cote J, Meylan S, Massot M (2009) Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecology Letters, 12, 197-209.
DOI PMID |
| [8] |
Coates BS, Sumerford DV, Miller NJ, Kim KS, Sappington TW, Siegfried BD, Lewis LC (2009) Comparative performance of single nucleotide polymorphism and microsatellite markers for population genetic analysis. Journal of Heredity, 100, 556-564.
DOI PMID |
| [9] | Collins CMT, Audusseau H, Hassall C, Keyghobadi N, Sinu PA, Saunders ME (2024) Insect ecology and conservation in urban areas: An overview of knowledge and needs. Insect Conservation and Diversity, 17, 169-181. |
| [10] | Dong TW, Huang ML, Wei X, Ma S, Yue Q, Liu WL, Zheng JX, Wang G, Ma R, Ding YZ, Bo SQ, Wang ZH (2023) Potential spatial distribution pattern and landscape connectivity of Pelophylax plancyi in Shanghai, China. Biodiversity Science, 31, 22692. (in Chinese with English abstract) |
|
[董廷玮, 黄美玲, 韦旭, 马硕, 岳衢, 刘文丽, 郑佳鑫, 王刚, 马蕊, 丁由中, 薄顺奇, 王正寰 (2023) 上海地区金线侧褶蛙种群的潜在空间分布格局及其景观连通性. 生物多样性, 31, 22692.]
DOI |
|
| [11] |
Dupont L, Torres-Leguizamon M, René-Corail P, Mathieu J (2017) Landscape features impact connectivity between soil populations: A comparative study of gene flow in earthworms. Molecular Ecology, 26, 3128-3140.
DOI PMID |
| [12] | Excoffier L, Laval G, Schneider S (2007) Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47-50. |
| [13] | Field SG, Lange M, Schulenburg H, Velavan TP, Michiels NK (2007) Genetic diversity and parasite defense in a fragmented urban metapopulation of earthworms. Animal Conservation, 10, 162-175. |
| [14] | Frankham R (2003) Genetics and conservation biology. Comptes Rendus Biologies, 326, 22-29. |
| [15] |
Grande C, Templado J, Zardoya R (2008) Evolution of gastropod mitochondrial genome arrangements. BMC Evolutionary Biology, 8, 61.
DOI PMID |
| [16] | Grant W, Bowen B (1998) Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89, 415-426. |
| [17] |
Grimm NB, Faeth SH, Golubiewski NE, Redman CL, Wu JG, Bai XM, Briggs JM (2008) Global change and the ecology of cities. Science, 319, 756-760.
DOI PMID |
| [18] |
Halpin LR, Terrington DI, Jones HP, Mott R, Wong WW, Dow DC, Carlile N, Clarke RH (2021) Arthropod predation of vertebrates structures trophic dynamics in island ecosystems. The American Naturalist, 198, 540-550.
DOI PMID |
| [19] | Han T, Lee YB, Kim SH, Yoon HJ, Park IG, Park H (2018) Genetic variation of COI gene of the Korean medicinal centipede Scolopendra mutilans Koch, 1878 (Scolopendromorpha: Scolopendridae). Entomological Research, 48, 559-566. |
| [20] | He YY, Liu YY, Zhang FB, Qin Q, Zeng Y, Lü ZY, Yang K (2023) Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River. Biodiversity Science, 31, 23160. (in Chinese with English abstract) |
|
[何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤 (2023) 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构. 生物多样性, 31, 23160.]
DOI |
|
| [21] | Hu CY, Wang SB, Huang BS, Liu HG, Xu L, Hu ZG, Liu YF (2020) The complete mitochondrial genome sequence of Scolopendra mutilans L. Koch, 1878 (Scolopendromorpha, Scolopendridae), with a comparative analysis of other centipede genomes. ZooKeys, 925, 73-88. |
| [22] | Hung KLJ, Ascher JS, Holway DA (2017) Urbanization- induced habitat fragmentation erodes multiple components of temporal diversity in a Southern California native bee assemblage. PLoS ONE, 12, e0184136. |
| [23] | Jenkins D, Brescacin C, Duxbury C, Elliott J, Evans J, Grablow K, Hillegass M, Lyon B, Metzger G, Olandese M, Pepe D, Silvers G, Suresch H, Thompson T, Trexler C, Williams G, Williams N, Williams S (2007) Does size matter for dispersal distance? Global Ecology and Biogeography, 16, 415-425. |
| [24] |
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587-589.
DOI PMID |
| [25] | Krauss J, Schmitt T, Seitz A, Steffan-Dewenter I, Tscharntke T (2004) Effects of habitat fragmentation on the genetic structure of the monophagous butterfly Polyommatus coridon along its northern range margin. Molecular Ecology, 13, 311-320. |
| [26] | Leigh JW, Bryant D (2015) Popart: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6, 1110-1116. |
| [27] | Li DY, Yin QQ, Hou N, Sun XW, Liang LQ (2009) Genetic diversity of different ecologo-geographical populations of yellow catfish Pelteobagrus eupogon. Oceanologia et Limnologia Sinica, 40, 460-469. (in Chinese with English abstract) |
| [李大宇, 殷倩茜, 侯宁, 孙效文, 梁利群 (2009) 黄颡鱼(Pelteobagrus eupogon)不同生态地理分布群体遗传多样性的微卫星分析. 海洋与湖沼, 40, 460-469.] | |
| [28] |
Li GD, Fang CL, Li YJ, Wang ZB, Sun SA, He SW, Qi W, Bao C, Ma HT, Fan YP, Feng YX, Liu XP (2022) Global impacts of future urban expansion on terrestrial vertebrate diversity. Nature Communications, 13, 1628.
DOI PMID |
| [29] |
Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452.
DOI PMID |
| [30] | Lin P, Yang L, Zhao SQ (2020) Urbanization effects on Chinese mammal and amphibian richness: A multi-scale study using the urban-rural gradient approach. Environmental Research Communications, 2, 125002. |
| [31] | Liordos V, Jokimäki J, Kaisanlahti-Jokimäki ML, Valsamidis E, Kontsiotis VJ (2021) Patch, matrix and disturbance variables negatively influence bird community structure in small-sized managed green spaces located in urban core areas. Science of the Total Environment, 801, 149617. |
| [32] |
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Research, 27, 209-220.
PMID |
| [33] |
McRae BH (2006) Isolation by resistance. Evolution, 60, 1551-1561.
PMID |
| [34] | McRae BH, Beier P (2007) Circuit theory predicts gene flow in plant and animal populations. Proceedings of the National Academy of Sciences, USA, 104, 19885-19890. |
| [35] |
Miles LS, Rivkin LR, Johnson MTJ, Munshi-South J, Verrelli BC (2019) Gene flow and genetic drift in urban environments. Molecular Ecology, 28, 4138-4151.
DOI PMID |
| [36] | Mukherjee S, Krishnan A, Tamma K, Home C, Navya R, Joseph S, Das A, Ramakrishnan U (2010) Ecology driving genetic variation: A comparative phylogeography of jungle cat (Felis chaus) and leopard cat (Prionailurus bengalensis) in India. PLoS ONE, 5, e13724. |
| [37] |
Munshi-South J, Zolnik CP, Harris SE (2016) Population genomics of the Anthropocene: Urbanization is negatively associated with genome-wide variation in white-footed mouse populations. Evolutionary Applications, 9, 546-564.
DOI PMID |
| [38] |
Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics, 28, 2537-2539.
PMID |
| [39] |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
DOI PMID |
| [40] | Pu JJ, Yang PJ, Dai Y, Tao KX, Gao L, Du YZ, Cao J, Yu XP, Yang QQ (2023) Species identification and population genetic structure of non-native apple snails (Ampullariidea: Pomacea) in the lower reaches of the Yangtze River. Biodiversity Science, 31, 22346. (in Chinese with English abstract) |
|
[蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩 (2023) 长江下游外来生物福寿螺的种类及其种群遗传结构. 生物多样性, 31, 22346.]
DOI |
|
| [41] | Ranta P, Blom T, Niemela J, Joensuu E, Siitonen M (1998) The fragmented Atlantic rain forest of Brazil: Size, shape and distribution of forest fragments. Biodiversity & Conservation, 7, 385-403. |
| [42] | Rivas N, Martínez-Hernández F, Antonio-Campos A, Sánchez-Cordero V, Alejandre-Aguilar R (2022) Genetic diversity in peridomiciliary populations of Triatoma mexicana (Hemiptera: Reduviidae: Triatominae) in central Mexico. Parasitology Research, 121, 2875-2886. |
| [43] | Sabtu FS, Ab Majid AH (2017) Genetic variation and population structure of the arboreal bicolored ant Tetraponera rufonigra Jerdon from selected urban locations in eastern Penang Island, Malaysia. Journal of Asia-Pacific Entomology, 20, 1350-1357. |
| [44] | Saka T, Nishita Y, Masuda R (2018) Low genetic variation in the MHC class II DRB gene and MHC-linked microsatellites in endangered island populations of the leopard cat (Prionailurus bengalensis) in Japan. Immunogenetics, 70, 115-124. |
| [45] | Semlitsch RD (2008) Differentiating migration and dispersal processes for pond-breeding amphibians. Journal of Wildlife Management, 72, 260-267. |
| [46] | Sharma R, Stuckas H, Bhaskar R, Khan I, Goyal SP, Tiedemann R (2011) Genetically distinct population of Bengal tiger (Panthera tigris tigris) in Terai Arc Landscape (TAL) of India. Mammalian Biology, 76, 484-490. |
| [47] |
Soanes K, Sievers M, Chee YE, Williams NSG, Bhardwaj M, Marshall AJ, Parris KM (2019) Correcting common misconceptions to inspire conservation action in urban environments. Conservation Biology, 33, 300-306.
DOI PMID |
| [48] |
Song X, Yin ZQ, Li L, Cheng AC, Jia RY, Xu J, Wang Y, Yao XP, Lv C, Zhao XH (2013) Antiviral activity of sulfated Chuanminshen violaceum polysaccharide against duck enteritis virus in vitro. Antiviral Research, 98, 344-351.
DOI PMID |
| [49] | Suárez D, Arribas P, Jiménez-García E, Emerson BC (2022) Dispersal ability and its consequences for population genetic differentiation and diversification. Proceedings of the Royal Society B: Biological Sciences, 289, 20220489. |
| [50] |
Takezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics, 144, 389-399.
DOI PMID |
| [51] | Taylor PD, Fahrig L, Henein K, Merriam G (1993) Connectivity is a vital element of landscape structure. Oikos, 68, 571-573. |
| [52] |
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882.
DOI PMID |
| [53] | Vessey SH, Vessey KB (2007) Linking behavior, life history and food supply with the population dynamics of white- footed mice (Peromyscus leucopus). Integrative Zoology, 2, 123-130. |
| [54] | Wang JJ, Li C, Chen JQ, Wang JJ, Jin JJ, Jiang SY, Yan LB, Lin HD, Zhao J (2021) Phylogeographic structure of the dwarf snakehead (Channa gachua) around Gulf of Tonkin: Historical biogeography and pronounced effects of sea-level changes. Ecology and Evolution, 11, 12583-12595. |
| [55] |
Wang JM, Zheng LJ, Liu H, Xu BW, Zou ZC (2020) The effects of habitat network construction and urban block unit structure on biodiversity in semiarid green spaces. Environmental Monitoring and Assessment, 192, 179.
DOI PMID |
| [56] |
Wang ST, Bai XM, Zhang XL, Reis S, Chen DL, Xu JM, Gu BJ (2021) Urbanization can benefit agricultural production with large-scale farming in China. Nature Food, 2, 183-191.
DOI PMID |
| [57] |
Weir BS, Cockerham CC (1984) Estimating f-statistics for the analysis of population structure. Evolution, 38, 1358-1370.
DOI PMID |
| [58] | Wood R, Kocher TD, Stepien CA (1998) Molecular systematics of fishes. Copeia, 1998, 530. |
| [59] |
Wright S (1943) Isolation by distance. Genetics, 28, 114-138.
DOI PMID |
| [60] |
Wu DH, Fu SL (2022) Soil animal biodiversity: Taxonomy and community ecology. Biodiversity Science, 30, 22680. (in Chinese)
DOI |
|
[吴东辉, 傅声雷 (2022) 土壤动物多样性: 物种与群落研究. 生物多样性, 30, 22680.]
DOI |
|
| [61] | Xu RF, Chen J, Pan Y, Wang JC, Chen L, Ruan HH, Wu YB, Xu HM, Wang GB, Liu HY (2022) Genetic diversity and population structure of Spirobolus bungii as revealed by mitochondrial DNA sequences. Insects, 13, 729. |
| [62] | Xu TJ, Zhou KX, Ye WT, Huang YL, Zhang YF, Zhu S, Ruan HH, Liu HY (2024) Genetic diversity and gene flow of the soil arthropod (Scolopendra mutilans) in urban landscapes: The roles of rivers, mountains and fragmentation. Insect Conservation and Diversity, 17, 203-214. |
| [63] | Yang R, Meng LH, Zhang Q, Liu JQ (2005) The mitochondrial DNA nad 1 sequence variation of Picea crassifolia (Pinaceae) in the Qinghai-Tibetan Plateau platform and adjacent populations. Acta Ecologica Sinica, 25, 3307-3313. (in Chinese with English abstrast) |
| [杨瑞, 孟丽华, 张茜, 刘建全 (2005) 线粒体DNA nad1序列在青海云杉青藏高原台面和周边地区种群中的变异. 生态学报, 25, 3307-3313.] | |
| [64] | Yuan J, Wang ZT, Duan PX, Xiao YS, Zhang HK, Huang ZX, Zhou RC, Wen H, Wang KX, Wang D (2021) Whistle signal variations among three Indo-Pacific humpback dolphin populations in the South China Sea: A combined effect of the Qiongzhou Strait’s geographical barrier function and local ambient noise? Integrative Zoology, 16, 499-511. |
| [65] | Yue WZ, Zhou QS, Li MM, van Vliet J (2023) Relocating built-up land for biodiversity conservation in an uncertain future. Journal of Environmental Management, 345, 118706. |
| [66] |
Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20, 348-355.
DOI PMID |
| [67] | Zhang SP, Suo ML, Liu SL, Liang W (2013) Do major roads reduce gene flow in urban bird populations? PLoS ONE, 8, e77026. |
| [68] | Zhao XY, Lu YF, Fan SM, Xu W, Wang JC, Wang GB, Liu HY (2022) The complete mitochondrial genome of Thereuopoda clunifera (Chilopoda: Scutigeridae) and phylogenetic implications within Chilopoda. Zootaxa, 5174, 165-180. |
| [1] | 刘淑琪, 崔东, 江智诚, 刘江慧, 闫江超. 短期氮、水添加和刈割减弱了苦豆子型退化草地土壤生物多样性与生态系统多功能性的联系[J]. 生物多样性, 2025, 33(3): 24305-. |
| [2] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [3] | 孙怡欣, 侯春雨, 周磊, 魏雪, 马金豪, 薛娟, 李小涵, 吴鹏飞. 青藏高原盆栽一年生和多年生豆科牧草对土壤线虫群落的影响[J]. 生物多样性, 2024, 32(7): 24040-. |
| [4] | 马骅, 李常青, 余品锋, 陈杰, 贺天耀, 王可洪. 澎溪河消落带大型土壤动物群落分布格局及其影响因素[J]. 生物多样性, 2024, 32(7): 24117-. |
| [5] | 王党军, 谢午阳, 林小元, 乔秀娟, 徐耀粘, 田秋香, 刘峰, 张娅妮, 左娟, 江明喜. 八大公山森林土壤动物群落与叶经济谱及凋落物分解速率的关系[J]. 生物多样性, 2024, 32(12): 24261-. |
| [6] | 李乐, 张承云, 裴男才, 高丙涛, 王娜, 李嘉睿, 武瑞琛, 郝泽周. 基于被动声学监测技术的城市绿地景观格局与鸟类多样性关联分析[J]. 生物多样性, 2024, 32(10): 24296-. |
| [7] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [8] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [9] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [10] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [11] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [12] | 谢致敬, 刘相钰, 孙晓铭, 刘继亮, 刘占锋, 张晓珂, 陈军, 杨效东, 朱波, 柯欣, 吴东辉. 中国土壤动物多样性监测网络建设、进展与展望[J]. 生物多样性, 2023, 31(12): 23365-. |
| [13] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [14] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [15] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn