



生物多样性 ›› 2018, Vol. 26 ›› Issue (12): 1318-1324. DOI: 10.17520/biods.2018184 cstr: 32101.14.biods.2018184
收稿日期:2018-07-03
接受日期:2018-12-21
出版日期:2018-12-20
发布日期:2019-02-11
通讯作者:
刘培贵
作者简介:# 共同第一作者
基金资助:
Xiaojuan Deng1, Jianli Liu1, Xingfu Yan1, Peigui Liu2,*( )
)
Received:2018-07-03
Accepted:2018-12-21
Online:2018-12-20
Published:2019-02-11
Contact:
Liu Peigui
About author:# 同等贡献作者 Contributed equally to this work
摘要:
块菌是重要的经济真菌, 在其生长发育过程中, 细菌扮演了重要角色。本文利用传统分离培养方法和高通量测序技术分析了印度块菌(Tuber indicum)子囊果内细菌的群落结构。共分离得到细菌532株, 根据物种累积曲线, 选取其中的112株细菌进行了16S rRNA基因序列分析, 共鉴定出4属40种, 其中假单胞菌属(Pseudomonas)菌株占所测菌株数的80%, 不动杆菌属(Acinetobacter)占12.5%, 链霉菌属(Streptomyces)占5%, 贪噬菌属(Variovorax)占2.5%。通过对印度块菌子囊果16S rRNA基因的V1-V3区高通量测序分析, 共获得细菌序列9,862条, 分属于7门43属220种, 其中变形菌门、拟杆菌门和放线菌门的物种占总物种数的99.7%, 是印度块菌子囊果内的优势细菌。黄杆菌属(Flavobacterium)、壤霉菌属(Agromyces)、微杆菌属(Microbacterium)、剑菌属(Ensifer)和寡养食单胞菌属(Stenotrophomonas)的物种数占总物种的86.3%, 是印度块菌子囊果内细菌的优势属。研究结果表明, 采用胰蛋白大豆培养基仅分离得到印度块菌子囊果内少数细菌物种, 而采用高通量测序技术分析发现, 印度块菌子囊果内细菌物种种类丰富, 群落结构复杂。
邓晓娟, 刘建利, 闫兴富, 刘培贵 (2018) 用传统分离培养法和高通量测序技术分析印度块菌子囊果内细菌的群落结构. 生物多样性, 26, 1318-1324. DOI: 10.17520/biods.2018184.
Xiaojuan Deng, Jianli Liu, Xingfu Yan, Peigui Liu (2018) Community composition of bacteria associated with ascocarps of Tuber indicum using traditional culture method and Roche 454 high-throughput sequencing. Biodiversity Science, 26, 1318-1324. DOI: 10.17520/biods.2018184.
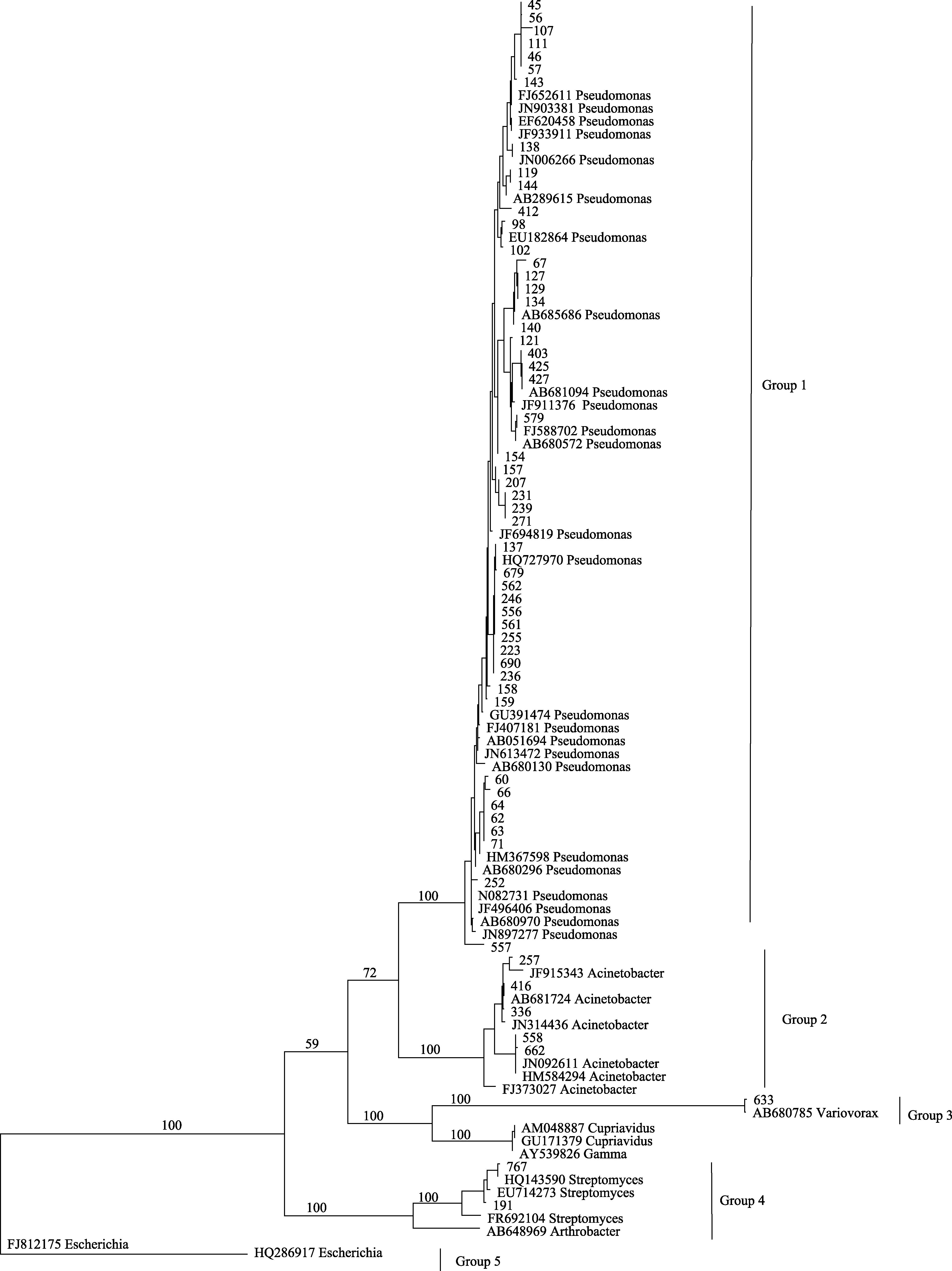
图2 基于印度块菌子囊果内可培养细菌16S rRNA基因序列构建的最大简约树。分支上的数字为≥ 50%的靴带值。
Fig. 2 One of most parsimonious trees based on the analysis of 16S rRNA gene sequences of culturable bacteria from Tuber indicum ascocarps. Numbers above branches indicate bootstrap support above 50%.
| 1 | Barbieri E, Bertini L, Rossi I, Ceccaroli P, Saltarelli R, Guidi C, Zambonelli A, Stocchi V (2005) New evidence for bacterial diversity in the ascoma of the ectomycorrhizal fungus Tuber borchii Vittad. FEMS Microbiology Letters, 247, 23-35. |
| 2 | Barbieri E, Guidi C, Bertaux J, Frey-Klett P, Garbaye J, Ceccaroli P, Saltarelli R, Zambonelli A, Stocchi V (2007) Occurrence and diversity of bacterial communities in Tuber magnatum during truffle maturation. Environmental Microbiology, 9, 2234-2246. |
| 3 | Citterio B, Cardoni P, Potenza L, Amicucci A, Stocchi V, Gola G, Nuti M (1995) >Isolation of bacteria from sporocarps of Tuber magnatum Pico, Tuber borchii Vitt. and Tuber maculatum Vitt. In: Biotechnology of Ectomycorrhizae (eds Stocchi V, Bonfante P, Nuti M), pp. 241-248. Plenum Press, New York. |
| 4 | Citterio B, Malatesta M, Battistelli S, Marcheggiani F, Baffone W, Saltarelli R, Stocchi V, Gazzanelli G (2001) Possible involvement of Pseudomonas fluorescens and Bacillaceae in structural modifications of Tuber borchii fruit bodies. Canadian Journal of Microbiology, 47, 264-268. |
| 5 | Deveau A, Antony-Babu S, Le Tacon F, Robin C, Frey-Klett P (2016) Temporal changes of bacterial communities in the Tuber melanosporum ectomycorrhizosphere during ascocarp development. Mycorrhiza, 26, 389-399. |
| 6 | Dib-Bellahouel S, Fortas Z (2014) Activity of the desert truffle Terfezia boudieri Chatin, against associated soil microflora. African Journal of Microbiology Research, 8, 3008-3016. |
| 7 | Dorofeev AG, Grigor’eva NV, Kozlov MN, Kevbrina MV, Aseeva VG, Nikolav YA (2014) Approaches to cultivation of “nonculturable” bacteria: Cyclic cultures. Microbiology, 83, 450-461. |
| 8 | Fu Y, Li X, Li Q, Wu H, Xiong C, Geng Q, Sun H, Sun Q (2016) Soil microbial communities of three major Chinese truffles in Southwest China. Canadian Journal of Microbiology, 62, 970-979. |
| 9 | Gryndler M, Soukupová L, Hršelová H, Gryndlerová H, Borovičika J, Streiblová E, Jansa J (2013) A quest for indigenous truffle helper prokaryotes. Environment Microbiology Reports, 5, 346-352. |
| 10 | Gurtler V, Stanisich VA (1996) New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology, 142, 3-16. |
| 11 | Hall TA (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98. |
| 12 | Liu PG, Chen J, Deng XJ, Wang XH, Qiao P, Zhang JP, Wan SP, Geng LY, Zhao FL, Zhao WQ, Wang XJ (2016) Truffles from China. Science Press, Beijing. (in Chinese) |
| [刘培贵, 陈娟, 邓晓娟, 王向华, 乔鹏, 张介平, 万山平, 耿丽英, 赵峰岚, 赵文青, 王晓进 (2016) 中国的块菌. 科学出版社, 北京.] | |
| 13 | Mavromatis K, Land M L, Brettin T S, Quest D J, Copeland A, Clum A, Goodwin L, Woyke T, Lapidus A, Klenk HP, Cottingham RW, Kyrpides NC (2012) The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation. PLoS ONE, 7, e48837. |
| 14 | Mello A, Miozzi L, Vizzini A, Napoli C, Kowalchuk G, Bonfante P (2010) Bacterial and fungal communities associated with Tuber magnatum—productive niches. Plant Biosystems, 144, 323-332. |
| 15 | Navarro-Ródenas A, Berná L M, Lozano-Carrillo C, Andrino A, Morte A (2016) Beneficial native bacteria improve survival and mycorrhization of desert truffle mycorrhizal plants in nursery conditions. Mycorrhiza, 26, 769-779. |
| 16 | Picceri GG, Leonardi P, Iotti M, Gallo M, Baldi F, Zambonelli A, Amicucci A, Vallorani L, Piccoli G, Ciccimarra G, Arshakyan M, Burattini S, Falcieri E, Chiarantini L (2018) Bacteria-produced ferric exopolysaccharide nanoparticles as iron delivery system for truffles (Tuber borchii). Applied Microbiology and Biotechnology, 102, 1429-1441. |
| 17 | Qiao P, Tian W, Liu PG, Yu GQ, Chen J, Deng XJ, Wan SP, Wang R, Wang Y, Guo HG (2018) Phylogeography and population genetic analyses reveal the speciation of the Tuber indicum complex. Fungal Genetics and Biology, 113, 14-23. |
| 18 | Sbrana C, Bagnoli G, Bedini S, Filippi C, Giovanetti M, Nuti MP (2000) Adhesion to hyphal matrix and antifungal activity of Pseudomonas strains isolated from Tuber borchii ascocarps. Canadian Journal of Microbiology, 46, 259-268. |
| 19 | Sbrana C, Agnolucci M, Bedini S, Lepera A, Toffanin A, Giovannetti M, Nuti MP (2002) Diversity of culturable bacterial populations associated to Tuber borchii ectomycorrhizas and their activity on T. borchii mycelial growth. FEMs Microbiology Letters, 211, 195-201. |
| 20 | Siqueira JF Jr, Fouad AF, Rôças IN (2012) Pyrosequencing as a tool for better understanding of human microbiomes. Journal of Oral Microbiology, 4, 10743. |
| 21 | Streiblová E, Gryndlerová H, Gryndler M (2012) Truffle brûlé: An efficient fungal life strategy. FEMS Microbiology Ecology, 80, 1-8. |
| 22 | Soudzilovskaia NA, Douma JC, Akhmetzhanova AA, Van Bodegom PM, Cornwell WK, Moens EJ, Treseder KK, Tibbett M, Wang YP, Cornelissen JHC (2015) Global patterns of plant root colonization intensity by mycorrhizal fungi explained by climate and soil chemistry. Global Ecology and Biogeography, 24, 371-382. |
| 23 | Splivallo R, Deveau A, Valdez N, Kirchhoff N, Frey-Klett P, Karlovsky P (2015) Bacteria associated with truffle fruiting bodies contribute to truffle aroma. Environmental Microbiology, 17, 2647-2660. |
| 24 | Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599. |
| 25 | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. |
| 26 | Wan SP, Liu PG (2014) Diversity of culturable bacteria associated with ascocarps of a Chinese white truffle. Plant Diversity and Resources, 36, 29-36. |
| 27 | Yan WR, Zhao TC, Xiao TB, Xiao M, Zhao ZX, Chen MC (2013) Applications of biocontrol bacterial in plant disease control. Genomics and Applied Biology, 32, 533-539. |
| [1] | 仝淼, 王欢, 张文双, 王超, 宋建潇. 重金属污染土壤中细菌抗生素抗性基因分布特征[J]. 生物多样性, 2025, 33(3): 24101-. |
| [2] | 魏诗雨, 宋天骄, 罗佳宜, 张燕, 赵子萱, 茹靖雯, 易华, 林雁冰. 秦岭火地塘针叶林土壤细菌群落的海拔分布格局[J]. 生物多样性, 2024, 32(9): 24180-. |
| [3] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [4] | 段晓敏, 李佳佳, 李靖宇, 李艳楠, 袁存霞, 王英娜, 刘建利. 腾格里沙漠东南缘藓结皮植物-土壤连续体不同粒径土壤微生物群落多样性[J]. 生物多样性, 2023, 31(9): 23131-. |
| [5] | 罗正明, 刘晋仙, 张变华, 周妍英, 郝爱华, 杨凯, 柴宝峰. 不同退化阶段亚高山草甸土壤原生生物群落多样性特征及驱动因素[J]. 生物多样性, 2023, 31(8): 23136-. |
| [6] | 张雅丽, 张丙昌, 赵康, 李凯凯, 刘燕晋. 毛乌素沙地不同类型生物结皮细菌群落差异及其驱动因子[J]. 生物多样性, 2023, 31(8): 23027-. |
| [7] | 吴春玲, 罗竹慧, 李意德, 许涵, 陈德祥, 丁琼. 热带山地雨林木本豆科和樟科植物叶内生细菌群落: 物种与功能群多样性及驱动因子[J]. 生物多样性, 2023, 31(8): 23146-. |
| [8] | 朱晓华, 高程, 王聪, 赵鹏. 尿素对土壤细菌与真菌多样性影响的研究进展[J]. 生物多样性, 2023, 31(6): 22636-. |
| [9] | 毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌. 毛竹扩张对杉木林土壤细菌群落的影响[J]. 生物多样性, 2023, 31(6): 22659-. |
| [10] | 赵雯, 王丹丹, 热依拉·木民, 黄开钏, 刘顺, 崔宝凯. 阿尔山地区兴安落叶松林土壤微生物群落结构[J]. 生物多样性, 2023, 31(2): 22258-. |
| [11] | 杨预展, 余建平, 钱海源, 陈小南, 陈声文, 袁志林. 钱江源国家公园体制试点区水稻田土壤微生物群落的格局及其驱动机制[J]. 生物多样性, 2023, 31(2): 22392-. |
| [12] | 杜芳, 荣晓莹, 徐鹏, 尹本丰, 张元明. 降水对古尔班通古特沙漠细菌群落多样性和构建过程的影响[J]. 生物多样性, 2023, 31(2): 22492-. |
| [13] | 刘金花, 李风, 田桃, 肖海峰. 土壤细菌和线虫对热带雨林优势植物凋落物特性和多样性的响应[J]. 生物多样性, 2023, 31(11): 23276-. |
| [14] | 夏凡, 杨婧, 李建, 史洋, 盖立新, 黄文华, 张经纬, 杨南, 高福利, 韩莹莹, 鲍伟东. 北京地区四个豹猫亚种群肠道菌群的组成[J]. 生物多样性, 2022, 30(9): 22103-. |
| [15] | 张旋, 杜薇, 徐颖, 王永龙. 包头市半干旱型森林公园土壤细菌多样性与功能[J]. 生物多样性, 2022, 30(7): 22245-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn