



生物多样性 ›› 2023, Vol. 31 ›› Issue (8): 23120. DOI: 10.17520/biods.2023120 cstr: 32101.14.biods.2023120
齐海玲1,2,3,4, 樊鹏振2,5, 王跃华3,4, 刘杰1,2,*( )
)
收稿日期:2023-04-16
接受日期:2023-06-25
出版日期:2023-08-20
发布日期:2023-07-03
通讯作者:
*E-mail: liujie@mail.kib.ac.cn
基金资助:
Hailing Qi1,2,3,4, Pengzhen Fan2,5, Yuehua Wang3,4, Jie Liu1,2,*( )
)
Received:2023-04-16
Accepted:2023-06-25
Online:2023-08-20
Published:2023-07-03
Contact:
*E-mail: liujie@mail.kib.ac.cn
摘要:
胡桃(Juglans regia)是重要的木本经济作物, 在我国北方广泛栽培。然而, 目前缺乏对北方地区胡桃的遗传多样性和群体结构的全面认识, 限制了胡桃资源的保护和利用。本研究以北方六省区的19个群体的491份胡桃样本为对象, 基于31对多态性微卫星引物的基因分型数据, 分析其遗传多样性、遗传分化和群体结构。遗传多样性估算结果表明胡桃群体遗传多样性较低(NA = 2.620, HO = 0.368, HE = 0.368), 遗传变异主要分布于群体内(84%), 群体间的遗传分化较低(FST = 0.16), 这可能与该地区胡桃是人为引入栽培、长期人工选择和扩散等有关。遗传结构分析发现胡桃包含东、西两个组, 其中青海海南州的一个群体构成一组, 其余省份的群体为另外一组, 组间具有较高水平的遗传分化(FST = 0.32), 但两个组交汇区的群体有基因渐渗的信号, 这种遗传格局可能由栽培历史和局域环境所塑造。基于上述结果, 我们建议对青海海南州和甘肃天水市的两个群体进行优先保护。本研究明晰了北方六省区胡桃的遗传多样性和群体结构, 提出胡桃遗传资源的保护策略, 有望为胡桃种质资源的利用提供科学依据。
齐海玲, 樊鹏振, 王跃华, 刘杰 (2023) 中国北方六省区胡桃的遗传多样性和群体结构. 生物多样性, 31, 23120. DOI: 10.17520/biods.2023120.
Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu (2023) Genetic diversity and population structure of Juglans regia from six provinces in northern China. Biodiversity Science, 31, 23120. DOI: 10.17520/biods.2023120.
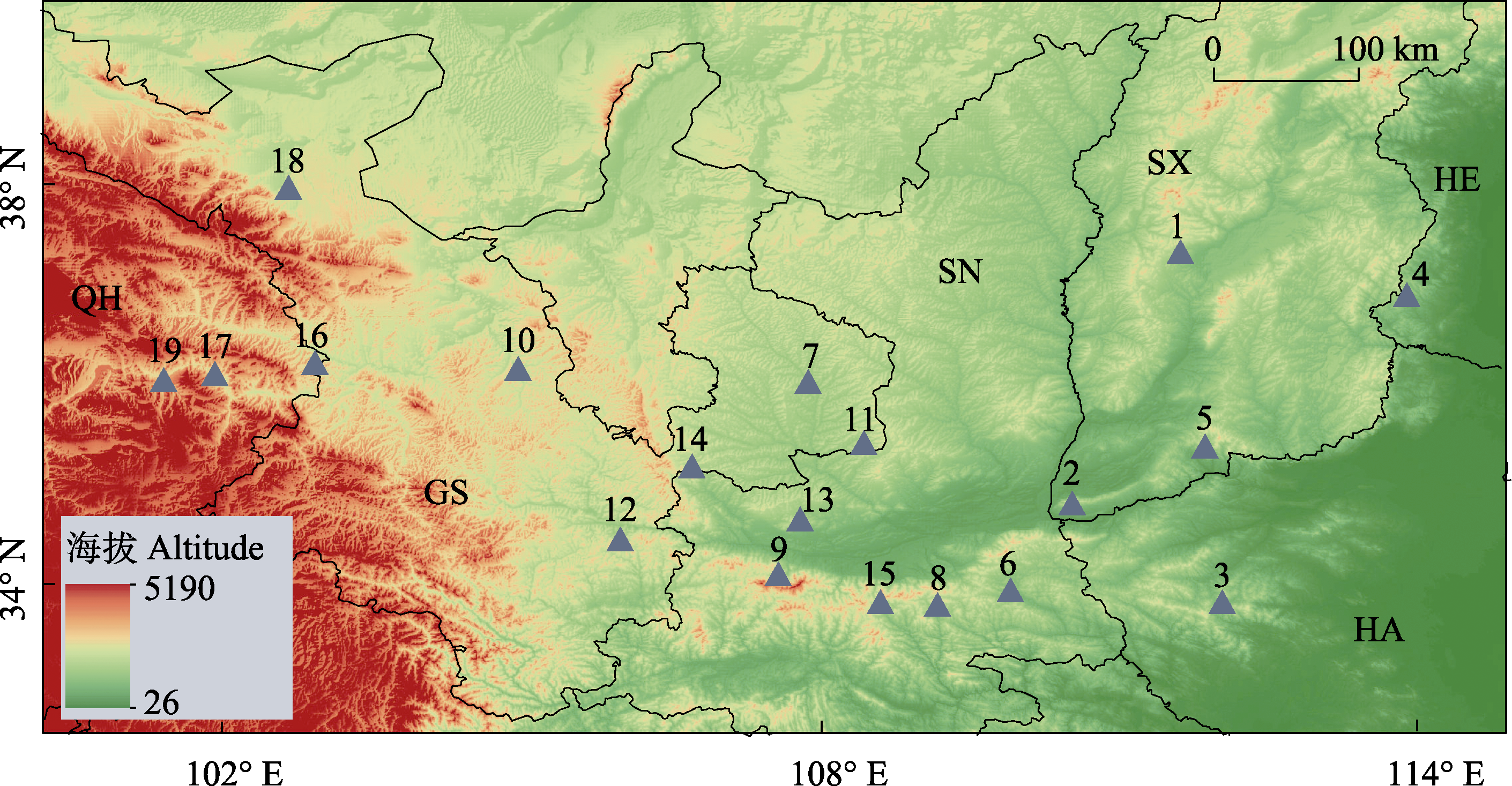
图1 中国北方六省区19个胡桃群体的地理分布。图中数字代表群体序号(见表1), 英文符号表示省份缩写(QH: 青海省; GS: 甘肃省; SN: 陕西省; SX: 山西省; HE: 河北省; HA: 河南省; 详见表1)。
Fig. 1 Geographic distribution of 19 Juglans regia populations from six provinces in northern China. The serial numbers on the map represent population IDs, and the English symbol indicates the abbreviation of province. QH, Qinghai Province; GS, Gansu Province; SN, Shaanxi Province; SX, Shanxi Province; HE, Hebei Province; HA, Henan Province; see details in Table 1.
| 群体 编号 Code | 群体 序号 ID | 样本大小Sample size | 地点 Locality | 纬度 Latitude (No) | 经度 Longitude (Eo) | 海拔 Altitude(m) | NT | NP | NA | NE | AR | HO | HE | FIS | 双相突变模型 TPM | 标准差异检测 Standardized difference test |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YFR | 1 | 30 | SX | 37.29 | 111.57 | 1,195 | 76 | 3 | 2.45 | 1.71 | 2.08 | 0.33 | 0.33 | 0.03 | 1.68 | 0.047 |
| RCR | 2 | 21 | SX | 34.77 | 110.49 | 1,418 | 77 | 3 | 2.48 | 1.77 | 2.10 | 0.34 | 0.35 | 0.07 | 1.76 | 0.039 |
| CCR | 3 | 30 | HA | 33.78 | 111.99 | 813 | 81 | 2 | 2.61 | 1.86 | 2.21 | 0.36 | 0.37 | 0.05 | 2.18 | 0.014 |
| WAR | 4 | 30 | HE | 36.86 | 113.83 | 805 | 80 | 0 | 2.58 | 1.79 | 2.12 | 0.36 | 0.36 | 0.03 | 2.00 | 0.023 |
| YQR | 5 | 30 | SX | 35.35 | 111.82 | 891 | 82 | 3 | 2.65 | 1.73 | 2.10 | 0.34 | 0.35 | 0.05 | 1.26 | 0.105 |
| LSR | 6 | 30 | SN | 33.91 | 109.87 | 804 | 91 | 1 | 2.94 | 1.88 | 2.27 | 0.38 | 0.39 | 0.04 | 0.62 | 0.269 |
| QCR | 7 | 30 | GS | 35.98 | 107.85 | 1,322 | 83 | 0 | 2.68 | 1.78 | 2.15 | 0.33 | 0.38 | 0.14 | 1.64 | 0.051 |
| GBR | 8 | 12 | SN | 33.76 | 109.14 | 1,433 | 72 | 0 | 2.32 | 1.68 | 2.10 | 0.41 | 0.34 | -0.18 | 1.27 | 0.102 |
| TBR | 9 | 30 | SN | 34.06 | 107.55 | 1,142 | 77 | 0 | 2.48 | 1.66 | 2.04 | 0.33 | 0.34 | 0.04 | 1.43 | 0.077 |
| Liuj11248 | 10 | 6 | GS | 36.12 | 104.94 | 1,578 | 65 | 0 | 2.10 | 1.56 | 2.21 | 0.27 | 0.29 | 0.17 | -0.63 | 0.266 |
| WQR | 11 | 30 | GS | 35.38 | 108.40 | 1,291 | 81 | 0 | 2.61 | 1.75 | 2.11 | 0.37 | 0.36 | -0.02 | 1.42 | 0.077 |
| GQR | 12 | 30 | GS | 34.42 | 105.97 | 1,271 | 92 | 6 | 2.97 | 1.80 | 2.27 | 0.42 | 0.39 | -0.06 | 0.39 | 0.348 |
| JCR | 13 | 27 | SN | 34.61 | 107.76 | 1,204 | 74 | 0 | 2.39 | 1.78 | 2.08 | 0.38 | 0.38 | 0.01 | 3.31 | 0.000 |
| SGR | 14 | 30 | GS | 35.15 | 106.68 | 1,550 | 77 | 3 | 2.48 | 1.70 | 2.07 | 0.35 | 0.36 | 0.06 | 2.30 | 0.011 |
| SXHT | 15 | 5 | SN | 33.79 | 108.57 | 1,876 | 69 | 0 | 2.23 | 1.82 | 2.39 | 0.50 | 0.38 | -0.20 | 1.78 | 0.038 |
| MLZR | 16 | 30 | QH | 36.18 | 102.91 | 1,898 | 87 | 0 | 2.81 | 1.80 | 2.19 | 0.36 | 0.37 | 0.06 | 0.99 | 0.161 |
| KYR | 17 | 30 | QH | 36.07 | 101.91 | 2,069 | 92 | 0 | 2.97 | 1.88 | 2.28 | 0.33 | 0.40 | 0.18 | 0.94 | 0.173 |
| WLZR | 18 | 30 | GS | 37.93 | 102.65 | 1,528 | 88 | 0 | 2.84 | 1.78 | 2.18 | 0.40 | 0.38 | -0.03 | 0.71 | 0.238 |
| HXR | 19 | 30 | QH | 36.01 | 101.40 | 2,259 | 100 | 5 | 3.23 | 2.14 | 2.58 | 0.43 | 0.47 | 0.10 | 2.72 | 0.003 |
| 平均值 Mean | - | - | - | - | - | - | 81 | 1 | 2.62 | 1.78 | 2.19 | 0.37 | 0.37 | 0.03 | - | - |
表1 本研究选用的19个胡桃群体的采集信息、遗传多样性和瓶颈效应分析
Table 1 Collection information, genetic diversity and bottleneck effect analysis of the19 Juglans regia populations in this study
| 群体 编号 Code | 群体 序号 ID | 样本大小Sample size | 地点 Locality | 纬度 Latitude (No) | 经度 Longitude (Eo) | 海拔 Altitude(m) | NT | NP | NA | NE | AR | HO | HE | FIS | 双相突变模型 TPM | 标准差异检测 Standardized difference test |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YFR | 1 | 30 | SX | 37.29 | 111.57 | 1,195 | 76 | 3 | 2.45 | 1.71 | 2.08 | 0.33 | 0.33 | 0.03 | 1.68 | 0.047 |
| RCR | 2 | 21 | SX | 34.77 | 110.49 | 1,418 | 77 | 3 | 2.48 | 1.77 | 2.10 | 0.34 | 0.35 | 0.07 | 1.76 | 0.039 |
| CCR | 3 | 30 | HA | 33.78 | 111.99 | 813 | 81 | 2 | 2.61 | 1.86 | 2.21 | 0.36 | 0.37 | 0.05 | 2.18 | 0.014 |
| WAR | 4 | 30 | HE | 36.86 | 113.83 | 805 | 80 | 0 | 2.58 | 1.79 | 2.12 | 0.36 | 0.36 | 0.03 | 2.00 | 0.023 |
| YQR | 5 | 30 | SX | 35.35 | 111.82 | 891 | 82 | 3 | 2.65 | 1.73 | 2.10 | 0.34 | 0.35 | 0.05 | 1.26 | 0.105 |
| LSR | 6 | 30 | SN | 33.91 | 109.87 | 804 | 91 | 1 | 2.94 | 1.88 | 2.27 | 0.38 | 0.39 | 0.04 | 0.62 | 0.269 |
| QCR | 7 | 30 | GS | 35.98 | 107.85 | 1,322 | 83 | 0 | 2.68 | 1.78 | 2.15 | 0.33 | 0.38 | 0.14 | 1.64 | 0.051 |
| GBR | 8 | 12 | SN | 33.76 | 109.14 | 1,433 | 72 | 0 | 2.32 | 1.68 | 2.10 | 0.41 | 0.34 | -0.18 | 1.27 | 0.102 |
| TBR | 9 | 30 | SN | 34.06 | 107.55 | 1,142 | 77 | 0 | 2.48 | 1.66 | 2.04 | 0.33 | 0.34 | 0.04 | 1.43 | 0.077 |
| Liuj11248 | 10 | 6 | GS | 36.12 | 104.94 | 1,578 | 65 | 0 | 2.10 | 1.56 | 2.21 | 0.27 | 0.29 | 0.17 | -0.63 | 0.266 |
| WQR | 11 | 30 | GS | 35.38 | 108.40 | 1,291 | 81 | 0 | 2.61 | 1.75 | 2.11 | 0.37 | 0.36 | -0.02 | 1.42 | 0.077 |
| GQR | 12 | 30 | GS | 34.42 | 105.97 | 1,271 | 92 | 6 | 2.97 | 1.80 | 2.27 | 0.42 | 0.39 | -0.06 | 0.39 | 0.348 |
| JCR | 13 | 27 | SN | 34.61 | 107.76 | 1,204 | 74 | 0 | 2.39 | 1.78 | 2.08 | 0.38 | 0.38 | 0.01 | 3.31 | 0.000 |
| SGR | 14 | 30 | GS | 35.15 | 106.68 | 1,550 | 77 | 3 | 2.48 | 1.70 | 2.07 | 0.35 | 0.36 | 0.06 | 2.30 | 0.011 |
| SXHT | 15 | 5 | SN | 33.79 | 108.57 | 1,876 | 69 | 0 | 2.23 | 1.82 | 2.39 | 0.50 | 0.38 | -0.20 | 1.78 | 0.038 |
| MLZR | 16 | 30 | QH | 36.18 | 102.91 | 1,898 | 87 | 0 | 2.81 | 1.80 | 2.19 | 0.36 | 0.37 | 0.06 | 0.99 | 0.161 |
| KYR | 17 | 30 | QH | 36.07 | 101.91 | 2,069 | 92 | 0 | 2.97 | 1.88 | 2.28 | 0.33 | 0.40 | 0.18 | 0.94 | 0.173 |
| WLZR | 18 | 30 | GS | 37.93 | 102.65 | 1,528 | 88 | 0 | 2.84 | 1.78 | 2.18 | 0.40 | 0.38 | -0.03 | 0.71 | 0.238 |
| HXR | 19 | 30 | QH | 36.01 | 101.40 | 2,259 | 100 | 5 | 3.23 | 2.14 | 2.58 | 0.43 | 0.47 | 0.10 | 2.72 | 0.003 |
| 平均值 Mean | - | - | - | - | - | - | 81 | 1 | 2.62 | 1.78 | 2.19 | 0.37 | 0.37 | 0.03 | - | - |
| 引物名称 Primer name | 等位基因 NA | 有效等位基因 NE | Shannon’s信息指数 I | 观察杂合度 HO | 期望杂合度 HE | 固定系数 F | 多态信息含量 PIC |
|---|---|---|---|---|---|---|---|
| JR02 | 6 | 2.11 | 0.86 | 0.46 | 0.53 | 0.12 | 0.42 |
| JR03 | 4 | 2.56 | 1.02 | 0.53 | 0.61 | 0.13 | 0.54 |
| JR04 | 6 | 2.76 | 1.13 | 0.59 | 0.64 | 0.08 | 0.57 |
| JR05 | 3 | 1.07 | 0.16 | 0.07 | 0.07 | -0.03 | 0.07 |
| JR06 | 4 | 1.68 | 0.76 | 0.35 | 0.40 | 0.14 | 0.37 |
| JR07 | 4 | 2.42 | 1.00 | 0.48 | 0.59 | 0.19 | 0.52 |
| JR08 | 4 | 1.03 | 0.10 | 0.03 | 0.03 | 0.11 | 0.03 |
| JR09 | 5 | 1.75 | 0.64 | 0.38 | 0.43 | 0.12 | 0.34 |
| JR10 | 3 | 2.22 | 0.92 | 0.57 | 0.55 | -0.03 | 0.48 |
| JR11 | 3 | 2.60 | 1.01 | 0.54 | 0.62 | 0.12 | 0.53 |
| JR12 | 5 | 2.33 | 0.93 | 0.51 | 0.57 | 0.10 | 0.48 |
| JS02 | 2 | 1.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| JS03 | 6 | 1.72 | 0.73 | 0.48 | 0.42 | -0.15 | 0.36 |
| JS04 | 3 | 2.35 | 0.96 | 0.51 | 0.57 | 0.12 | 0.50 |
| JS05 | 6 | 2.11 | 0.97 | 0.50 | 0.53 | 0.05 | 0.47 |
| JS06 | 6 | 1.46 | 0.64 | 0.12 | 0.31 | 0.60 | 0.29 |
| JS07 | 6 | 1.90 | 0.94 | 0.34 | 0.47 | 0.27 | 0.43 |
| JS09 | 5 | 1.20 | 0.38 | 0.17 | 0.17 | -0.01 | 0.16 |
| JS12 | 7 | 2.90 | 1.26 | 0.48 | 0.65 | 0.27 | 0.60 |
| JS13 | 4 | 1.93 | 0.78 | 0.43 | 0.48 | 0.10 | 0.40 |
| JS14 | 4 | 1.59 | 0.60 | 0.36 | 0.37 | 0.04 | 0.31 |
| JS15 | 3 | 2.48 | 0.98 | 0.51 | 0.60 | 0.14 | 0.51 |
| JS22 | 5 | 1.68 | 0.66 | 0.41 | 0.41 | -0.01 | 0.33 |
| JS28 | 2 | 1.07 | 0.14 | 0.05 | 0.06 | 0.16 | 0.06 |
| BFU-Jr277 | 4 | 2.45 | 0.99 | 0.48 | 0.59 | 0.18 | 0.52 |
| BFU-Jr38 | 6 | 3.18 | 1.31 | 0.57 | 0.69 | 0.16 | 0.63 |
| CUJRD102 | 3 | 1.51 | 0.54 | 0.31 | 0.34 | 0.10 | 0.28 |
| CUJRD462 | 7 | 2.39 | 0.98 | 0.51 | 0.58 | 0.13 | 0.49 |
| JM5446 | 2 | 1.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| SSR18 | 8 | 2.59 | 1.17 | 0.52 | 0.61 | 0.15 | 0.55 |
| ZMZ7 | 4 | 1.04 | 0.12 | 0.03 | 0.04 | 0.37 | 0.04 |
| 平均值 Mean | 4.52 | 1.94 | 0.73 | 0.36 | 0.42 | 0.12 | 0.37 |
表2 本研究选用的31个微卫星位点的遗传多样性特征
Table 2 The characteristics of genetic diversity of 31 microsatellite loci used in this study
| 引物名称 Primer name | 等位基因 NA | 有效等位基因 NE | Shannon’s信息指数 I | 观察杂合度 HO | 期望杂合度 HE | 固定系数 F | 多态信息含量 PIC |
|---|---|---|---|---|---|---|---|
| JR02 | 6 | 2.11 | 0.86 | 0.46 | 0.53 | 0.12 | 0.42 |
| JR03 | 4 | 2.56 | 1.02 | 0.53 | 0.61 | 0.13 | 0.54 |
| JR04 | 6 | 2.76 | 1.13 | 0.59 | 0.64 | 0.08 | 0.57 |
| JR05 | 3 | 1.07 | 0.16 | 0.07 | 0.07 | -0.03 | 0.07 |
| JR06 | 4 | 1.68 | 0.76 | 0.35 | 0.40 | 0.14 | 0.37 |
| JR07 | 4 | 2.42 | 1.00 | 0.48 | 0.59 | 0.19 | 0.52 |
| JR08 | 4 | 1.03 | 0.10 | 0.03 | 0.03 | 0.11 | 0.03 |
| JR09 | 5 | 1.75 | 0.64 | 0.38 | 0.43 | 0.12 | 0.34 |
| JR10 | 3 | 2.22 | 0.92 | 0.57 | 0.55 | -0.03 | 0.48 |
| JR11 | 3 | 2.60 | 1.01 | 0.54 | 0.62 | 0.12 | 0.53 |
| JR12 | 5 | 2.33 | 0.93 | 0.51 | 0.57 | 0.10 | 0.48 |
| JS02 | 2 | 1.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| JS03 | 6 | 1.72 | 0.73 | 0.48 | 0.42 | -0.15 | 0.36 |
| JS04 | 3 | 2.35 | 0.96 | 0.51 | 0.57 | 0.12 | 0.50 |
| JS05 | 6 | 2.11 | 0.97 | 0.50 | 0.53 | 0.05 | 0.47 |
| JS06 | 6 | 1.46 | 0.64 | 0.12 | 0.31 | 0.60 | 0.29 |
| JS07 | 6 | 1.90 | 0.94 | 0.34 | 0.47 | 0.27 | 0.43 |
| JS09 | 5 | 1.20 | 0.38 | 0.17 | 0.17 | -0.01 | 0.16 |
| JS12 | 7 | 2.90 | 1.26 | 0.48 | 0.65 | 0.27 | 0.60 |
| JS13 | 4 | 1.93 | 0.78 | 0.43 | 0.48 | 0.10 | 0.40 |
| JS14 | 4 | 1.59 | 0.60 | 0.36 | 0.37 | 0.04 | 0.31 |
| JS15 | 3 | 2.48 | 0.98 | 0.51 | 0.60 | 0.14 | 0.51 |
| JS22 | 5 | 1.68 | 0.66 | 0.41 | 0.41 | -0.01 | 0.33 |
| JS28 | 2 | 1.07 | 0.14 | 0.05 | 0.06 | 0.16 | 0.06 |
| BFU-Jr277 | 4 | 2.45 | 0.99 | 0.48 | 0.59 | 0.18 | 0.52 |
| BFU-Jr38 | 6 | 3.18 | 1.31 | 0.57 | 0.69 | 0.16 | 0.63 |
| CUJRD102 | 3 | 1.51 | 0.54 | 0.31 | 0.34 | 0.10 | 0.28 |
| CUJRD462 | 7 | 2.39 | 0.98 | 0.51 | 0.58 | 0.13 | 0.49 |
| JM5446 | 2 | 1.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| SSR18 | 8 | 2.59 | 1.17 | 0.52 | 0.61 | 0.15 | 0.55 |
| ZMZ7 | 4 | 1.04 | 0.12 | 0.03 | 0.04 | 0.37 | 0.04 |
| 平均值 Mean | 4.52 | 1.94 | 0.73 | 0.36 | 0.42 | 0.12 | 0.37 |
| 变异来源 Source of variation | 自由度 df | 平方和 Sum of square | 均方差 Mean square error | 变异百分比 Percentage of variation (%) |
|---|---|---|---|---|
| 群体间 Among populations | 18 | 1,366.09 | 75.89 | 16 |
| 群体内 Within populations | 472 | 5,901.66 | 12.50 | 84 |
| 总和 Total | 490 | 7,267.75 | 100 | |
| 组间 Among groups | 1 | 528.63 | 528.63 | 32 |
| 组内 Within groups | 468 | 6,366.60 | 13.60 | 68 |
| 总和 Total | 469 | 6,895.24 | 100 |
表3 胡桃19个群体及其2个分组的分子方差分析(AMOVA)结果
Table 3 Analysis of molecular variance (AMOVA) for 19 populations and two groups of Juglans regia
| 变异来源 Source of variation | 自由度 df | 平方和 Sum of square | 均方差 Mean square error | 变异百分比 Percentage of variation (%) |
|---|---|---|---|---|
| 群体间 Among populations | 18 | 1,366.09 | 75.89 | 16 |
| 群体内 Within populations | 472 | 5,901.66 | 12.50 | 84 |
| 总和 Total | 490 | 7,267.75 | 100 | |
| 组间 Among groups | 1 | 528.63 | 528.63 | 32 |
| 组内 Within groups | 468 | 6,366.60 | 13.60 | 68 |
| 总和 Total | 469 | 6,895.24 | 100 |
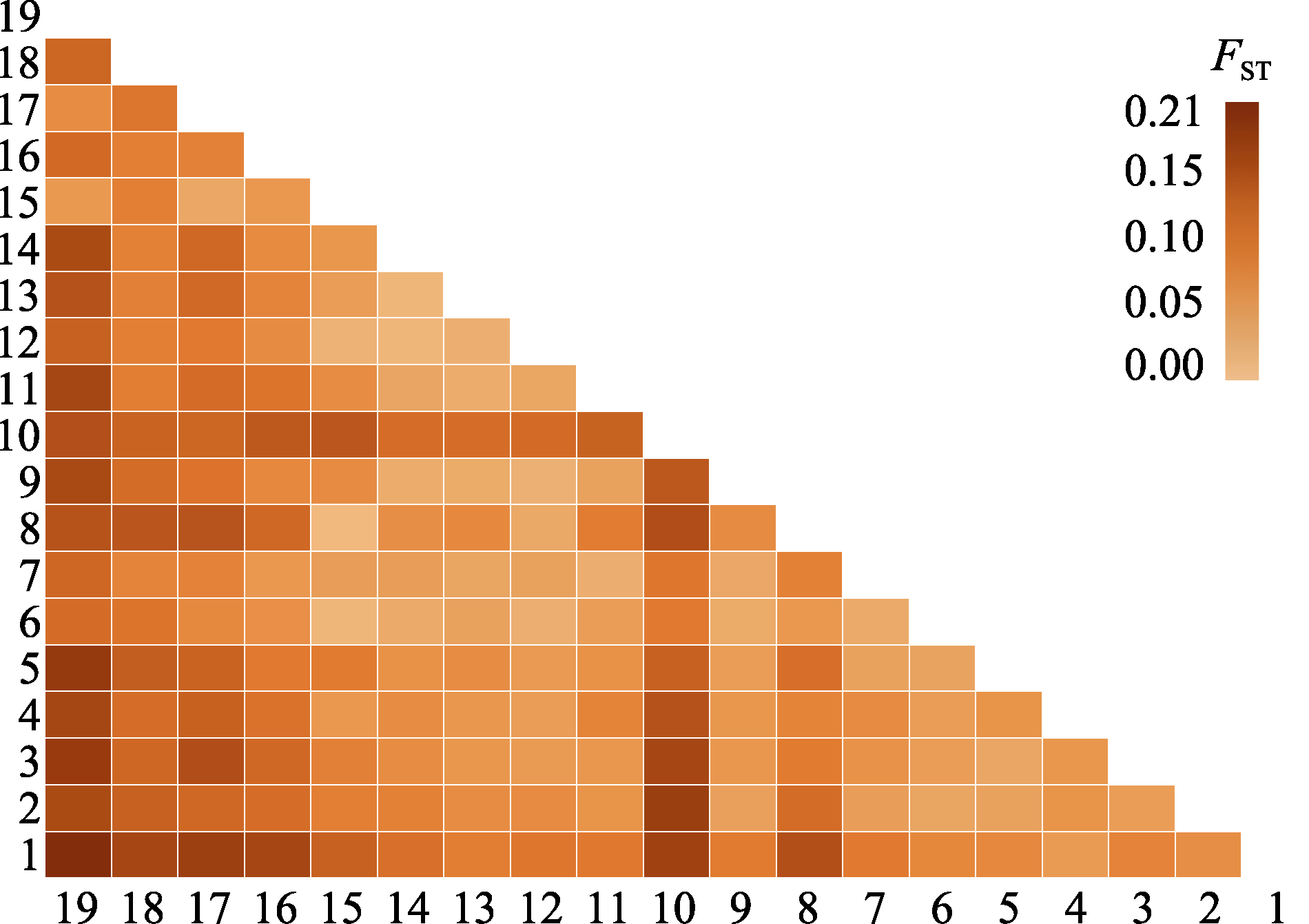
图2 胡桃成对群体间的遗传分化(FST)热图。1-19代表群体序号(详见表1)。
Fig. 2 Heat map depicting genetic differentiation (FST) among 19 Juglans regia populations. 1-19 indicate the population IDs (see details in Table 1).
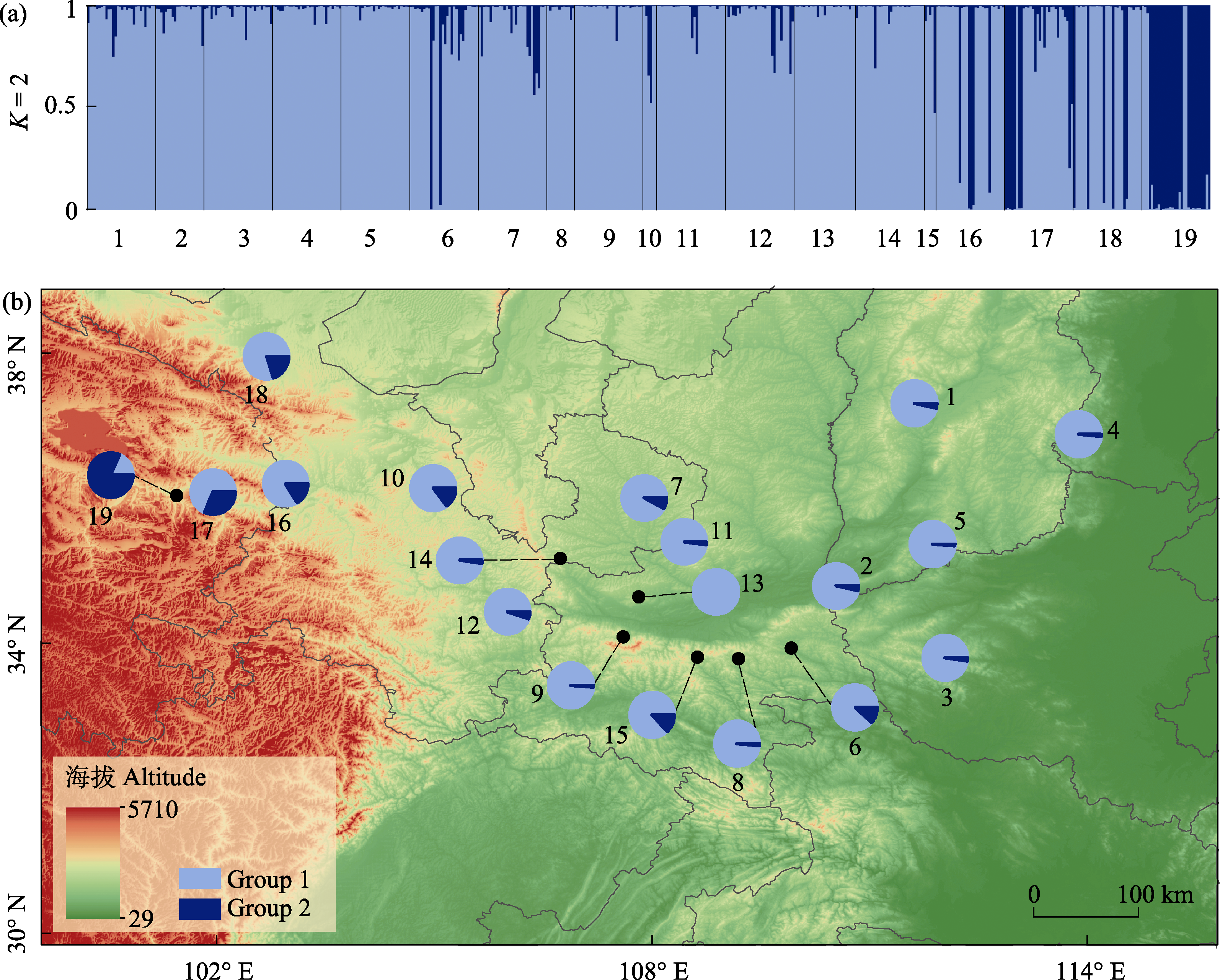
图3 胡桃的遗传结构及其地理分布。(a) 19个胡桃群体491份样本在K = 2时的STRUCTURE图。浅蓝色为Group 1, 深蓝色为Group 2, 1-19表示群体序号(表1); (b)基于STRUCTURE K = 2的19个胡桃群体的遗传结构地理分布图, 分组颜色同(a)。
Fig. 3 Genetic structure and geographical distribution of Juglans regia. (a) STRUCTURE analysis of 491 individuals with a total of 19 populations. The color of light blue represents Group 1, deep blue is Group 2, 1-19 indicate the population IDs (see details in Table 1); (b) Geographical distribution of the genetic structure of 19 J. regia populations with STRUCTURE at K = 2. The color scheme is same to (a).
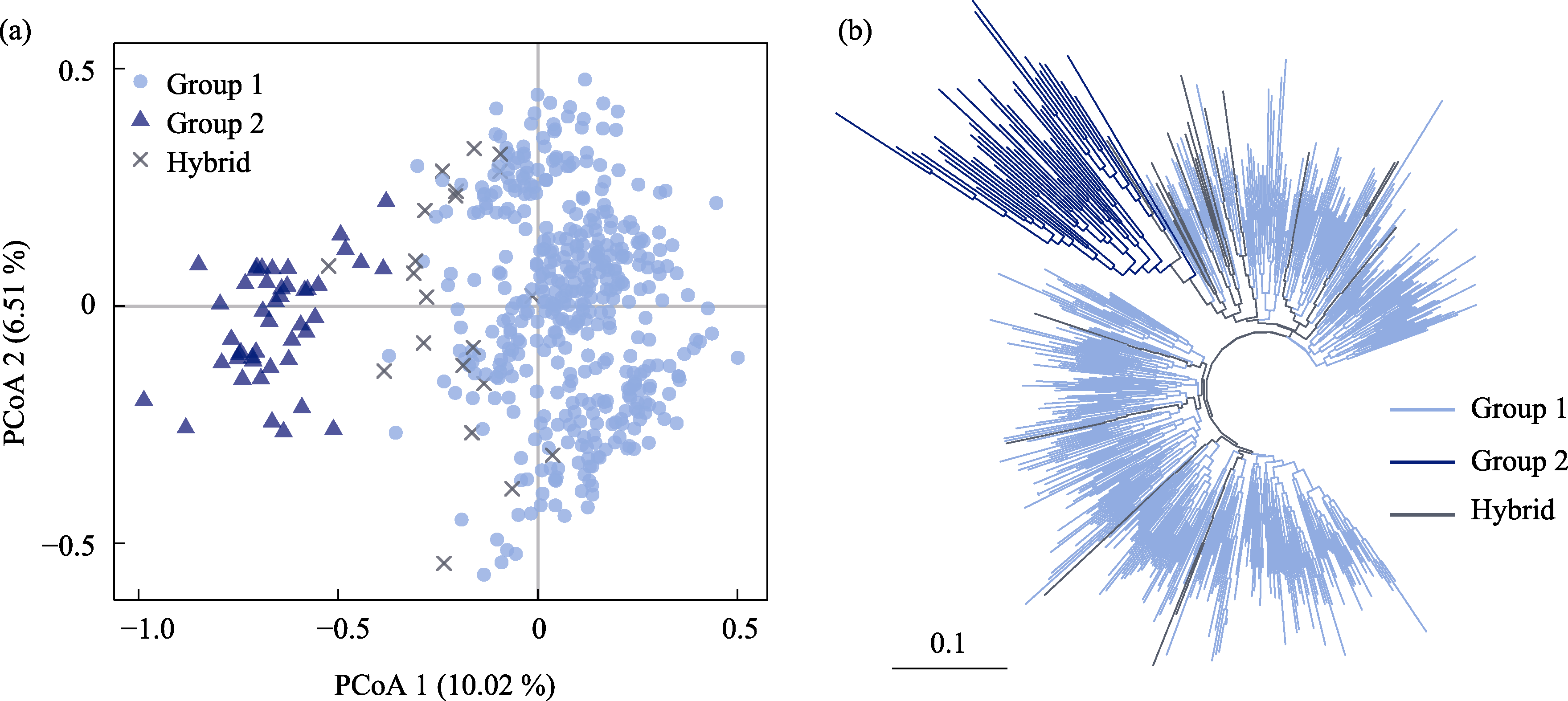
图4 基于19个胡桃群体491个个体的Nei’s遗传距离的主坐标(a)和邻接树(b)聚类结果。浅蓝色圆形表示Group 1, 深蓝色三角形表示Group 2, 黑色叉号表示杂交个体(Hybrid)。左下角的标尺表示枝长。
Fig. 4 The clustering results based on Nei’s genetic distance of 491 individuals of 19 Juglans regia populations. (a) Principal coordinates analysis (PCoA). Light blue circles represent Group 1, dark blue triangles represent Group 2, and black crosses represent hybridized individual (Hybrids). The left-lower scale represents branch length.
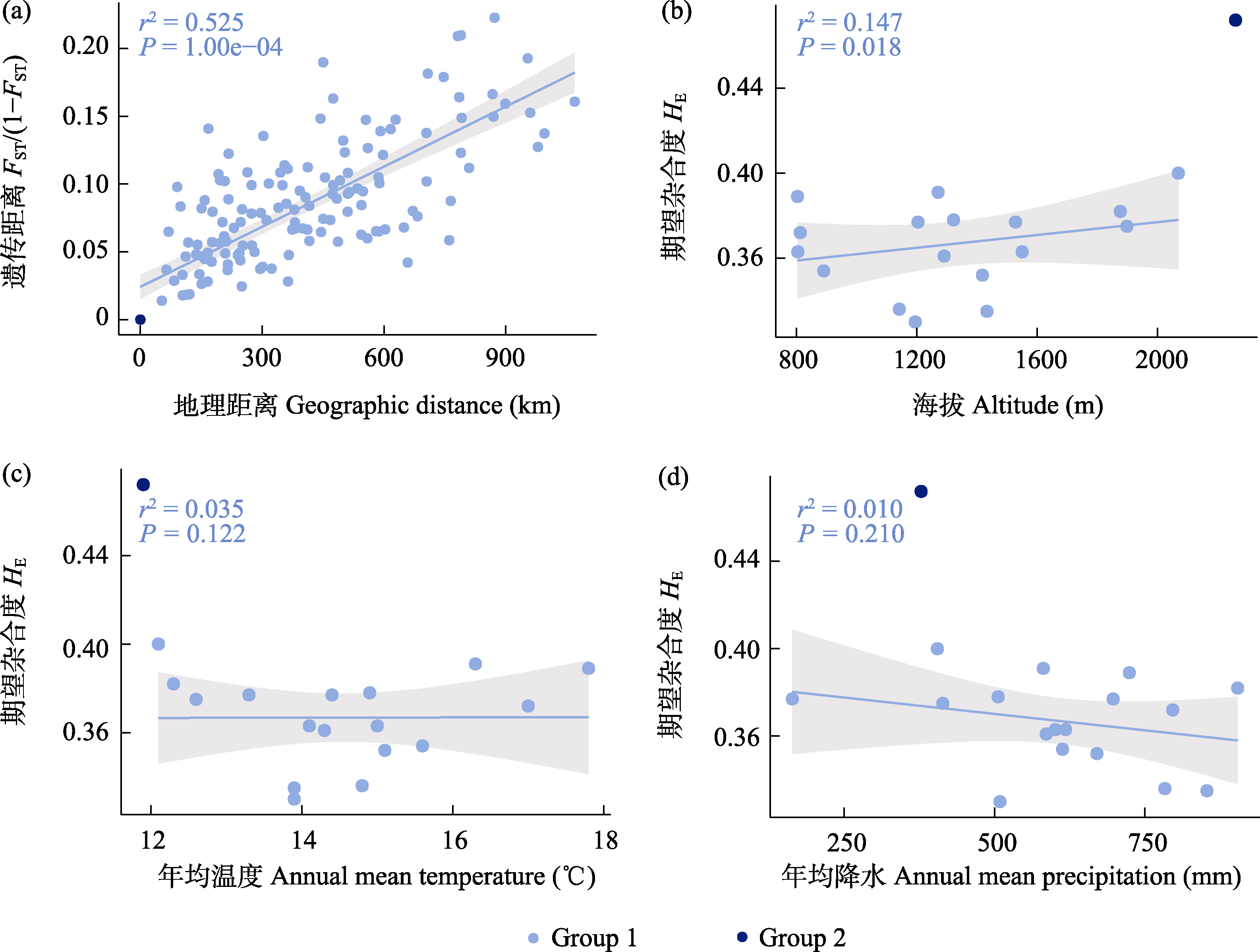
图5 胡桃19个群体的遗传分化和多样性与地理和环境的相关性。(a)遗传距离和地理距离的相关性。浅蓝色表示Group 1, 深蓝色表示Group 2, 仅计算了Group 1的r2和P值; (b-d)遗传多样性(HE)和海拔、年均温、年平均降水的相关性。
Fig. 5 The correlations between genetic differentiation, genetic diversity, and environment variables of 19 Juglans regia populations. (a) Mantel test between pairwise genetic differentiation (FST/(1-FST)) and geographic distance. (b-d) Correlation between genetic diversity (HE) and altitude, annual mean temperature, and annual mean precipitation.
| [1] |
Ahmad N, Tian RZ, Lu JD, Li GH, Sun J, Lin RX, Zhao CZ, Zhou CS, Chang HX, Zhao SZ, Wang XJ (2023) DNA fingerprinting and genetic diversity analysis in Asparagus officinalis L. cultivars using microsatellite molecular markers. Genetic Resources and Crop Evolution, 70, 1163-1177.
DOI |
| [2] | Ahmed HGMD, Rizwan M, Naeem M, Khan MA, Baloch FS, Sun SM, Chung G (2022) Molecular characterization and validation of sunflower (Helianthus annuus L.) hybrids through SSR markers. PLoS ONE, 17, e0267383. |
| [3] |
Amaral JS, Casal S, Pereira JA, Seabra RM, Oliveira BPP (2003) Determination of sterol and fatty acid compositions, oxidative stability, and nutritional value of six walnut (Juglans regia L.) cultivars grown in Portugal. Journal of Agricultural and Food Chemistry, 51, 7698-7702.
PMID |
| [4] |
Bai WN, Zeng YF, Zhang DY (2007) Mating patterns and pollen dispersal in a heterodichogamous tree, Juglans mandshurica (Juglandaceae). New Phytologist, 176, 699-707.
DOI URL |
| [5] |
Barkley NA, Roose ML, Krueger RR, Federici CT (2006) Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theoretical and Applied Genetics, 112, 1519-1531.
DOI PMID |
| [6] | Bernard A, Barreneche T, Lheureux F, Dirlewanger E (2018) Analysis of genetic diversity and structure in a worldwide walnut (Juglans regia L.) germplasm using SSR markers. PLoS ONE, 13, e0208021. |
| [7] |
Bhattarai G, Shi AN, Kandel DR, Solís-Gracia N, da Silva JA, Avila CA (2021) Genome-wide simple sequence repeats (SSR) markers discovered from whole-genome sequence comparisons of multiple spinach accessions. Scientific Reports, 11, 9999.
DOI PMID |
| [8] |
Bourguiba H, Audergon JM, Krichen L, Trifi-Farah N, Mamouni A, Trabelsi S, D’Onofrio C, Asma BM, Santoni S, Khadari B (2012) Loss of genetic diversity as a signature of apricot domestication and diffusion into the Mediterranean Basin. BMC Plant Biology, 12, 49.
DOI PMID |
| [9] |
Chauhan A, Chauhan V (2020) Beneficial effects of walnuts on cognition and brain health. Nutrients, 12, 550.
DOI URL |
| [10] |
Dang M, Yue M, Zhang M, Zhao GF, Zhao P (2019) Gene introgression among closely related species in sympatric populations: A case study of three walnut (Juglans) species. Forests, 10, 965.
DOI URL |
| [11] |
Dang M, Zhou HJ, Woeste KE, Yue M, Zhang Y, Zhao GF, Zhang SX, Zhao P (2021) Comparative phylogeography of Juglans regia and J. mandshurica combining organellar and nuclear DNA markers to assess genetic diversity and introgression in regions of sympatry. Trees, 35, 1993-2007.
DOI |
| [12] |
Ding YM, Cao Y, Zhang WP, Chen J, Liu J, Li P, Renner SS, Zhang DY, Bai WN (2022) Population-genomic analyses reveal bottlenecks and asymmetric introgression from Persian into iron walnut during domestication. Genome Biology, 23, 145.
DOI |
| [13] | Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19, 11-15. |
| [14] |
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359-361.
DOI URL |
| [15] | Ebrahimi A, Zarei A, Lawson S, Woeste KE, Smulders MJM (2016) Genetic diversity and genetic structure of Persian walnut (Juglans regia) accessions from 14 European, African, and Asian countries using SSR markers. Tree Genetics & Genomes, 12, 114. |
| [16] |
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater JM, Zyprian E, Moreira FM, Grando MS (2013) Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biology, 13, 39.
DOI PMID |
| [17] |
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI PMID |
| [18] |
Fukuda T, Ito H, Yoshida T (2003) Antioxidative polyphenols from walnuts (Juglans regia L.). Phytochemistry, 63, 795-801.
DOI URL |
| [19] |
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica, 113, 163-185.
DOI URL |
| [20] | Hartl DL, Clark AG, Clark AG (1997) Principles of Population Genetics. Sinauer Associates, Sunderland, Massachusetts, USA. |
| [21] | Itoo H, Ahmad Shah R, Qurat S, Jeelani A, Khursheed S, Bhat ZA, Mir MA, Rather GH, Zargar SM, Shah MD, Padder BA (2023) Genome-wide characterization and development of SSR markers for genetic diversity analysis in northwestern Himalayas walnut (Juglans regia L.). 3 Biotech, 13, 136. |
| [22] |
Jakobsson M, Rosenberg NA (2007) CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23, 1801-1806.
DOI PMID |
| [23] |
Khounani Z, Hosseinzadeh-Bandbafha H, Nizami AS, Sulaiman A, Goli SAH, Tavassoli-Kafrani E, Ghaffari A, Ali Rajaeifar M, Kim KH, Talebi AF, Aghbashlo M, Tabatabaei M (2020) Unlocking the potential of walnut husk extract in the production of waste cooking oil-based biodiesel. Renewable and Sustainable Energy Reviews, 119, 109588.
DOI URL |
| [24] | Kris Etherton PM (2014) Walnuts decrease risk of cardiovascular disease: A summary of efficacy and biologic mechanisms. The Journal of Nutrition, 144, 547-554. |
| [25] |
Kumar M, Choi JY, Kumari N, Pareek A, Kim SR (2015) Molecular breeding in Brassica for salt tolerance: Importance of microsatellite (SSR) markers for molecular breeding in Brassica. Frontiers in Plant Science, 6, 688.
DOI PMID |
| [26] | Langella O (1999) Populations version 1.2.31: Population Genetic Software (Individuals or Populations Distances, Phylogenetic Trees). http://bioinformatics.org/-tryphon/pop ulations. (accessed on 2022-12-12) |
| [27] | Leslie CA, Mcgranahan GH (1998) The Origin of the Walnut. University of California Agriculture and Natural Resources, Oakland, California, USA. |
| [28] |
Liang W, Dondini L, De Franceschi P, Paris R, Sansavini S, Tartarini S (2015) Genetic diversity, population structure and construction of a core collection of apple cultivars from Italian germplasm. Plant Molecular Biology Reporter, 33, 458-473.
DOI URL |
| [29] |
Liao JQ, Nai YF, Feng L, Chen YM, Li M, Xu HD (2020) Walnut oil prevents scopolamine-induced memory dysfunction in a mouse model. Molecules, 25, 1630.
DOI URL |
| [30] | Liu J, Gao LM (2011) Comparative analysis of three different methods of total DNA extraction used in Taxus. Guihaia, 31, 244-249, 159. (in Chinese with English abstract) |
| [ 刘杰, 高连明 (2011) 红豆杉属植物三种不同总DNA提取方法的分析比较. 广西植物, 31, 244-249, 159.] | |
| [31] | Lu AM, Stone DE, Grauke LJ (1999) Juglandaceae. In: Flora of China (eds Wu ZY, Peter RH), pp. 277-285. Science Press & Missouri Botanical Garden Press, Beijing, China & St. Science Press & Missouri Botanical Garden Press, Beijing, China & St. Louis, Missouri, USA. |
| [32] | Luo KF (1954) A draft for physical geography regionalization of China. Acta Geographica Sinica, 20, 379-394. (in Chinese) |
|
[ 罗开富 (1954) 中国自然地理分区草案. 地理学报, 20, 379-394.]
DOI |
|
| [33] |
Magige EA, Fan PZ, Wambulwa MC, Milne R, Wu ZY, Luo YH, Khan R, Wu HY, Qi HL, Zhu GF, Maity D, Khan I, Gao LM, Liu J (2022) Genetic diversity and structure of Persian walnut (Juglans regia L.) in Pakistan: Implications for conservation. Plants, 11, 1652.
DOI URL |
| [34] | Miao FJ, Shan CL, Ma T, Geng SX, Ning DL (2021) Walnut oil alleviates DSS-induced colitis in mice by inhibiting NLRP 3 inflammasome activation and regulating gut microbiota. Microbial Pathogenesis, 154, 104866. |
| [35] | National Bureau of Statistics (2021) China Statistical Yearbook. China Statistics Press, Beijing. (in Chinese) |
| [ 中华人民共和国国家统计局 (2021) 中国统计年鉴. 中国统计出版社, 北京.] | |
| [36] | Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Vegan: Community Ecology Package. https://CRAN.R-project.org/package=vegan. (accessed on 2023-03-16) |
| [37] |
Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics, 28, 2537-2539.
PMID |
| [38] |
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. Journal of Heredity, 90, 502-503.
DOI URL |
| [39] |
Pollegioni P, Lungo SD, Müller R, Woeste KE, Chiocchini F, Clark J, Hemery GE, Mapelli S, Villani F, Malvolti ME, Mattioni C (2020) Biocultural diversity of common walnut (Juglans regia L.) and sweet chestnut (Castanea sativa Mill.) across Eurasia. Ecology and Evolution, 10, 11192-11216.
DOI PMID |
| [40] |
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends in Plant Science, 1, 215-222.
DOI URL |
| [41] |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
DOI PMID |
| [42] |
Rosenberg NA (2004) Distruct: A program for the graphical display of population structure. Molecular Ecology Notes, 4, 137-138.
DOI URL |
| [43] |
Shah AR, Baksi P, Jasrotia A, Bhat DJ, Gupta R, Bakshi M (2020) Genetic diversity of walnut (Juglans regia L.) seedlings through SSR markers in north-western Himalayan region of Jammu. Bangladesh Journal of Botany, 49, 1003-1012.
DOI URL |
| [44] |
Shahi Shavvon R, Qi HL, Mafakheri M, Fan PZ, Wu HY, Bazdid Vahdati F, Al Shmgani HS, Wang YH, Liu J (2023) Unravelling the genetic diversity and population structure of common walnut in the Iranian Plateau. BMC Plant Biology, 23, 201.
DOI PMID |
| [45] |
Sharma H, Bhandawat A, Kumar P, Rahim MS, Parveen A, Kumar P, Madhawan A, Rishi V, Roy J (2020) Development and characterization of bZIP transcription factor based SSRs in wheat. Gene, 756, 144912.
DOI URL |
| [46] |
Shen HY, Hou YJ, Xi MH, Cai YY, Ao JF, Wang J, Li M, Luo AW (2022) Electron beam irradiation enhanced extraction and antioxidant activity of active compounds in green walnut husk. Food Chemistry, 373, 131520.
DOI URL |
| [47] |
Su M, Zhang CY, Feng SC (2022) Identification and genetic diversity analysis of hybrid offspring of Azalea based on EST-SSR markers. Scientific Reports, 12, 15239.
DOI PMID |
| [48] |
Sun YW, Hou N, Woeste K, Zhang CC, Yue M, Yuan XY, Zhao P (2019) Population genetic structure and adaptive differentiation of iron walnut Juglans regia subsp. sigillata in southwestern China. Ecology and Evolution, 9, 14154-14166.
DOI URL |
| [49] |
Tapia MI, Sánchez-Morgado JR, García-Parra J, Ramírez R, Hernández T, González-Gómez D (2013) Comparative study of the nutritional and bioactive compounds content of four walnut (Juglans regia L.) cultivars. Journal of Food Composition and Analysis, 31, 232-237.
DOI URL |
| [50] |
This P, Lacombe T, Thomas MR (2006) Historical origins and genetic diversity of wine grapes. Trends in Genetics, 22, 511-519.
DOI PMID |
| [51] | Torokeldiev N, Ziehe M, Gailing O, Finkeldey R (2019) Genetic diversity and structure of natural Juglans regia L. populations in the southern Kyrgyz Republic revealed by nuclear SSR and EST-SSR markers. Tree Genetics & Genomes, 15, 5. |
| [52] |
Wambulwa MC, Fan PZ, Milne R, Wu ZY, Luo YH, Wang YH, Wang H, Gao LM, Xiahou ZY, Jin YC, Ye LJ, Xu ZC, Yang ZC, Li DZ, Liu J (2022) Genetic analysis of walnut cultivars from southwest China: Implications for germplasm improvement. Plant Diversity, 44, 530-541.
DOI |
| [53] | Wang H, Pan G, Ma QG, Zhang JP, Pei D (2015) The genetic diversity and introgression of Juglans regia and Juglans sigillata in Tibet as revealed by SSR markers. Tree Genetics & Genomes, 11, 804. |
| [54] | Wang ZL, Wang Y, Cao X, Wu D, Hui M, Han X, Yao F, Li YH, Li H, Wang H (2022) Screening and validation of SSR molecular markers for identification of downy mildew resistance in intraspecific hybrid F 1 progeny (V. vinifera). Horticulturae, 8, 706. |
| [55] | Wickham H (2016) Ggplot2:Elegant Graphics for Data Analysis, 2nd edn. Springer Nature, New York. |
| [56] |
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics, 163, 1177-1191.
DOI PMID |
| [57] |
Wojdyło A, Turkiewicz IP, Tkacz K, Nowicka P, Bobak Ł (2022) Nuts as functional foods: Variation of nutritional and phytochemical profiles and their in vitro bioactive properties. Food Chemistry: X, 15, 100418.
DOI URL |
| [58] | Xi RT, Zhang YP (1996) China Fruit-plant Monograph (Walnut Flora). China Forestry Publishing House, Beijing. (in Chinese) |
| [ 郗荣庭, 张毅萍 (1996) 中国果树志(核桃卷). 中国林业出版社, 北京.] | |
| [59] | Xu YJ, Han HB, Wang H, Chen LN, Ma QG, Pei D (2016) Phenotypic and genetic diversities of nuts of walnut (Juglans regia) populations originated from seedlings in Daba Mountains. Scientia Silvae Sinicae, 52, 111-119. (in Chinese with English abstract) |
| [ 徐永杰, 韩华柏, 王滑, 陈凌娜, 马庆国, 裴东 (2016) 大巴山区核桃实生居群的坚果表型和遗传多样性. 林业科学, 52, 111-119.] | |
| [60] | Xu ZC, Jin YC, Milne RI, Xiahou ZY, Qin HT, Ye LJ, Gao LM, Liu J, Li DZ (2020) Development of 32 novel microsatellite loci in Juglans sigillata using genomic data. Applications in Plant Sciences, 8, e11328. |
| [61] |
Yang WL, Bai ZY, Wang FQ, Zou MZ, Wang XR, Xie JK, Zhang FT (2022) Analysis of the genetic diversity and population structure of Monochasma savatieri Franch. ex Maxim using novel EST-SSR markers. BMC Genomics, 23, 597.
DOI |
| [62] |
Yu GC, Smith DK, Zhu HC, Guan Y, Lam TTY (2017) Ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution, 8, 28-36.
DOI URL |
| [63] |
Yuan XY, Sun YW, Bai XR, Dang M, Feng XJ, Zulfiqar S, Zhao P (2018) Population structure, genetic diversity, and gene introgression of two closely related walnuts (Juglans regia and J. sigillata) in southwestern China revealed by EST-SSR markers. Forests, 9, 646.
DOI URL |
| [64] |
Zhang HM, Chu W, Zhang ZB (2017) Cultivated walnut trees showed earlier but not final advantage over its wild relatives in competing for seed dispersers. Integrative Zoology, 12, 12-25.
DOI PMID |
| [65] |
Zhao P, Zhou HJ, Potter D, Hu YH, Feng XJ, Dang M, Feng L, Zulfiqar S, Liu WZ, Zhao GF, Woeste K (2018) Population genetics, phylogenomics and hybrid speciation of Juglans in China determined from whole chloroplast genomes, transcriptomes, and genotyping-by-sequencing (GBS). Molecular Phylogenetics and Evolution, 126, 250-265.
DOI URL |
| [66] |
Zhou HJ, Zhao P, Woeste K, Zhang SX (2021) Gene flow among wild and cultivated common walnut (Juglans regia) trees in the Qinling Mountains revealed by microsatellite markers. Journal of Forestry Research, 32, 2189-2201.
DOI |
| [67] |
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics, 20, 176-183.
DOI PMID |
| [68] | Zohary D, Hopf M, Weiss E (2012) Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin, 4th edn. Oxford University Press, Oxford. |
| [69] | Zong JW, Zhao TT, Ma QH, Liang LS, Wang GX (2015) Assessment of genetic diversity and population genetic structure of Corylus mandshurica in China using SSR markers. PLoS ONE, 10, e0137528. |
| [1] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [2] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [3] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [4] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [5] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [6] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [7] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [8] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [9] | 崔静, 徐明芳, 章群, 李瑶, 曾晓舒, 李莎. 基于3种线粒体标记探讨中日沿海角木叶鲽遗传多样性差异[J]. 生物多样性, 2022, 30(5): 21485-. |
| [10] | 孙军, 宋煜尧, 施义锋, 翟键, 燕文卓. 近十年中国海洋生物多样性研究进展[J]. 生物多样性, 2022, 30(10): 22526-. |
| [11] | 栗冬梅, 杨卫红, 李庆多, 韩茜, 宋秀平, 潘虹, 冯云. 巴尔通体在滇西南蝙蝠中高度流行并具有丰富的遗传变异特征[J]. 生物多样性, 2021, 29(9): 1245-1255. |
| [12] | 姚志, 郭军, 金晨钟, 刘勇波. 中国纳入一级保护的极小种群野生植物濒危机制[J]. 生物多样性, 2021, 29(3): 394-408. |
| [13] | 叶俊伟, 田斌. 中国西南地区重要木本油料植物扁核木的遗传结构及成因[J]. 生物多样性, 2021, 29(12): 1629-1637. |
| [14] | 向登高, 李跃飞, 李新辉, 陈蔚涛, 马秀慧. 多基因联合揭示海南鲌的遗传结构与遗传多样性[J]. 生物多样性, 2021, 29(11): 1505-1512. |
| [15] | 苏金源, 燕语, 李冲, 李丹, 杜芳. 通过遗传多样性探讨极小种群野生植物的致濒机理及保护策略: 以裸子植物为例[J]. 生物多样性, 2020, 28(3): 376-384. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn