



生物多样性 ›› 2012, Vol. 20 ›› Issue (2): 199-206. DOI: 10.3724/SP.J.1003.2012.08159 cstr: 32101.14.SP.J.1003.2012.08159
何晓红1,2, 韩秀丽1,2, 关伟军1,2, 田可川3, 张文彬4, 马月辉1,2,*( )
)
收稿日期:2011-09-09
接受日期:2012-03-27
出版日期:2012-03-20
发布日期:2012-04-09
通讯作者:
马月辉
作者简介:* E-mail: yuehui.ma@263.net基金资助:
Xiaohong He1,2, Xiuli Han1,2, Weijun Guan1,2, Kechuan Tian3, Wenbin Zhang4, Yuehui Ma1,2,*( )
)
Received:2011-09-09
Accepted:2012-03-27
Online:2012-03-20
Published:2012-04-09
Contact:
Yuehui Ma
摘要:
为从分子水平上对我国双峰驼(Camelus bactrianus)群体的遗传多样性、群体间遗传关系、群体遗传分化及近交情况进行全面、系统地研究, 为双峰驼种质资源保护和新品种培育提供基础数据, 本文利用18对微卫星引物, 分析了我国9个双峰驼群体和1个蒙古双峰驼群体的遗传多样性和遗传关系。结果显示: 10个群体均具有较高的遗传多样性, 共检测到242个等位基因, 平均等位基因数为13.44, 平均有效等位基因数为4.18, 平均观察杂合度(Ho)为0.5528。10个群体间存在显著的遗传分化, 有9.6%的遗传变异来自群体间, 90.4%的遗传变异来自群体内部的个体间。聚类分析、主成分分析和群体遗传结构分析结果都表明10个群体被分成2个明显的分支, 新疆4个群体单独聚为一类, 剩下的6个群体聚为一类。这一结果可能与它们的地理分布和群体间的地理屏障有关。
何晓红, 韩秀丽, 关伟军, 田可川, 张文彬, 马月辉 (2012) 采用微卫星标记分析10个双峰驼群体的遗传变异和群体间的遗传关系. 生物多样性, 20, 199-206. DOI: 10.3724/SP.J.1003.2012.08159.
Xiaohong He, Xiuli Han, Weijun Guan, Kechuan Tian, Wenbin Zhang, Yuehui Ma (2012) Genetic variability and relationship of 10 Bactrian camel populations revealed by microsatellite markers. Biodiversity Science, 20, 199-206. DOI: 10.3724/SP.J.1003.2012.08159.
| 代码 Code | 群体名称 Population name | 样本数 Sample size | 采集地 Sampling location |
|---|---|---|---|
| QH | 青海双峰驼 Qinghai Bactrian camel | 95 | 青海省海西州 Haixi Autonomous Prefecture, Qinghai, China |
| AS | 阿拉善双峰驼-沙漠型 Alashan Bactrian camel-desert type | 50 | 内蒙古阿拉善左旗 Alashan Prefecture, Inner Mongolia, China |
| AG | 阿拉善双峰驼-戈壁型 Alashan Bactrian camel-Gobi type | 50 | 内蒙古阿拉善左旗 Alashan Prefecture, Inner Mongolia, China |
| GS | 甘肃双峰驼 Gansu Bactrian camel | 51 | 甘肃省酒泉 Jiuquan city, Gansu, China |
| SU | 苏尼特双峰驼 Sunite Bactrian camel | 27 | 内蒙古锡林郭勒盟 Silinghol League, Inner Mongolia, China |
| ALT | 阿勒泰双峰驼 Altay Bactrian camel | 46 | 新疆阿勒泰市 Altay City, Xinjiang, China |
| KKZ | 柯尔克孜双峰驼 Kirgizi Bactrian camel | 30 | 新疆克孜勒苏柯尔克孜自治州 Kizilsu Kirghiz Autonomous Prefecture, Xinjiang, China |
| ML | 木垒长眉驼 Mulei Bactrian camel | 34 | 新疆木垒 Mulei Kazak Autonomous Prefecture, Xinjiang, China |
| YL | 伊犁州双峰驼 Yili Bactrian camel | 34 | 新疆伊犁哈萨克自治州 Yili Kazak Autonomous Prefecture, Xinjiang, China |
| MG | 蒙古双峰驼 Mongolian Bactrian camel | 38 | 蒙古国南戈壁省 South Gobi Province, Mongolia |
表1 10个双峰驼群体代码、名称、采集地点和样本数量
Table 1 Code, name, sample size and sampling locations of 10 Bactrian camel populations
| 代码 Code | 群体名称 Population name | 样本数 Sample size | 采集地 Sampling location |
|---|---|---|---|
| QH | 青海双峰驼 Qinghai Bactrian camel | 95 | 青海省海西州 Haixi Autonomous Prefecture, Qinghai, China |
| AS | 阿拉善双峰驼-沙漠型 Alashan Bactrian camel-desert type | 50 | 内蒙古阿拉善左旗 Alashan Prefecture, Inner Mongolia, China |
| AG | 阿拉善双峰驼-戈壁型 Alashan Bactrian camel-Gobi type | 50 | 内蒙古阿拉善左旗 Alashan Prefecture, Inner Mongolia, China |
| GS | 甘肃双峰驼 Gansu Bactrian camel | 51 | 甘肃省酒泉 Jiuquan city, Gansu, China |
| SU | 苏尼特双峰驼 Sunite Bactrian camel | 27 | 内蒙古锡林郭勒盟 Silinghol League, Inner Mongolia, China |
| ALT | 阿勒泰双峰驼 Altay Bactrian camel | 46 | 新疆阿勒泰市 Altay City, Xinjiang, China |
| KKZ | 柯尔克孜双峰驼 Kirgizi Bactrian camel | 30 | 新疆克孜勒苏柯尔克孜自治州 Kizilsu Kirghiz Autonomous Prefecture, Xinjiang, China |
| ML | 木垒长眉驼 Mulei Bactrian camel | 34 | 新疆木垒 Mulei Kazak Autonomous Prefecture, Xinjiang, China |
| YL | 伊犁州双峰驼 Yili Bactrian camel | 34 | 新疆伊犁哈萨克自治州 Yili Kazak Autonomous Prefecture, Xinjiang, China |
| MG | 蒙古双峰驼 Mongolian Bactrian camel | 38 | 蒙古国南戈壁省 South Gobi Province, Mongolia |
| 位点 Locus | 引物(5′-3′) Primer(5′-3′) | 等位基因 Allele (bp) | 荧光标记 Fluorescent label | 退火温度 Tm (℃) | FIT | FST | FIS | 基因流 Nm |
|---|---|---|---|---|---|---|---|---|
| CMS09 | TGCTTTAGACGACTTTTACTTTAC ATTTCACTTTCTTCATACTTGTGAT | 233-256 | FAM | 55 | 0.128*** | 0.042*** | 0.090** | 4.150 |
| CMS17 | TATAAAGGATCACTGCCTTC AAAATGAACCTCCATAAAGTTAG | 144-149 | FAM | 55 | 0.184*** | 0.146*** | 0.045 | 1.600 |
| CMS18 | GAACGACCCTTGAAGACGAA AGCAGCTGGTTTTAGGTCCA | 157-186 | HEX | 60 | 0.231*** | 0.153*** | 0.092*** | 1.851 |
| CMS32 | ACGGACAAGAACTGCTCATA ACAACCAATAAATCCCCATT | 198-204 | FAM | 51 | 0.420*** | 0.019*** | 0.408*** | 7.381 |
| CMS50 | TTTATAGTCAGAGAGAGTGCTG TGTAGGGTTCATTGTAACA | 154-183 | FAM | 55 | 0.042 | 0.050*** | -0.008 | 3.180 |
| CMS121 | CAAGAGAACTGGTGAGGATTTTC AGTTGATAAAAATACAGCTGGAAAG | 151-159 | FAM | 59 | 0.163*** | 0.062*** | 0.107*** | 4.029 |
| CVRL01 | GAAGAGGTTGGGGCACTAC CAGGCAGATATCCATTGAA | 188-253 | HEX | 60 | 0.590*** | 0.105*** | 0.542*** | 2.001 |
| CVRL05 | CCTTGGACCTCCTTGCTCTG GCCACTGGTCCCTGTCATT | 148-174 | HEX | 56 | 0.090** | 0.031*** | 0.061* | 5.612 |
| CVRL06 | TTTTAAAAATTCTGACCAGGAGTCTG CATAATAGCCAAAACATGGAAACAAC | 185-205 | HEX | 59 | 0.243*** | 0.045*** | 0.208*** | 4.061 |
| CVRL07 | AATACCCTAGTTGAAGCTCTGTCCT GAGTGCCTTTATAAATATGGGTCTG | 255-263 | FAM | 56 | 0.366*** | 0.224*** | 0.183*** | 0.662 |
| LCA66 | GTGCAGCGTCCAAATAGTCA CCAGCATCGTCCAGTATTCA | 212-242 | HEX | 55 | 0.434*** | 0.166*** | 0.321*** | 1.163 |
| VOLP08 | CCATTCACCCCATCTCTC TCGCCAGTGACCTTATTTAGA | 142-180 | FAM | 52 | 0.127*** | 0.073*** | 0.059* | 2.536 |
| VOLP10 | CTTTCTCCTTTCCTCCCTACT CGTCCACTTCCTTCATTTC | 232-260 | FAM | 58 | 0.117*** | 0.048*** | 0.073*** | 3.973 |
| VOLP32 | GTGATCGGAATGGCTTGAAA CAGCGAGCACCTGAAAGAA | 256-262 | HEX | 55 | 0.399*** | 0.207*** | 0.241*** | 0.882 |
| VOLP67 | TTAGAGGGTCTATCCAGTTTC TGGACCTAAAAGAGTGGAG | 142-172 | FAM | 60 | 0.399*** | 0.034*** | 0.378*** | 5.698 |
| YWLL08 | ATCAAGTTTGAGGTGCTTTCC CCATGGCATTGTGTTGAAGAC | 154-180 | HEX | 59 | 0.208*** | 0.119*** | 0.101*** | 1.685 |
| YWLL38 | GGCCTAAATCCTACTAGAC CCTCTCACTCTTGTTCTCCTC | 180-192 | FAM | 58 | 0.080*** | 0.049*** | 0.033 | 4.118 |
| YWLL59 | TGTGCAGGAGTTAGGTGTA CCATGTCTCTGAAGCTCTGGA | 109-135 | FAM | 58 | 0.277*** | 0.112*** | 0.186*** | 1.667 |
| 所有位点 All loci | 0.240*** | 0.096*** | 0.159*** | |||||
表2 本研究所采用的18个微卫星位点的相关信息和F-统计量分析结果
Table 2 The information of 18 microsatellite loci employed in this study and F-statistics
| 位点 Locus | 引物(5′-3′) Primer(5′-3′) | 等位基因 Allele (bp) | 荧光标记 Fluorescent label | 退火温度 Tm (℃) | FIT | FST | FIS | 基因流 Nm |
|---|---|---|---|---|---|---|---|---|
| CMS09 | TGCTTTAGACGACTTTTACTTTAC ATTTCACTTTCTTCATACTTGTGAT | 233-256 | FAM | 55 | 0.128*** | 0.042*** | 0.090** | 4.150 |
| CMS17 | TATAAAGGATCACTGCCTTC AAAATGAACCTCCATAAAGTTAG | 144-149 | FAM | 55 | 0.184*** | 0.146*** | 0.045 | 1.600 |
| CMS18 | GAACGACCCTTGAAGACGAA AGCAGCTGGTTTTAGGTCCA | 157-186 | HEX | 60 | 0.231*** | 0.153*** | 0.092*** | 1.851 |
| CMS32 | ACGGACAAGAACTGCTCATA ACAACCAATAAATCCCCATT | 198-204 | FAM | 51 | 0.420*** | 0.019*** | 0.408*** | 7.381 |
| CMS50 | TTTATAGTCAGAGAGAGTGCTG TGTAGGGTTCATTGTAACA | 154-183 | FAM | 55 | 0.042 | 0.050*** | -0.008 | 3.180 |
| CMS121 | CAAGAGAACTGGTGAGGATTTTC AGTTGATAAAAATACAGCTGGAAAG | 151-159 | FAM | 59 | 0.163*** | 0.062*** | 0.107*** | 4.029 |
| CVRL01 | GAAGAGGTTGGGGCACTAC CAGGCAGATATCCATTGAA | 188-253 | HEX | 60 | 0.590*** | 0.105*** | 0.542*** | 2.001 |
| CVRL05 | CCTTGGACCTCCTTGCTCTG GCCACTGGTCCCTGTCATT | 148-174 | HEX | 56 | 0.090** | 0.031*** | 0.061* | 5.612 |
| CVRL06 | TTTTAAAAATTCTGACCAGGAGTCTG CATAATAGCCAAAACATGGAAACAAC | 185-205 | HEX | 59 | 0.243*** | 0.045*** | 0.208*** | 4.061 |
| CVRL07 | AATACCCTAGTTGAAGCTCTGTCCT GAGTGCCTTTATAAATATGGGTCTG | 255-263 | FAM | 56 | 0.366*** | 0.224*** | 0.183*** | 0.662 |
| LCA66 | GTGCAGCGTCCAAATAGTCA CCAGCATCGTCCAGTATTCA | 212-242 | HEX | 55 | 0.434*** | 0.166*** | 0.321*** | 1.163 |
| VOLP08 | CCATTCACCCCATCTCTC TCGCCAGTGACCTTATTTAGA | 142-180 | FAM | 52 | 0.127*** | 0.073*** | 0.059* | 2.536 |
| VOLP10 | CTTTCTCCTTTCCTCCCTACT CGTCCACTTCCTTCATTTC | 232-260 | FAM | 58 | 0.117*** | 0.048*** | 0.073*** | 3.973 |
| VOLP32 | GTGATCGGAATGGCTTGAAA CAGCGAGCACCTGAAAGAA | 256-262 | HEX | 55 | 0.399*** | 0.207*** | 0.241*** | 0.882 |
| VOLP67 | TTAGAGGGTCTATCCAGTTTC TGGACCTAAAAGAGTGGAG | 142-172 | FAM | 60 | 0.399*** | 0.034*** | 0.378*** | 5.698 |
| YWLL08 | ATCAAGTTTGAGGTGCTTTCC CCATGGCATTGTGTTGAAGAC | 154-180 | HEX | 59 | 0.208*** | 0.119*** | 0.101*** | 1.685 |
| YWLL38 | GGCCTAAATCCTACTAGAC CCTCTCACTCTTGTTCTCCTC | 180-192 | FAM | 58 | 0.080*** | 0.049*** | 0.033 | 4.118 |
| YWLL59 | TGTGCAGGAGTTAGGTGTA CCATGTCTCTGAAGCTCTGGA | 109-135 | FAM | 58 | 0.277*** | 0.112*** | 0.186*** | 1.667 |
| 所有位点 All loci | 0.240*** | 0.096*** | 0.159*** | |||||
| 群体 Population | Ho | He | PIC | 平均等位基因数 MNA | 有效等位基因 NE | 基因丰富度 AR | 所有位点FIS FIS of all loci |
|---|---|---|---|---|---|---|---|
| QH | 0.572 | 0.687 | 0.639 | 8.78 | 3.63 | 4.91 | 0.168* |
| AS | 0.519 | 0.646 | 0.597 | 6.83 | 3.28 | 4.63 | 0.199* |
| AG | 0.518 | 0.638 | 0.587 | 6.94 | 3.18 | 4.57 | 0.191* |
| GS | 0.552 | 0.679 | 0.632 | 6.44 | 3.67 | 4.72 | 0.189* |
| SU | 0.574 | 0.649 | 0.589 | 5.56 | 3.13 | 4.43 | 0.118* |
| MG | 0.538 | 0.671 | 0.622 | 6.56 | 3.47 | 4.71 | 0.201* |
| ALT | 0.561 | 0.649 | 0.595 | 8.00 | 3.20 | 4.70 | 0.138* |
| KKZ | 0.589 | 0.648 | 0.591 | 6.22 | 3.05 | 4.48 | 0.093 |
| ML | 0.533 | 0.594 | 0.543 | 6.06 | 2.72 | 4.18 | 0.105* |
| YL | 0.573 | 0.654 | 0.600 | 6.39 | 3.19 | 4.66 | 0.125* |
表3 家养双峰驼遗传变异系数
Table 3 Genetic variation information of the Bactrian camel populations
| 群体 Population | Ho | He | PIC | 平均等位基因数 MNA | 有效等位基因 NE | 基因丰富度 AR | 所有位点FIS FIS of all loci |
|---|---|---|---|---|---|---|---|
| QH | 0.572 | 0.687 | 0.639 | 8.78 | 3.63 | 4.91 | 0.168* |
| AS | 0.519 | 0.646 | 0.597 | 6.83 | 3.28 | 4.63 | 0.199* |
| AG | 0.518 | 0.638 | 0.587 | 6.94 | 3.18 | 4.57 | 0.191* |
| GS | 0.552 | 0.679 | 0.632 | 6.44 | 3.67 | 4.72 | 0.189* |
| SU | 0.574 | 0.649 | 0.589 | 5.56 | 3.13 | 4.43 | 0.118* |
| MG | 0.538 | 0.671 | 0.622 | 6.56 | 3.47 | 4.71 | 0.201* |
| ALT | 0.561 | 0.649 | 0.595 | 8.00 | 3.20 | 4.70 | 0.138* |
| KKZ | 0.589 | 0.648 | 0.591 | 6.22 | 3.05 | 4.48 | 0.093 |
| ML | 0.533 | 0.594 | 0.543 | 6.06 | 2.72 | 4.18 | 0.105* |
| YL | 0.573 | 0.654 | 0.600 | 6.39 | 3.19 | 4.66 | 0.125* |
| QH | AS | AG | GS | SU | MG | ALT | KKZ | ML | YL | |
|---|---|---|---|---|---|---|---|---|---|---|
| QH | 0.0603** | 0.0746** | 0.0734** | 0.0691** | 0.0645** | 0.0966** | 0.1253** | 0.1284** | 0.1238** | |
| AS | 3.8959 | 0.0537** | 0.0960** | 0.0873** | 0.0624** | 0.0775** | 0.1170** | 0.1276** | 0.1266** | |
| AG | 3.1012 | 4.4055 | 0.0729** | 0.1043** | 0.0790** | 0.0914** | 0.1400** | 0.1204** | 0.1415** | |
| GS | 3.1560 | 2.3542 | 3.1794 | 0.1169** | 0.0678** | 0.1088** | 0.1493** | 0.1382** | 0.1394** | |
| SU | 3.3679 | 2.6137 | 2.1469 | 1.8886 | 0.0852 | 0.1278** | 0.1694** | 0.1623** | 0.1455** | |
| MG | 3.6260 | 3.7564 | 2.9146 | 3.4373 | 2.6843 | 0.0805** | 0.1280* | 0.1058** | 0.1235* | |
| ALT | 2.3380 | 2.9758 | 2.4852 | 2.0478 | 1.7062 | 2.8556 | 0.0486** | 0.0346** | 0.0449** | |
| KKZ | 1.7452 | 1.8868 | 1.5357 | 1.4245 | 1.2258 | 1.7031 | 4.8940 | 0.1004** | 0.0269** | |
| ML | 1.6970 | 1.7092 | 1.8264 | 1.5590 | 1.2904 | 2.1129 | 6.9754 | 2.2400 | 0.0918** | |
| YL | 1.7694 | 1.7247 | 1.5168 | 1.5434 | 1.4682 | 1.7743 | 5.3179 | 9.0437 | 2.4733 |
表4 10个双峰驼群体的遗传分化程度(FST, 对角线上)和基因流(Nm, 对角线下)
Table 4 Fixation index resulting from comparing subpopulations to the total population (FST , above the diagonal) and gene flow (Nm, below the diagonal) among 10 Bactrain camel populations
| QH | AS | AG | GS | SU | MG | ALT | KKZ | ML | YL | |
|---|---|---|---|---|---|---|---|---|---|---|
| QH | 0.0603** | 0.0746** | 0.0734** | 0.0691** | 0.0645** | 0.0966** | 0.1253** | 0.1284** | 0.1238** | |
| AS | 3.8959 | 0.0537** | 0.0960** | 0.0873** | 0.0624** | 0.0775** | 0.1170** | 0.1276** | 0.1266** | |
| AG | 3.1012 | 4.4055 | 0.0729** | 0.1043** | 0.0790** | 0.0914** | 0.1400** | 0.1204** | 0.1415** | |
| GS | 3.1560 | 2.3542 | 3.1794 | 0.1169** | 0.0678** | 0.1088** | 0.1493** | 0.1382** | 0.1394** | |
| SU | 3.3679 | 2.6137 | 2.1469 | 1.8886 | 0.0852 | 0.1278** | 0.1694** | 0.1623** | 0.1455** | |
| MG | 3.6260 | 3.7564 | 2.9146 | 3.4373 | 2.6843 | 0.0805** | 0.1280* | 0.1058** | 0.1235* | |
| ALT | 2.3380 | 2.9758 | 2.4852 | 2.0478 | 1.7062 | 2.8556 | 0.0486** | 0.0346** | 0.0449** | |
| KKZ | 1.7452 | 1.8868 | 1.5357 | 1.4245 | 1.2258 | 1.7031 | 4.8940 | 0.1004** | 0.0269** | |
| ML | 1.6970 | 1.7092 | 1.8264 | 1.5590 | 1.2904 | 2.1129 | 6.9754 | 2.2400 | 0.0918** | |
| YL | 1.7694 | 1.7247 | 1.5168 | 1.5434 | 1.4682 | 1.7743 | 5.3179 | 9.0437 | 2.4733 |
| QH | AS | AG | GS | SU | MG | ALT | KKZ | ML | YL | |
|---|---|---|---|---|---|---|---|---|---|---|
| QH | 0.1102 | 0.1299 | 0.1660 | 0.1587 | 0.1508 | 0.1985 | 0.2443 | 0.2249 | 0.2385 | |
| AS | 0.1404 | 0.0986 | 0.1842 | 0.1695 | 0.1386 | 0.1586 | 0.2276 | 0.1991 | 0.2390 | |
| AG | 0.1763 | 0.1118 | 0.1426 | 0.1820 | 0.1538 | 0.1736 | 0.2631 | 0.1946 | 0.2615 | |
| GS | 0.1915 | 0.2385 | 0.1684 | 0.2374 | 0.1980 | 0.2228 | 0.2933 | 0.2443 | 0.2782 | |
| SU | 0.1694 | 0.1997 | 0.2423 | 0.3143 | 0.1955 | 0.2701 | 0.3203 | 0.2853 | 0.3016 | |
| MG | 0.1632 | 0.1423 | 0.1818 | 0.1710 | 0.2061 | 0.1723 | 0.2607 | 0.1914 | 0.2458 | |
| ALT | 0.2470 | 0.1712 | 0.2044 | 0.2804 | 0.3208 | 0.1898 | 0.1298 | 0.0897 | 0.1223 | |
| KKZ | 0.3499 | 0.2840 | 0.3529 | 0.4361 | 0.4786 | 0.3422 | 0.1027 | 0.1680 | 0.0825 | |
| ML | 0.3149 | 0.2785 | 0.2538 | 0.3359 | 0.3821 | 0.2280 | 0.0617 | 0.2027 | 0.1453 | |
| YL | 0.3484 | 0.3190 | 0.3632 | 0.4003 | 0.3894 | 0.3309 | 0.0957 | 0.0566 | 0.1843 |
表5 10个双峰驼群体的Nei’s遗传距离(DA, 对角线上)和标准遗传距离(DS, 对角线下)
Table 5 Nei’s genetic distance (DA, above the diagonal) and Nei’s standard genetic distance (DS, below the diagonal) among 10 Bactrain camel populations
| QH | AS | AG | GS | SU | MG | ALT | KKZ | ML | YL | |
|---|---|---|---|---|---|---|---|---|---|---|
| QH | 0.1102 | 0.1299 | 0.1660 | 0.1587 | 0.1508 | 0.1985 | 0.2443 | 0.2249 | 0.2385 | |
| AS | 0.1404 | 0.0986 | 0.1842 | 0.1695 | 0.1386 | 0.1586 | 0.2276 | 0.1991 | 0.2390 | |
| AG | 0.1763 | 0.1118 | 0.1426 | 0.1820 | 0.1538 | 0.1736 | 0.2631 | 0.1946 | 0.2615 | |
| GS | 0.1915 | 0.2385 | 0.1684 | 0.2374 | 0.1980 | 0.2228 | 0.2933 | 0.2443 | 0.2782 | |
| SU | 0.1694 | 0.1997 | 0.2423 | 0.3143 | 0.1955 | 0.2701 | 0.3203 | 0.2853 | 0.3016 | |
| MG | 0.1632 | 0.1423 | 0.1818 | 0.1710 | 0.2061 | 0.1723 | 0.2607 | 0.1914 | 0.2458 | |
| ALT | 0.2470 | 0.1712 | 0.2044 | 0.2804 | 0.3208 | 0.1898 | 0.1298 | 0.0897 | 0.1223 | |
| KKZ | 0.3499 | 0.2840 | 0.3529 | 0.4361 | 0.4786 | 0.3422 | 0.1027 | 0.1680 | 0.0825 | |
| ML | 0.3149 | 0.2785 | 0.2538 | 0.3359 | 0.3821 | 0.2280 | 0.0617 | 0.2027 | 0.1453 | |
| YL | 0.3484 | 0.3190 | 0.3632 | 0.4003 | 0.3894 | 0.3309 | 0.0957 | 0.0566 | 0.1843 |
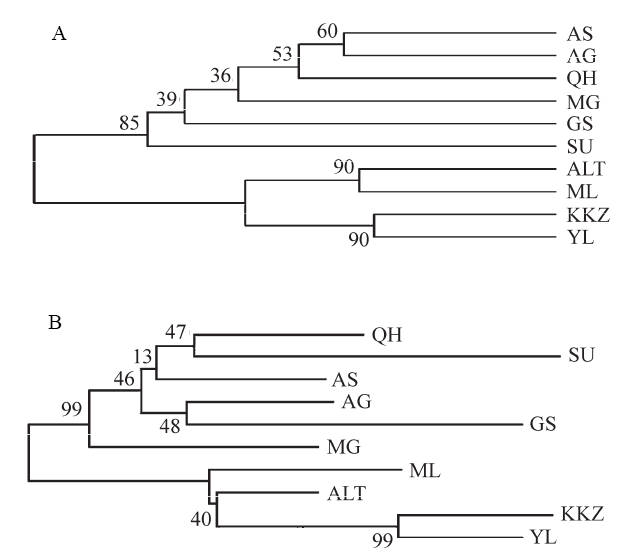
图1 基于遗传距离(DA)构建的10个双峰驼群体的UPGMA聚类树(A)和NJ树(B) (群体代号同表1)
Fig. 1 UPGMA tree (A) and neighbor-joining tree (B) of 10 Bactrian camel populations based on DA。Population codes are the same as Table 1.
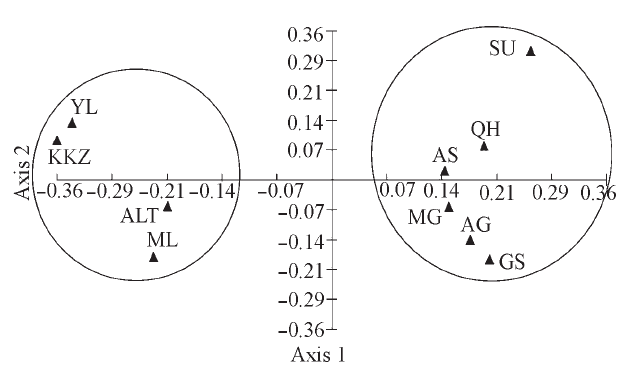
图2 10个双峰驼群体的第一、二主成分二维散点图(群体代号同表1)
Fig. 2 Scatter plot of the first and second factors for 10 Bactrian camel populations。Population codes are the same as Table 1.
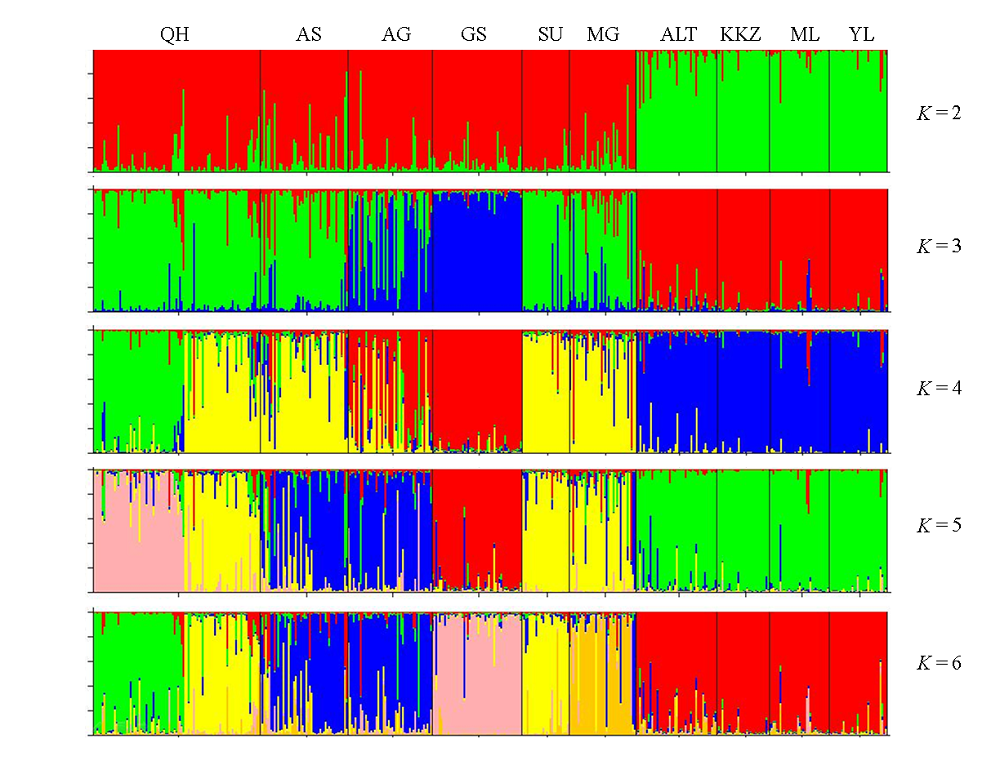
图3 用Structure推导的10个双峰驼群体遗传结构(K=2, 3, 4, 5, 6。群体代号同表1)
Fig. 3 Structure analysis of 10 Bactrian camel populations. K = 2, 3, 4, 5, 6. Population codes are the same as Table 1.
| [1] |
Balmus G, Trifonov VA, Biltueva LS, O'Brien PC, Alkalaeva ES, Fu B, Skidmore JA, Allen T, Graphodatsky AS, Yang F, Ferguson-Smith MA (2007) Cross-species chromosome painting among camel, cattle, pig and human: further insights into the putative Cetartiodactyla ancestral karyotype. Chromosome Research, 15, 499-514.
DOI URL PMID |
| [2] | Chen YC (陈幼春), Wang YY (王毓英), Chang H (常洪), Cao HH (曹红鹤), Pang ZH (庞之洪), Zhang Y (张跃) (1990) The classification of Chinese yellow cattle. In: Characteristics of Chinese Yellow Cattle Ecospecies and Their Course of Utilization) (中国黄牛生态种特征及其利用方向)(ed. Chen YC (陈幼春)), pp. 89-93. China Agricultural Press, Beijing. (in Chinese) |
| [3] | Di R (狄冉) (2008) The Microsatellite and SNPs Study of Chinese Cashmere Goats (中国产绒山羊微卫星和单核苷酸多态性研究). PhD dissertation, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing. (in Chinese) |
| [4] | Evdotchenko D, Han Y, Bartenschlager H, Preuss S, Geldermann H (2003) New polymorphic microsatellite loci for different camel species. Molecular Ecology Notes, 3, 431-434. |
| [5] | Gao HW (高宏巍), Wang J (王晶), He JX (何俊霞), Chen LY (陈鲁勇), Ji R (吉日木图), Meng H (孟和) (2009) Analysis on the genetic diversity and evolution in the Camelus bactrianus using SSR markers. Journal of Shanghai Jiaotong University (Agriculture Science) (上海交通大学学报) (农业科学版), 27, 89-95. (in Chinese with English abstract) |
| [6] | Han JL (韩建林) (2000) Origin, Evolution and Genetic Diversity of Old World Genus of Camelus (旧世界驼属动物的起源、演化及其遗传多样性). PhD dissertation, Lanzhou University, Lanzhou. (in Chinese) |
| [7] | Han JL, Ochieng JW, Lkhagva B, Hanotte O (2004) Genetic diversity and relationship of domestic Bactrian camels (Camelus bactrianus) in China and Mongolia. Journal of Camel Practice and Research, 11, 97-99. |
| [8] | Hoffmann I, Marsan PA, Barker JSF, Cothran EG, Hanotte O, Lenstra JA, Milan D, Weigend S, Simianer H(2004) New MoDAD marker sets to be used in diversity studies for the major farm animal species: recommendations of a joint ISAG/FAO working group. In: Proc. 29th International Conference on Animal Genetics. Tokyo, Japan.Lang KDM, Wang Y, Plante Y (1996) Fifteen polymorphic dinuleotide microsatellites in llamas and alpacas. Animal Genetics, 27, 293. |
| [9] | Men ZM (门正明), Han JL (韩建林), Zhang YZ (张月周) (1989) A study on the polymorphous heredity of blood proteins in Bactrian camel. Acta Veterinariaet Zootechnica Sinica (畜牧兽医学报), 20, 313-314. (in Chinese with English abstract) |
| [10] |
Obreque V, Coogle L, Henney PJ, Bailey E, Mancilla R, García-Huidobro J, Hinrichsen P, Cothran EG (1998) Characterisation of 10 polymorphic alpaca dinucleotide microsatellites. Animal Genetics, 29, 461-462.
URL PMID |
| [11] |
Penedo MCT, Caetano AR, Cordova K (1999) Eight microsatellite markers for south American camelids. Animal Genetics, 30, 166-167.
DOI URL PMID |
| [12] |
Prichard JK, Stephens M, Donnelly P (2000) Inference structure using multilocus genotype data. Genetics, 155, 945-959.
URL PMID |
| [13] | Raymond M, Rousset F (1995) GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86, 248-249. |
| [14] | Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York. |
| [15] |
Taylor KM, Hungerford DA, Snyder RL, Ulmer FA Jr (1968) Uniformity of kryotypes in the Camelidae. Cytogenetics, 7, 8-15.
DOI URL PMID |
| [16] | Wang L (王乐), Ren ZJ (任战军), Cheng J (程佳), Wang YX (王咏絮), Wang HL (王洪亮) (2010) Genetic diversity of five domesticated Bactrian camel populations in China. Acta Agriculturae Boreali-occidentalis Sinica (西北农业学报), 9(11), 18-23. (in Chinese with English abstract) |
| [17] | Yang SL, Wang ZG, Liu B, Zhang GX, Zhao SH, Yu M, Fan B, Li MH, Xiong TA, Li K (2003) Genetic variation and relationships of eighteen Chinese indigenous pig breeds. Genetics, Selection, Evolution, 35, 657-671. |
| [18] | Zhao QJ (赵倩君) (2007) Origin, Genetic Diversity and Conservation of Chinese Sheep Populations (中国部分绵羊群体的起源、遗传多样性及保护研究). PhD dissertation, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing. (in Chinese) |
| [1] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [2] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [3] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [4] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [5] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [6] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [7] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [8] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [9] | 牛晓锋, 王晓梅, 张研, 赵志鹏, 樊恩源. 鲟鱼分子鉴定方法的整合应用[J]. 生物多样性, 2022, 30(6): 22034-. |
| [10] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [11] | 崔静, 徐明芳, 章群, 李瑶, 曾晓舒, 李莎. 基于3种线粒体标记探讨中日沿海角木叶鲽遗传多样性差异[J]. 生物多样性, 2022, 30(5): 21485-. |
| [12] | 孙军, 宋煜尧, 施义锋, 翟键, 燕文卓. 近十年中国海洋生物多样性研究进展[J]. 生物多样性, 2022, 30(10): 22526-. |
| [13] | 栗冬梅, 杨卫红, 李庆多, 韩茜, 宋秀平, 潘虹, 冯云. 巴尔通体在滇西南蝙蝠中高度流行并具有丰富的遗传变异特征[J]. 生物多样性, 2021, 29(9): 1245-1255. |
| [14] | 姚志, 郭军, 金晨钟, 刘勇波. 中国纳入一级保护的极小种群野生植物濒危机制[J]. 生物多样性, 2021, 29(3): 394-408. |
| [15] | 叶俊伟, 田斌. 中国西南地区重要木本油料植物扁核木的遗传结构及成因[J]. 生物多样性, 2021, 29(12): 1629-1637. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2026 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()