



生物多样性 ›› 2022, Vol. 30 ›› Issue (5): 21031. DOI: 10.17520/biods.2021031 cstr: 32101.14.biods.2021031
• 研究报告: 遗传多样性 • 下一篇
陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来( ), 韩海格(
), 韩海格( )
)
收稿日期:2021-07-20
接受日期:2022-04-29
出版日期:2022-05-20
发布日期:2022-05-20
通讯作者:
芒来,韩海格
作者简介:hanhaige@imau.edu.cn基金资助:
Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin( ), Haige Han(
), Haige Han( )
)
Received:2021-07-20
Accepted:2022-04-29
Online:2022-05-20
Published:2022-05-20
Contact:
Manglai Dugarjaviin,Haige Han
摘要:
家马地方品种的遗传多样性是动态演变的, 并与其育种模式息息相关; 在过去约300年中, 育种者培育了具突出表型特征的标准化培育品种, 该类型品种对家马地方品种的繁育产生了巨大影响, 这是导致家马地方品种遗传多样性下降及遗传分化的主要因素之一。本研究采集了5个东亚家马地方品种(蒙古马、哈萨克马、河曲马、藏马和西南马)、2个西亚家马品种(阿拉伯马和阿哈尔捷金马)以及2个欧洲家马品种(设特兰矮马和克莱斯黛尔马)的70个样品, 并整合之前所发表的100匹内蒙古地区家马不同群体单核苷酸多态性(SNPs)数据集, 通过全基因组重测序和生物信息学方法分析了东亚家马不同群体的遗传多样性。研究发现东亚家马地方品种具有较丰富的遗传多样性, 相比欧洲和西亚品种产生了显著的遗传分化, 其中蒙古高原群体的遗传多样性最为丰富; 受杂交改良影响, 内蒙古不同群体间产生了一定程度的遗传分化; 河曲马和藏马的遗传背景最为单一, 受引种杂交繁育影响较小。本研究评估了东亚家马地方品种的遗传多样性分布格局和演化特征, 可为建立家马地方种质核心保护群体以及培育新品种提供遗传学理论支持。
陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格 (2022) 基于基因组SNPs对东亚家马不同群体遗传多样性的评估. 生物多样性, 30, 21031. DOI: 10.17520/biods.2021031.
Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han (2022) Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses. Biodiversity Science, 30, 21031. DOI: 10.17520/biods.2021031.
| 品种/群体 Breed/Population | 样品代号 Code | 采样地 Sampling region | 样本量 Sample size | 近交系数 Inbreeding coefficient (F) | 期望杂合度 Expected heterozygosity (He) | 数据来源 Data source |
|---|---|---|---|---|---|---|
| 藏马 Tibetan horse | TIB | 西藏那曲 Nagqu, Tibet, China | 7 | 0.031 | 0.246 | 本研究 Present study |
| 河曲马 Hequ horse | HQ | 青海省河南蒙古族自治县 Henan Mongol Autonomous County, Qinghai, China | 6 | 0.045 | 0.239 | 本研究 Present study |
| 西南马 Southwest horse | SW | 云南丽江 Lijiang, Yunnan, China | 7 | 0.043 | 0.247 | 本研究 Present study |
| 蒙古马 Mongolian horse | MN | 乌兰巴托市, 蒙古国 Ulaanbaatar, Mongolia | 13 | 0.017 | 0.261 | 本研究 Present study |
| 乌珠穆沁马 Ujimqin horse | UJM | 内蒙古锡林郭勒盟 Xilingol League, Inner Mongolia, China | 3 21 | 0.029 | 0.266 | 本研究 Present study; Han et al, |
| 乌审马 Uxin horse | UX | 内蒙古鄂尔多斯市乌审旗 Uxin Banner, Inner Mongolia, China | 3 22 | 0.042 | 0.266 | 本研究 Present study; Han et al, |
| 百岔铁蹄马 Baicha iron hoof horse | BCIH | 内蒙古赤峰市克什克腾旗 Hexigten Banner, Inner Mongolia, China | 2 19 | 0.019 | 0.264 | 本研究 Present study; Han et al, |
| 三河马 Sanhe horse | SH | 内蒙古呼伦贝尔市 Hulunbuir, Inner Mongolia, China | 23 | -0.003 | 0.269 | Han et al, |
| 阿巴嘎黑马 Abag horse | AB | 内蒙古锡林郭勒盟阿巴嘎旗 Abag Banner, Inner Mongolia, China | 15 | 0.027 | 0.251 | Han et al, |
| 哈萨克马 Kazakh horse | KZ | 新疆塔城地区 Tacheng, Xinjiang, China | 6 | 0.00028 | 0.252 | 本研究 Present study |
| 阿拉伯马 Arabian horse | ARR | 北京 Beijing, China | 5 | 0.108 | 0.213 | 本研究 Present study |
| 设特兰矮马 Shetland pony | STL | 内蒙古鄂尔多斯市可汗御马苑 Ordos, Inner Mongolia, China | 5 | 0.120 | 0.221 | 本研究 Present study |
| 阿哈尔捷金马 Akhal-Teke horse | AKTK | 内蒙古太仆寺旗 Taibus Banner, Inner Mongolia, China | 5 | 0.07 | 0.223 | 本研究 Present study |
| 克莱斯戴尔马 Clydesdale horse | DRT | 内蒙古鄂尔多斯市可汗御马苑 Ordos, Inner Mongolia, China | 5 | 0.24 | 0.227 | 本研究 Present study |
| 总计 Total | 14群体 14 populations | 167个体 167 individuals |
表1 本研究所使用样品来源及其近交系数和期望杂合度信息
Table 1 Information on collection source, inbreeding coefficient (F) and expected heterozygosity (He) of samples used in this study
| 品种/群体 Breed/Population | 样品代号 Code | 采样地 Sampling region | 样本量 Sample size | 近交系数 Inbreeding coefficient (F) | 期望杂合度 Expected heterozygosity (He) | 数据来源 Data source |
|---|---|---|---|---|---|---|
| 藏马 Tibetan horse | TIB | 西藏那曲 Nagqu, Tibet, China | 7 | 0.031 | 0.246 | 本研究 Present study |
| 河曲马 Hequ horse | HQ | 青海省河南蒙古族自治县 Henan Mongol Autonomous County, Qinghai, China | 6 | 0.045 | 0.239 | 本研究 Present study |
| 西南马 Southwest horse | SW | 云南丽江 Lijiang, Yunnan, China | 7 | 0.043 | 0.247 | 本研究 Present study |
| 蒙古马 Mongolian horse | MN | 乌兰巴托市, 蒙古国 Ulaanbaatar, Mongolia | 13 | 0.017 | 0.261 | 本研究 Present study |
| 乌珠穆沁马 Ujimqin horse | UJM | 内蒙古锡林郭勒盟 Xilingol League, Inner Mongolia, China | 3 21 | 0.029 | 0.266 | 本研究 Present study; Han et al, |
| 乌审马 Uxin horse | UX | 内蒙古鄂尔多斯市乌审旗 Uxin Banner, Inner Mongolia, China | 3 22 | 0.042 | 0.266 | 本研究 Present study; Han et al, |
| 百岔铁蹄马 Baicha iron hoof horse | BCIH | 内蒙古赤峰市克什克腾旗 Hexigten Banner, Inner Mongolia, China | 2 19 | 0.019 | 0.264 | 本研究 Present study; Han et al, |
| 三河马 Sanhe horse | SH | 内蒙古呼伦贝尔市 Hulunbuir, Inner Mongolia, China | 23 | -0.003 | 0.269 | Han et al, |
| 阿巴嘎黑马 Abag horse | AB | 内蒙古锡林郭勒盟阿巴嘎旗 Abag Banner, Inner Mongolia, China | 15 | 0.027 | 0.251 | Han et al, |
| 哈萨克马 Kazakh horse | KZ | 新疆塔城地区 Tacheng, Xinjiang, China | 6 | 0.00028 | 0.252 | 本研究 Present study |
| 阿拉伯马 Arabian horse | ARR | 北京 Beijing, China | 5 | 0.108 | 0.213 | 本研究 Present study |
| 设特兰矮马 Shetland pony | STL | 内蒙古鄂尔多斯市可汗御马苑 Ordos, Inner Mongolia, China | 5 | 0.120 | 0.221 | 本研究 Present study |
| 阿哈尔捷金马 Akhal-Teke horse | AKTK | 内蒙古太仆寺旗 Taibus Banner, Inner Mongolia, China | 5 | 0.07 | 0.223 | 本研究 Present study |
| 克莱斯戴尔马 Clydesdale horse | DRT | 内蒙古鄂尔多斯市可汗御马苑 Ordos, Inner Mongolia, China | 5 | 0.24 | 0.227 | 本研究 Present study |
| 总计 Total | 14群体 14 populations | 167个体 167 individuals |
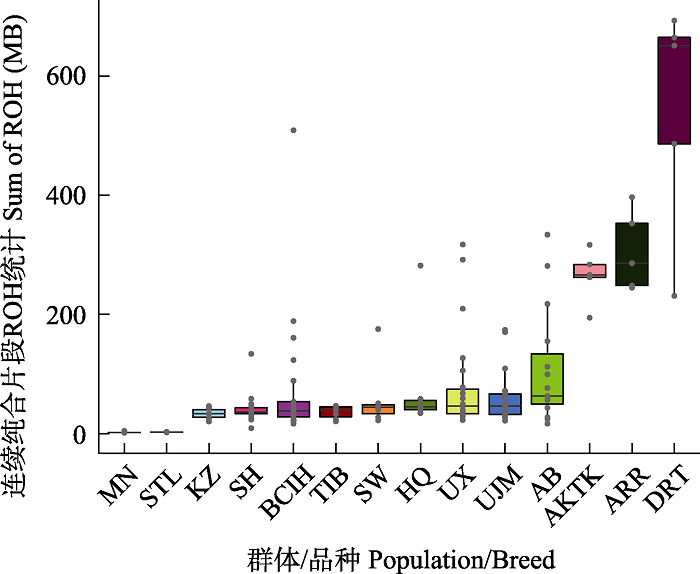
图1 样品基因组ROH长度(MB)统计。MN: 蒙古马; STL:设特兰矮马; KZ: 哈萨克马; SH: 三河马; BCIH: 百岔铁蹄马; TIB: 藏马; SW: 西南马; HQ: 河曲马; UX: 乌审马; UJM: 乌珠穆沁马; AB: 阿巴嘎黑马; AKTK: 阿哈尔捷金马; ARR: 阿拉伯马; DRT: 克莱斯戴尔马。
Fig. 1 The sum of ROH length (MB) per individual genome in each population of horse samples. MN, Mongolian horse; STL, Shetland pony; KZ, Kazakh horse; SH, Sanhe horse; BCIH, Baicha iron hoof horse; TIB, Tibetan horse; SW, Southwest horse; HQ, Hequ horse; UX, Uxin horse; UJM, Ujimqin horse; AB, Abag horse; AKTK, Akhal-Teke horse; ARR, Arabian horse; DRT, Clydesdale horse.
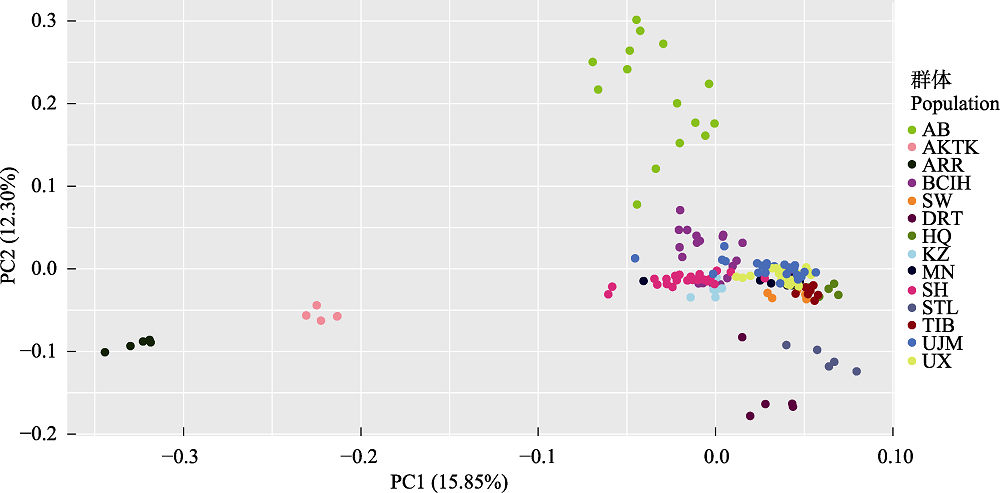
图2 167个样品PC1和PC2的主成分分析(PCA)。AB: 阿巴嘎黑马; AKTK: 阿哈尔捷金马; ARR: 阿拉伯马; BCIH: 百岔铁蹄马; SW: 西南马; DRT: 克莱斯戴尔马; HQ: 河曲马; KZ: 哈萨克马; MN: 蒙古马; SH: 三河马; STL: 设特兰矮马; TIB: 藏马; UJM: 乌珠穆沁马; UX: 乌审马。
Fig. 2 Principal component analysis (PCA) plot for PC1 and PC2 comprising 167 individuals. AB, Abag horse; AKTK, Akhal-Teke horse; ARR, Arabian horse; BCIH, Baicha iron hoof horse; SW, Southwest horse; DRT, Clydesdale horse; HQ, Hequ horse; KZ, Kazakh horse; MN, Mongolian horse; SH, Sanhe horse; STL, Shetland pony; TIB, Tibetan horse; UJM, Ujimqin horse; UX, Uxin horse.
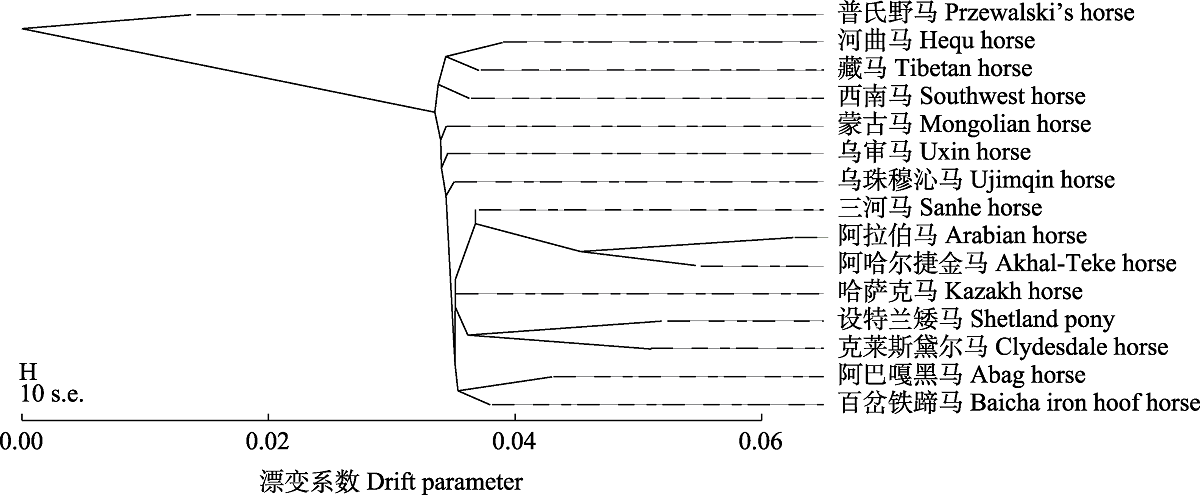
图3 基于最大似然法构建的14个家马群体系统发育树。比例尺展示了样品协方差矩阵中元素的10倍平均标准差(10 s.e.)。
Fig. 3 Phylogenetic tree of 14 horse populations based on maximum likelihood (ML). The scale bar shows 10 times the average standard error of the entries in the sample covariance matrix (10 s.e.).
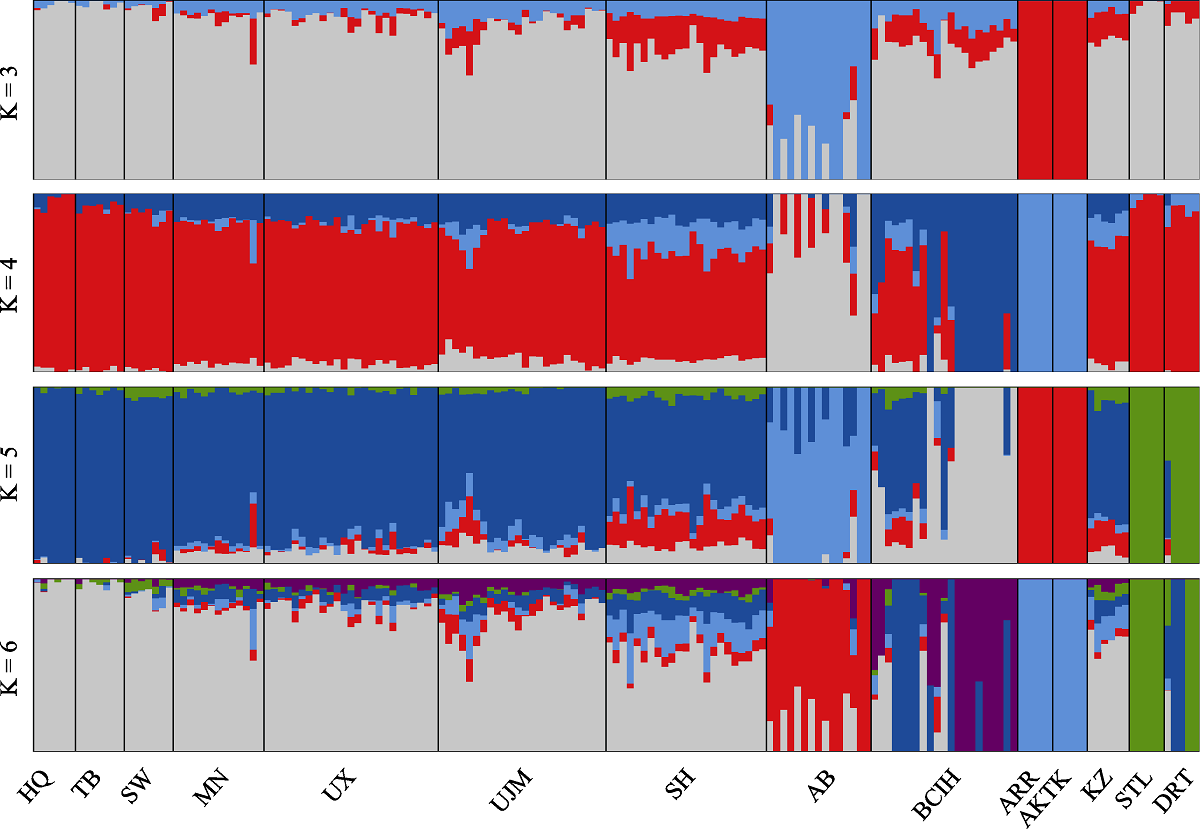
图4 基于最大似然法构建的14个家马群体遗传结构。HQ: 河曲马; TIB: 藏马; SW: 西南马; MN: 蒙古马; UX: 乌审马; UJM: 乌珠穆沁马; SH: 三河马; AB: 阿巴嘎黑马; BCIH: 百岔铁蹄马; ARR: 阿拉伯马; AKTK: 阿哈尔捷金马; KZ: 哈萨克马; STL: 设特兰矮马; DRT: 克莱斯黛尔马。
Fig. 4 Genetic structure of 14 horse populations based on maximum likelihood (ML). HQ, Hequ horse; TIB, Tibetan horse; SW, Southwest horse; MN, Mongolian horse; UX, Uxin horse; UJM, Ujimqin horse; SH, Sanhe horse; AB, Abag horse; BCIH, Baicha iron hoof horse; ARR, Arabian horse; AKTK, Akhal-Teke horse; KZ, Kazakh horse; STL, Shetland pony; DRT, Clydesdale horse.
| [1] |
Anthony DW, Brown DR (2000) Eneolithic horse exploitation in the Eurasian steppes: Diet, ritual and riding. Antiquity, 74, 75-86.
DOI URL |
| [2] | Anthony DW, Telegin DY, Brown D (1991) The origin of horseback riding. Scientific American, 265, 94-101. |
| [3] |
Arbuckle BS (2012) Animals and inequality in Chalcolithic central Anatolia. Journal of Anthropological Archaeology, 31, 302-313.
DOI URL |
| [4] |
Bendrey R (2011) Identification of metal residues associated with bit-use on prehistoric horse teeth by scanning electron microscopy with energy dispersive X-ray microanalysis. Journal of Archaeological Science, 38, 2989-2994.
DOI URL |
| [5] | Benecke N (2006) On the beginning of horse husbandry in the southern Balkan Peninsula: The horse bones from Kirklareli-Kanhgecit (Turkish Thrace). In: Equids in Time and Space: Papers in Honour of Véra Eisenmann (eds Albarella U, Dobney K, Rowley-Conwy P). Oxbow Books, Oxford. |
| [6] |
Berthouly-Salazar C, Thévenon S, Van TN, Nguyen BT, Pham LD, Chi CV, Maillard JC (2012) Uncontrolled admixture and loss of genetic diversity in a local Vietnamese pig breed. Ecology and Evolution, 2, 962-975.
DOI PMID |
| [7] | Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience, 4, s13742-015. |
| [8] | Chysyma RB, Khrabrova LA, Zaitsev АM, Мakarova ЕY, Fedorov YN, Ludu BM (2017) Genetic diversity in Tyva horses derived from polymorphism of blood systems and microsatellite DNA. Agricultural Biology, 52, 679-685. |
| [9] |
Cosgrove EJ, Sadeghi R, Schlamp F, Holl HM, Moradi- Shahrbabak M, Miraei-Ashtiani SR, Abdalla S, Shykind B, Troedsson M, Stefaniuk-Szmukier M, Prabhu A, Bucca S, Bugno-Poniewierska M, Wallner B, Malek J, Miller DC, Clark AG, Antczak DF, Brooks SA (2020) Genome diversity and the origin of the Arabian horse. Scientific Reports, 10, 9702.
DOI PMID |
| [10] | Cunliffe B, Cunliffe BW (2015) By Steppe, Desert, and Ocean: The Birth of Eurasia. Oxford University Press, New York. |
| [11] |
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics, 43, 491-498.
DOI PMID |
| [12] |
Erdemsurakh O, Ochirbat K, Gombosuren U, Tserendorj B, Purevdorj B, Vanaabaatar B, Aoshima K, Kobayashi A, Kimura T (2020) Seroprevalence of equine glanders in horses in the central and eastern parts of Mongolia. The Journal of Veterinary Medical Science, 82, 1247-1252.
DOI URL |
| [13] |
Fages A, Hanghøj K, Khan N, Gaunitz C, Seguin-Orlando A, Leonardi M, Constantz CM, Gamba C, Al-Rasheid KAS, Albizuri S, Alfarhan AH, Allentoft M, Alquraishi S, Anthony D, Baimukhanov N, Barrett JH, Bayarsaikhan J, Benecke N, Bernáldez-Sánchez E, Berrocal-Rangel L, Biglari F, Boessenkool S, Boldgiv B, Brem G, Brown D, Burger J, Crubézy E, Daugnora L, Davoudi H, de Barros Damgaard P, de Los Ángeles de Chorro y de Villa-Ceballos M, Deschler-Erb S, Detry C, Dill N, Oom MDM, Dohr A, Ellingvåg S, Erdenebaatar D, Fathi H, Felkel S, Fernández-Rodríguez C, García-Viñas E, Germonpré M, Granado JD, Hallsson JH, Hemmer H, Hofreiter M, Kasparov A, Khasanov M, Khazaeli R, Kosintsev P, Kristiansen K, Kubatbek T, Kuderna L, Kuznetsov P, Laleh H, Leonard JA, Lhuillier J, von Lettow-Vorbeck CL, Logvin A, Lõugas L, Ludwig A, Luis C, Arruda AM, Marques-Bonet T, Silva RM, Merz V, Mijiddorj E, Miller BK, Monchalov O, Mohaseb FA, Morales A, Nieto-Espinet A, Nistelberger H, Onar V, Pálsdóttir AH, Pitulko V, Pitskhelauri K, Pruvost M, Sikanjic PR, Papeša AR, Roslyakova N, Sardari A, Sauer E, Schafberg R, Scheu A, Schibler J, Schlumbaum A, Serrand N, Serres-Armero A, Shapiro B, Seno SS, Shevnina I, Shidrang S, Southon J, Star B, Sykes N, Taheri K, Taylor W, Teegen WR, Vukičević TT, Trixl S, Tumen D, Undrakhbold S, Usmanova E, Vahdati A, Valenzuela-Lamas S, Viegas C, Wallner B, Weinstock J, Zaibert V, Clavel B, Lepetz S, Mashkour M, Helgason A, Stefánsson K, Barrey E, Willerslev E, Outram AK, Librado P, Orlando L (2019) Tracking five millennia of horse management with extensive ancient genome time series. Cell, 177, 1419-1435.
DOI URL |
| [14] | FAO (2015) The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture. FAO Commission on Genetic Resources for Food and Agriculture Assessments, Rome. |
| [15] |
Felkel S, Vogl C, Rigler D, Jagannathan V, Leeb T, Fries R, Neuditschko M, Rieder S, Velie B, Lindgren G, Rubin CJ, Schlötterer C, Rattei T, Brem G, Wallner B (2018) Asian horses deepen the MSY phylogeny. Animal Genetics, 49, 90-93.
DOI PMID |
| [16] |
Gaunitz C, Fages A, Hanghøj K, Albrechtsen A, Khan N, Schubert M, Seguin-Orlando A, Owens IJ, Felkel S, Bignon-Lau O, de Barros Damgaard P, Mittnik A, Mohaseb AF, Davoudi H, Alquraishi S, Alfarhan AH, Al-Rasheid KAS, Crubézy E, Benecke N, Olsen S, Brown D, Anthony D, Massy K, Pitulko V, Kasparov A, Brem G, Hofreiter M, Mukhtarova G, Baimukhanov N, Lõugas L, Onar V, Stockhammer PW, Krause J, Boldgiv B, Undrakhbold S, Erdenebaatar D, Lepetz S, Mashkour M, Ludwig A, Wallner B, Merz V, Merz I, Zaibert V, Willerslev E, Librado P, Outram AK, Orlando L (2018) Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science, 360, 111-114.
DOI URL |
| [17] |
Guimaraes S, Arbuckle BS, Peters J, Adcock SE, Buitenhuis H, Chazin H, Manaseryan N, Uerpmann HP, Grange T, Geigl EM (2020) Ancient DNA shows domestic horses were introduced in the southern Caucasus and Anatolia during the Bronze Age. Science Advances, 6, eabb0030.
DOI URL |
| [18] | Han HG, Bryan K, Shiraigol W, Bai DY, Zhao YP, Bao W, Yang SQ, Zhang WG, MacHugh DE, Dugarjaviin M, Hill EW (2019a) Refinement of global domestic horse biogeography using historic Landrace Chinese Mongolian populations. Journal of Heredity, 110, 769-781. |
| [19] |
Han HG, Wallner B, Rigler D, MacHugh DE, Manglai D, Hill EW (2019b) Chinese Mongolian horses may retain early domestic male genetic lineages yet to be discovered. Animal Genetics, 50, 399-402.
DOI URL |
| [20] |
Hill EW, Gu JJ, Eivers SS, Fonseca RG, McGivney BA, Govindarajan P, Orr N, Katz LM, MacHugh DE (2010) A sequence polymorphism in MSTN predicts sprinting ability and racing stamina in thoroughbred horses. PLoS ONE, 5, e8645.
DOI URL |
| [21] | Jansen T, Forster P, Levine MA, Oelke H, Hurles M, Renfrew C, Weber J, Olek K (2002) Mitochondrial DNA and the origins of the domestic horse. Proceedings of the National Academy of Sciences, USA, 99, 10905-10910. |
| [22] |
Kalbfleisch TS, Rice ES, DePriest MS, Walenz BP, Hestand MS, Vermeesch JR, Brendan L, Fiddes IT, Vershinina AO, Saremi NF (2018) Improved reference genome for the domestic horse increases assembly contiguity and composition. Communications Biology, 1, 197.
DOI PMID |
| [23] |
Khanshour A, Conant E, Juras R, Cothran EG (2013) Microsatellite analysis of genetic diversity and population structure of Arabian horse populations. Journal of Heredity, 104, 386-398.
DOI PMID |
| [24] |
Levine MA (1990) Dereivka and the problem of horse domestication. Antiquity, 64, 727-740.
DOI URL |
| [25] | Levine MA (2005) Domestication and early history of the horse. In: The Domestic Horse: The Origins, Development, and Management of Its Behaviour (eds Mills DS, McDonnell SM, McDonnell S), pp. 5-22. Cambridge University Press, Cambridge. |
| [26] |
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics, 26, 589-595.
DOI URL |
| [27] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078-2079.
DOI URL |
| [28] | Librado P, der Sarkissian C, Ermini L, Schubert M, Jónsson H, Albrechtsen A, Fumagalli M, Yang MA, Gamba C, Seguin-Orlando A, Mortensen CD, Petersen B, Hoover CA, Lorente-Galdos B, Nedoluzhko A, Boulygina E, Tsygankova S, Neuditschko M, Jagannathan V, Thèves C, Alfarhan AH, Alquraishi SA, Al-Rasheid KAS, Sicheritz-Ponten T, Popov R, Grigoriev S, Alekseev AN, Rubin EM, McCue M, Rieder S, Leeb T, Tikhonov A, Crubézy E, Slatkin M, Marques-Bonet T, Nielsen R, Willerslev E, Kantanen J, Prokhortchouk E, Orlando L (2015) Tracking the origins of Yakutian horses and the genetic basis for their fast adaptation to subarctic environments. Proceedings of the National Academy of Sciences, USA, 112, E6889-E6897. |
| [29] |
Librado P, Fages A, Gaunitz C, Leonardi M, Wagner S, Khan N, Hanghøj K, Alquraishi SA, Alfarhan AH, Al-Rasheid KA, der Sarkissian C, Schubert M, Orlando L (2016) The evolutionary origin and genetic makeup of domestic horses. Genetics, 204, 423-434.
PMID |
| [30] |
Librado P, Khan N, Fages A, Kusliy MA, Suchan T, Tonasso-Calvière L, Schiavinato S, Alioglu D, Fromentier A, Perdereau A, Aury JM et al (2021) The origins and spread of domestic horses from the Western Eurasian steppes. Nature, 598, 634-640.
DOI URL |
| [31] |
Liu XX, Zhang YL, Li YF, Pan JF, Wang DD, Chen WH, Zheng ZQ, He XH, Zhao QJ, Pu YB, Guan WJ, Han JL, Orlando L, Ma YH, Jiang L (2019) EPAS 1 gain-of-function mutation contributes to high-altitude adaptation in Tibetan horses. Molecular Biology and Evolution, 36, 2591-2603.
DOI URL |
| [32] |
Lopes MS, Mendonça D, Cymbron T, Valera M, da Costa-Ferreira J, Machado A (2005) The Lusitano horse maternal lineage based on mitochondrial D-loop sequence variation. Animal Genetics, 36, 196-202.
PMID |
| [33] |
McCue ME, Valberg SJ, Miller MB, Wade C, DiMauro S, Akman HO, Mickelson JR (2008) Glycogen synthase (GYS1) mutation causes a novel skeletal muscle glycogenosis. Genomics, 91, 458-466.
DOI URL |
| [34] |
Mileto S, Kaiser E, Rassamakin Y, Evershed RP (2017) New insights into the subsistence economy of the Eneolithic Dereivka culture of the Ukrainian North-Pontic region through lipid residues analysis of pottery vessels. Journal of Archaeological Science: Reports, 13, 67-74.
DOI URL |
| [35] |
Orlando L (2020) The evolutionary and historical foundation of the modern horse: Lessons from ancient genomics. Annual Review of Genetics, 54, 563-581.
DOI URL |
| [36] |
Orlando L, Ginolhac A, Zhang GJ, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, Johnson PLF, Fumagalli M, Vilstrup JT, Raghavan M, Korneliussen T, Malaspinas AS, Vogt J, Szklarczyk D, Kelstrup CD, Vinther J, Dolocan A, Stenderup J, Velazquez AMV, Cahill J, Rasmussen M, Wang XL, Min JM, Zazula GD, Seguin-Orlando A, Mortensen C, Magnussen K, Thompson JF, Weinstock J, Gregersen K, Røed KH, Eisenmann V, Rubin CJ, Miller DC, Antczak DF, Bertelsen MF, Brunak S, Al-Rasheid KAS, Ryder O, Andersson L, Mundy J, Krogh A, Gilbert MTP, Kjær K, Sicheritz-Ponten T, Jensen LJ, Olsen JV, Hofreiter M, Nielsen R, Shapiro B, Wang J, Willerslev E (2013) Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature, 499, 74-78.
DOI URL |
| [37] |
Orlando L, Librado P (2019) Origin and evolution of deleterious mutations in horses. Genes, 10, 649.
DOI URL |
| [38] |
Outram AK, Stear NA, Bendrey R, Olsen S, Kasparov A, Zaibert V, Thorpe N, Evershed RP (2009) The earliest horse harnessing and milking. Science, 323, 1332-1335.
DOI PMID |
| [39] |
Petersen JL, Mickelson JR, Cothran EG, Andersson LS, Axelsson J, Bailey E, Bannasch D, Binns MM, Borges AS, Brama P, da Câmara Machado A, Distl O, Felicetti M, Fox-Clipsham L, Graves KT, Guérin G, Haase B, Hasegawa T, Hemmann K, Hill EW, Leeb T, Lindgren G, Lohi H, Lopes MS, McGivney BA, Mikko S, Orr N, Penedo MCT, Piercy RJ, Raekallio M, Rieder S, Røed KH, Silvestrelli M, Swinburne J, Tozaki T, Vaudin M, Wade CM, McCue ME (2013a) Genetic diversity in the modern horse illustrated from genome-wide SNP data. PLoS ONE, 8, e54997.
DOI URL |
| [40] |
Petersen JL, Mickelson JR, Rendahl AK, Valberg SJ, Andersson LS, Axelsson J, Bailey E, Bannasch D, Binns MM, Borges AS (2013b) Genome-wide analysis reveals selection for important traits in domestic horse breeds. PLoS Genetics, 9, e1003211.
DOI URL |
| [41] |
Pickrell J, Pritchard J (2012) Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genetics, 8, e1002967.
DOI URL |
| [42] |
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 38, 904-909.
DOI URL |
| [43] |
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Daly MJ, Sham PC (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics, 81, 559-575.
DOI PMID |
| [44] |
Raj A, Stephens M, Pritchard JK (2014) fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics, 197, 573-589.
DOI URL |
| [45] |
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science, 298, 2381-2385.
PMID |
| [46] |
Sommer RS, Benecke N, Lõugas L, Nelle O, Schmölcke U (2011) Holocene survival of the wild horse in Europe: A matter of open landscape? Journal of Quaternary Science, 26, 805-812.
DOI URL |
| [47] |
Srikanth K, Kim NY, Park W, Kim JM, Kim KD, Lee KT, Son JH, Chai HH, Choi JW, Jang GW, Kim H, Ryu YC, Nam JW, Park JE, Kim JM, Lim D (2019) Comprehensive genome and transcriptome analyses reveal genetic relationship, selection signature, and transcriptome landscape of small-sized Korean native Jeju horse. Scientific Reports, 9, 16672.
DOI URL |
| [48] | Stetter MG, Gates DJ, Mei WB, Ross-Ibarra J (2017) How to make a domesticate. Current Biology, 27, R896-R900. |
| [49] |
Tozaki T, Kikuchi M, Kakoi H, Hirota K, Nagata S, Yamashita D, Ohnuma T, Takasu M, Kobayashi I, Hobo S, Manglai D, Petersen JL (2019) Genetic diversity and relationships among native Japanese horse breeds, the Japanese Thoroughbred and horses outside of Japan using genome-wide SNP data. Animal Genetics, 50, 449-459.
DOI PMID |
| [50] |
Tozaki T, Takezaki N, Hasegawa T, Ishida N, Kurosawa M, Tomita M, Saitou N, Mukoyama H (2003) Microsatellite variation in Japanese and Asian horses and their phylogenetic relationship using a European horse outgroup. Journal of Heredity, 94, 374-380.
PMID |
| [51] |
Wallner B, Palmieri N, Vogl C, Rigler D, Bozlak E, Druml T, Jagannathan V, Leeb T, Fries R, Tetens J, Thaller G, Metzger J, Distl O, Lindgren G, Rubin CJ, Andersson L, Schaefer R, McCue M, Neuditschko M, Rieder S, Schlotterer C, Brem G (2017) Y chromosome uncovers the recent oriental origin of modern stallions. Current Biology, 27, 2029-2035.
DOI URL |
| [1] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [2] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [3] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [4] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [5] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [6] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [7] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [8] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [9] | 崔静, 徐明芳, 章群, 李瑶, 曾晓舒, 李莎. 基于3种线粒体标记探讨中日沿海角木叶鲽遗传多样性差异[J]. 生物多样性, 2022, 30(5): 21485-. |
| [10] | 孙军, 宋煜尧, 施义锋, 翟键, 燕文卓. 近十年中国海洋生物多样性研究进展[J]. 生物多样性, 2022, 30(10): 22526-. |
| [11] | 栗冬梅, 杨卫红, 李庆多, 韩茜, 宋秀平, 潘虹, 冯云. 巴尔通体在滇西南蝙蝠中高度流行并具有丰富的遗传变异特征[J]. 生物多样性, 2021, 29(9): 1245-1255. |
| [12] | 姚志, 郭军, 金晨钟, 刘勇波. 中国纳入一级保护的极小种群野生植物濒危机制[J]. 生物多样性, 2021, 29(3): 394-408. |
| [13] | 叶俊伟, 田斌. 中国西南地区重要木本油料植物扁核木的遗传结构及成因[J]. 生物多样性, 2021, 29(12): 1629-1637. |
| [14] | 向登高, 李跃飞, 李新辉, 陈蔚涛, 马秀慧. 多基因联合揭示海南鲌的遗传结构与遗传多样性[J]. 生物多样性, 2021, 29(11): 1505-1512. |
| [15] | 陶克涛, 韩海格, 赵若阳, 图格琴, 芒来, 白东义. 家马的驯化起源与遗传演化特征[J]. 生物多样性, 2020, 28(6): 734-748. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn