



生物多样性 ›› 2022, Vol. 30 ›› Issue (9): 22103. DOI: 10.17520/biods.2022103 cstr: 32101.14.biods.2022103
夏凡1, 杨婧1, 李建2, 史洋3, 盖立新4, 黄文华5, 张经纬4, 杨南6, 高福利1, 韩莹莹1, 鲍伟东1,*( )
)
收稿日期:2022-03-09
接受日期:2022-07-11
出版日期:2022-09-20
发布日期:2022-09-09
通讯作者:
鲍伟东
作者简介:* E-mail: wdbao@bjfu.edu.cn基金资助:
Fan Xia1, Jing Yang1, Jian Li2, Yang Shi3, Lixin Gai4, Wenhua Huang5, Jingwei Zhang4, Nan Yang6, Fuli Gao1, Yingying Han1, Weidong Bao1,*( )
)
Received:2022-03-09
Accepted:2022-07-11
Online:2022-09-20
Published:2022-09-09
Contact:
Weidong Bao
About author:First author contact:# Co-first author
摘要:
野生动物的肠道微生物组成受食物资源和遗传属性的影响较大, 为了解北京地区小型猫科动物肠道菌群组成特点及其影响因素, 本研究通过扩增细菌16S rRNA的V3‒V4高变区进行高通量测序, 对云蒙山、云峰山、松山、百花山4个区域的豹猫(Prionailurus bengalensis)亚种群进行肠道菌群组成分析。结果表明, 在门水平上, 豹猫的肠道优势菌群主要由厚壁菌门(相对多度52.40%)、变形菌门(25.18%)、放线菌门(9.07%)、拟杆菌门(8.17%)和梭杆菌门(4.74%)组成。在属水平上, 相对多度最高的前5个属为假单胞菌属(Pseudomonas, 13.37%)、布劳特氏菌属(Blautia, 11.20%)、梭菌属(Clostridium_sensu_stricto_1, 9.10%)、消化梭菌属(Peptoclostridium, 8.62%)、乳杆菌属(Lactobacillus, 6.08%), 共约占总多度的50%。各区域豹猫亚种群的肠道菌群β多样性不存在显著差异, 松山区域的ACE指数和Chao 1指数与云蒙山和云峰山存在显著差异。鉴于松山亚种群的遗传结构与其他区域有所不同, 而4个区域的气候类型和豹猫食物构成相似性高, 推测该亚种群的肠道菌群主要受遗传结构分化的影响, 对此还需进一步研究。
夏凡, 杨婧, 李建, 史洋, 盖立新, 黄文华, 张经纬, 杨南, 高福利, 韩莹莹, 鲍伟东 (2022) 北京地区四个豹猫亚种群肠道菌群的组成. 生物多样性, 30, 22103. DOI: 10.17520/biods.2022103.
Fan Xia, Jing Yang, Jian Li, Yang Shi, Lixin Gai, Wenhua Huang, Jingwei Zhang, Nan Yang, Fuli Gao, Yingying Han, Weidong Bao (2022) Gut bacterial composition of four leopard cat subpopulations in Beijing. Biodiversity Science, 30, 22103. DOI: 10.17520/biods.2022103.
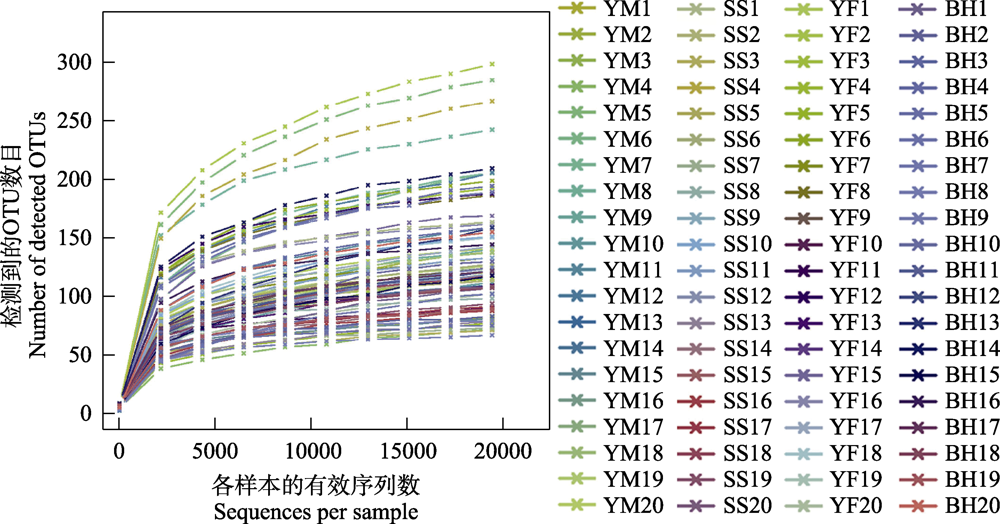
图2 北京地区豹猫肠道菌群分析粪样序列稀释曲线。图中每条曲线代表1个样本, 用不同颜色标记。
Fig. 2 Rarefaction curves of all samples for analysis of gut bacteria of leopard cat in Beijing. Each curve represents a sample and is marked with different color.
| 亚种群 Subpopulation | Q20碱基百分比 Q20 (%) | Q30碱基百分比 Q30 (%) | GC碱基百分比 GC (%) | 测序深度 Good’s coverage |
|---|---|---|---|---|
| 松山 SS | 94.32 | 91.52 | 53.749 | 0.999 |
| 云蒙山 YM | 92.58 | 89.07 | 54.014 | 0.999 |
| 云峰山 YF | 92.91 | 89.63 | 52.819 | 0.999 |
| 百花山 BH | 91.69 | 88.12 | 53.125 | 0.999 |
表1 北京地区豹猫肠道菌群分析测序质量参数(平均值)
Table 1 Sequencing quality parameters in gut bacterial analysis of leopard cat in Beijing (average value). SS, Songshan; YM, Yunmengshan; YF, Yunfengshan; BH, Baihuashan.
| 亚种群 Subpopulation | Q20碱基百分比 Q20 (%) | Q30碱基百分比 Q30 (%) | GC碱基百分比 GC (%) | 测序深度 Good’s coverage |
|---|---|---|---|---|
| 松山 SS | 94.32 | 91.52 | 53.749 | 0.999 |
| 云蒙山 YM | 92.58 | 89.07 | 54.014 | 0.999 |
| 云峰山 YF | 92.91 | 89.63 | 52.819 | 0.999 |
| 百花山 BH | 91.69 | 88.12 | 53.125 | 0.999 |
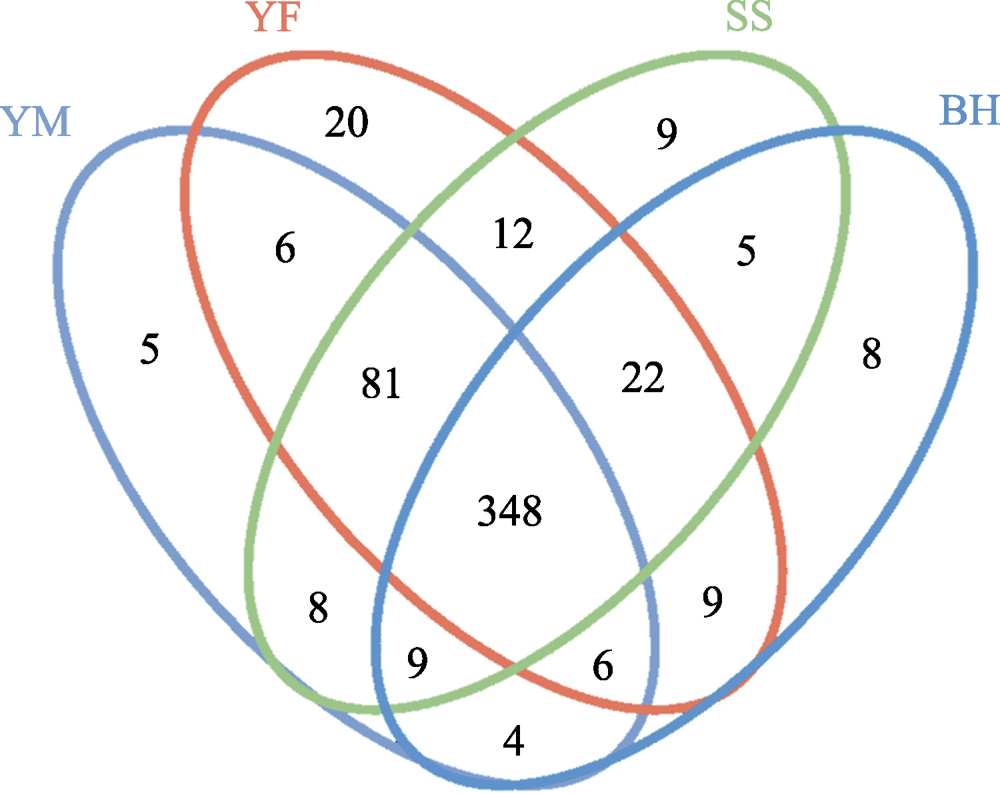
图3 北京四个地区豹猫肠道菌群OTU的Venn图。图中4种颜色的椭圆表示4个采样区域。YM: 云蒙山, YF: 云峰山, SS: 松山, BH: 百花山。数字分别代表4个区域特有或共有的OTU数目。
Fig. 3 Venn diagram of the OTU numbers of gut bacteria of leopard cat at four sampling areas in Beijing. The ellipses of four colors in the figure represent four sampling areas. YM, Yunmengshan; YF, Yunfengshan; SS, Songshan; BH, Baihuashan. The numbers represent the unique or shared number of OTUs of the four sampling areas.
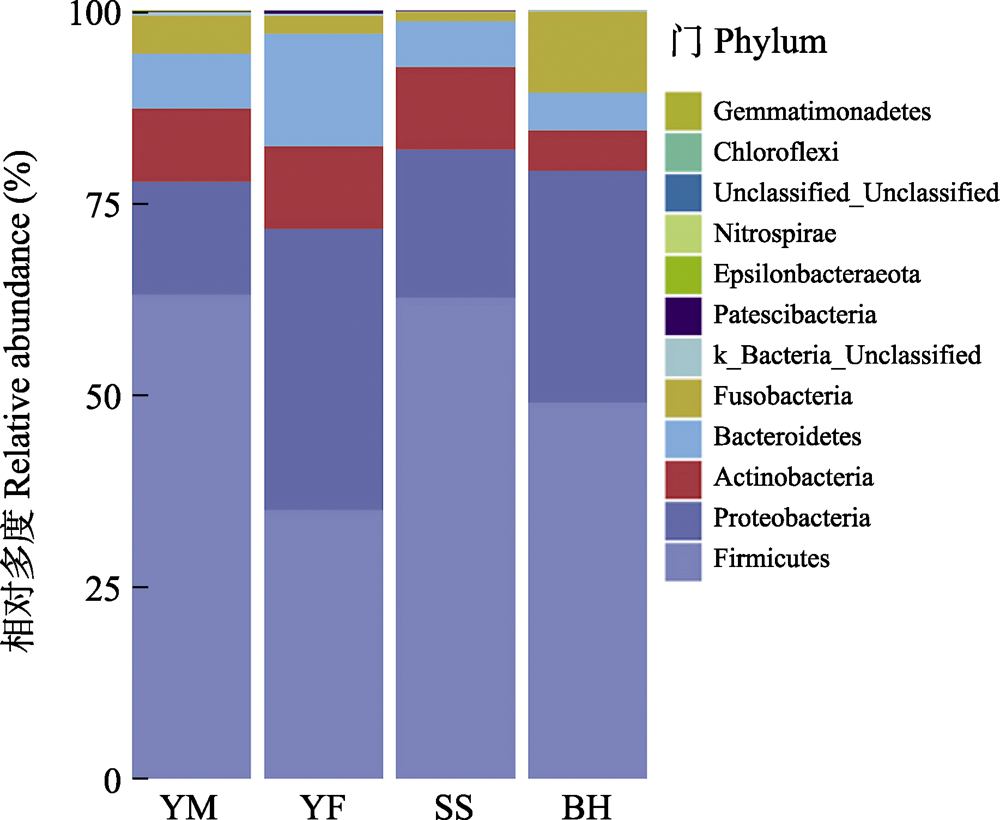
图4 北京四个地区豹猫肠道菌群门水平的相对多度。不同颜色代表细菌门水平分类。四个采样区域全称见图3。
Fig. 4 The relative abundance of gut bacteria of leopard cat at four sampling areas in Beijing at the phylum level. The abscissa is the four sampling areas, and different colors represent the classification of bacteria at the phylum level. Full names of the four sampling areas see Fig. 3.
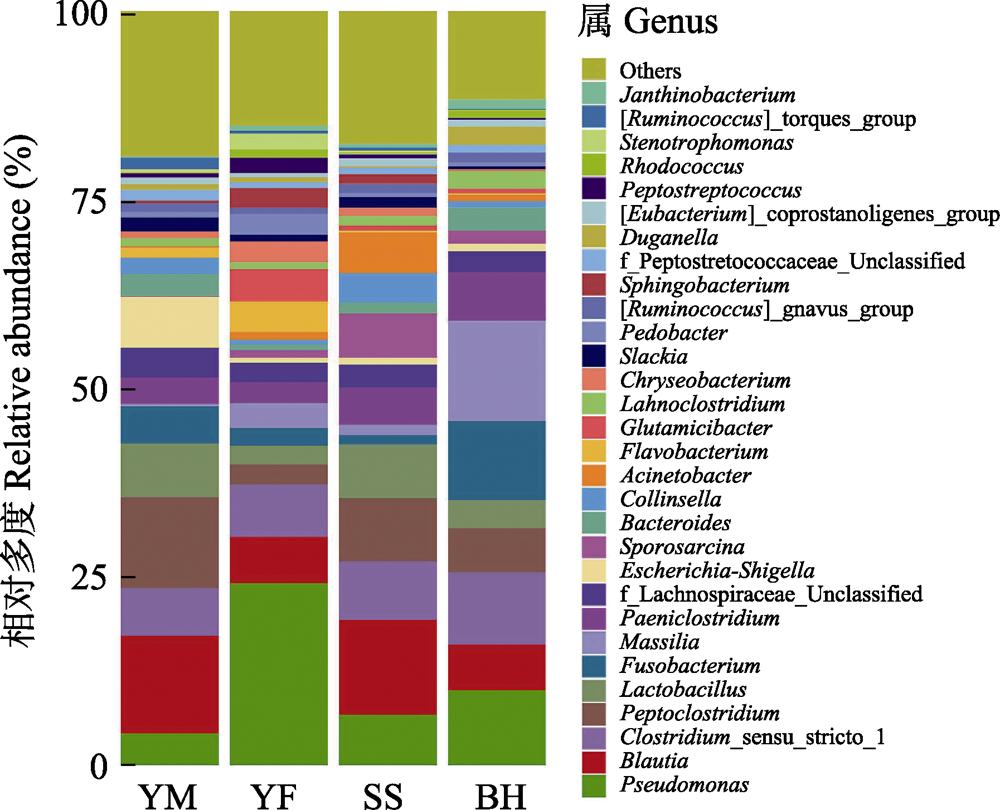
图5 北京四个地区豹猫肠道菌群属水平的相对多度。不同颜色代表细菌属。四个采样区域全称见图3。
Fig. 5 The relative abundance of gut bacteria of leopard cat at four sampling areas in Beijing at genus level. Different colors represent the classification of bacteria at the genus level. Full names of the four sampling areas see Fig. 3.
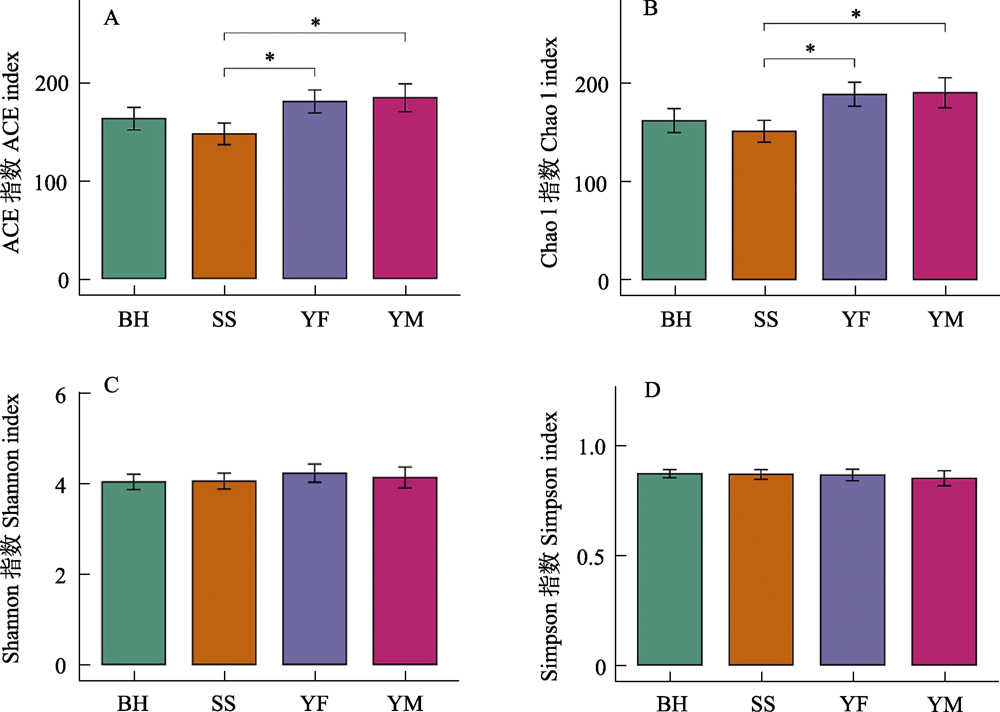
图6 北京四个地区豹猫肠道菌群α多样性。* P < 0.05。四个采样区域全称见图3。
Fig. 6 Differences in α-diversity of gut bacteria of leopard cat at four sampling areas in Beijing. * P < 0.05. Full names of the four sampling areas see Fig. 3.
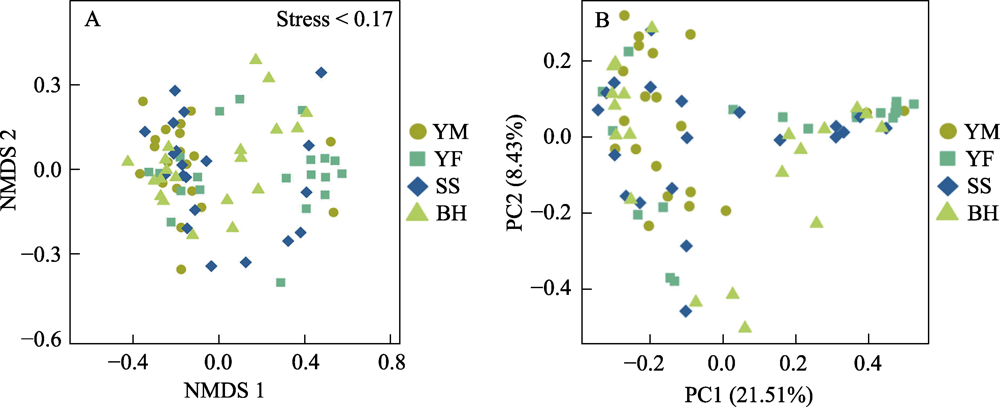
图7 基于非度量多维尺度NMDS和主坐标PCoA对北京豹猫种群肠道菌群的距离分析。A图为NMDS分析, 每个点表示1个样本, 点与点之间的距离表示差异程度; B图为PCoA分析, 样点间的距离越近, 相似度越高。四个采样区域全称见图3。
Fig. 7 Analysis of gut bacteria distance based on non-metric multidimensional scaling (NMDS) and principal coordinates analysis (PCoA) of leopard cat at four sampling areas in Beijing. In NMDS analysis, each point represents one sample, and the distance between points represents difference degree. In PCoA analysis, the closer the distance between the sample points is, the higher the similarity is. Full names of the four sampling areas see Fig. 3.
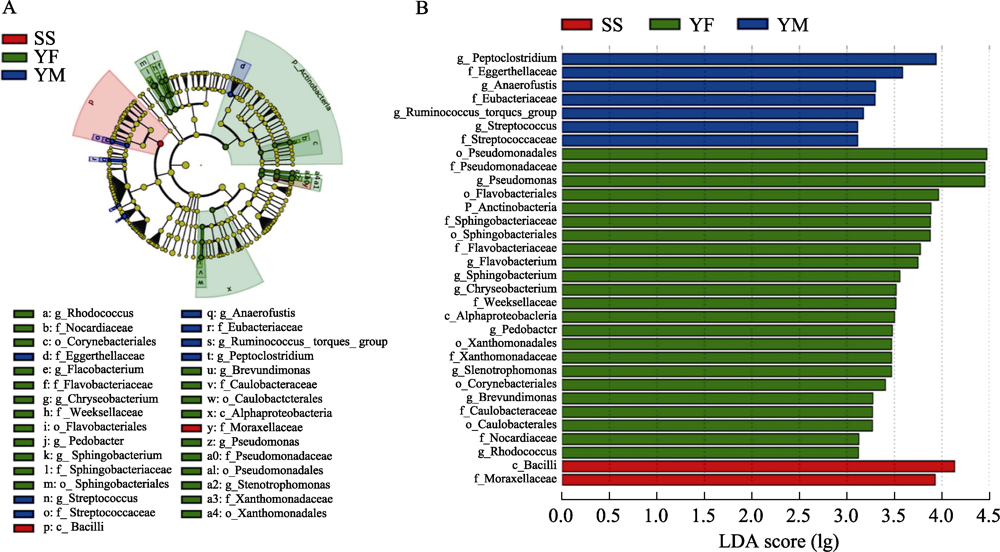
图8 北京3个豹猫亚种群之间肠道菌群组成的线性判别分析效应量(LEfSe)。A图为3个豹猫亚种群肠道菌群的聚类进化树, B图为不同分类级别下各类细菌的线性判别分析(LDA)评分。不同颜色表示不同采样区域, 红色代表松山, 绿色代表云峰山, 蓝色代表云蒙山。
Fig. 8 LEfSe analysis of gut bacterial composition among three leopard cat subpopulations in Beijing. Figure A is the cluster evolutionary tree of gut bacteria of three leopard cat subpopulations, and figure B shows the LDA scores of various bacteria under different classification levels. Different colors indicate sampling areas with red for Songshan, green for Yunfengshan, and blue for Yunmengshan.
| [1] |
Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M, Gillis M, Wilson BA, Nelson KE, White BA, Leigh SR (2013) Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. The ISME Journal, 7, 1344-1353.
DOI URL |
| [2] |
An C, Okamoto Y, Xu SY, Eo KY, Kimura J, Yamamoto N (2017) Comparison of fecal microbiota of three captive carnivore species inhabiting Korea. The Journal of Veterinary Medical Science, 79, 542-546.
DOI URL |
| [3] |
Arboleya S, Watkins C, Stanton C, Ross RP (2016) Gut bifidobacteria populations in human health and aging. Frontiers in Microbiology, 7, 1204.
DOI PMID |
| [4] | Bao WD, Li XJ, Shi Y (2005) Comparative analysis of food habits in carnivores from three areas of Beijing. Zoological Research, 26, 118-122. (in Chinese with English abstract) |
| [鲍伟东, 李晓京, 史阳 (2005) 北京市三个区域食肉类动物食性的比较分析. 动物学研究, 26, 118-122.] | |
| [5] | Chen W, Gao W, Fu BQ (2002) Mammal Records of Beijing. Beijing Press, Beijing. (in Chinese) |
| [陈卫, 高武, 傅必谦 (2002) 北京兽类志. 北京出版社, 北京.] | |
| [6] | Clarke K, Gorley RN (2006) Primer v6: User Manual/Tutorial. Plymouth Marine Laboratory, Plymouth. |
| [7] |
Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, Gougis S, Rizkalla S, Batto JM, Renault P, Doré J, Zucker JD, Clément K, Ehrlich SD (2013) Dietary intervention impact on gut microbial gene richness. Nature, 500, 585-588.
DOI URL |
| [8] |
Doré J, Blottière H (2015) The influence of diet on the gut microbiota and its consequences for health. Current Opinion in Biotechnology, 32, 195-199.
DOI PMID |
| [9] | Du LH, Wang XP, Chen JQ, Liu GL, Wu JG, Zhang ZX (2012) Comprehensive Scientific Investigation Report of Beijing Songshan Nature Reserve. China Forestry Publishing House, Beijing. (in Chinese) |
| [杜连海, 王小平, 陈峻崎, 刘桂林, 吴记贵, 张志翔 (2012) 北京松山自然保护区综合科学考察报告. 中国林业出版社, 北京.] | |
| [10] | Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science, 308, 1635-1638. |
| [11] |
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27, 2194-2200.
DOI PMID |
| [12] | Eo SH, Ko BJ, Lee BJ, Seomun H, Kim S, Kim MJ, Kim JH, An J (2016) A set of microsatellite markers for population genetics of leopard cat (Prionailurus bengalensis) and cross-species amplification in other felids. Biochemical Systematics & Ecology, 66, 196-200. |
| [13] | Fan YQ, Yang J, Zhang HL, Jiang J, Jiang WJ, Tang SP, Bao WD (2020) Food composition of medium sized carnivores in Beijing Songshan Nature Reserve. Journal of Biology, 37(1), 59-62. (in Chinese with English abstract) |
| [范雅倩, 杨婧, 张洪亮, 蒋健, 蒋万杰, 唐书培, 鲍伟东 (2020) 北京松山自然保护区中型捕食动物的食物构成分析. 生物学杂志, 37(1), 59-62.] | |
| [14] |
Godoy-Vitorino F, Goldfarb KC, Karaoz U, Leal S, Garcia-Amado MA, Hugenholtz P, Tringe SG, Brodie EL, Dominguez-Bello MG (2012) Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. The ISME Journal, 6, 531-541.
DOI URL |
| [15] | Han SY, Guan Y, Dou HL, Yang HT, Yao M, Ge JP, Feng LM (2019) Comparison of the fecal microbiota of two free-ranging Chinese subspecies of the leopard (Panthera pardus) using high-throughput sequencing. PeerJ, 7, e6684. |
| [16] |
Hooper LV, Littman DR, MacPherson AJ (2012) Interactions between the microbiota and the immune system. Science, 336, 1268-1273.
DOI PMID |
| [17] |
Kaoutari AE, Armougom F, Gordon JI, Raoult D, Henrissat B (2013) The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nature Reviews Microbiology, 11, 497-504.
DOI PMID |
| [18] | Ko BJ, An J, Seomun H, Lee MY, Eo SH (2018) Microsatellite DNA analysis reveals lower than expected genetic diversity in the threatened leopard cat (Prionailurus bengalensis) in South Korea. Genes & Genomics, 40, 521-530. |
| [19] | Kong FL, Zhao JC, Han SS, Zeng B, Yang JD, Si XH, Yang BQ, Yang MY, Xu HL, Li Y (2014) Characterization of the gut microbiota in the red panda (Ailurus fulgens). PLoS ONE, 9, e87885. |
| [20] |
Konopka A (2009) What is microbial community ecology? The ISME Journal, 3, 1223-1230.
DOI URL |
| [21] |
Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U (2011) Genotype is a stronger determinant than sex of the mouse gut microbiota. Microbial Ecology, 61, 423-428.
DOI PMID |
| [22] |
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI (2008) Worlds within worlds: Evolution of the vertebrate gut microbiota. Nature Reviews Microbiology, 6, 776-788.
DOI PMID |
| [23] | Luan XF, Li DQ, Li GL (2011) Study on Biodiversity and Protection of Beijing Yunfengshan Nature Reserve. China Land Press, Beijing. (in Chinese) |
| [栾晓峰, 李迪强, 李广良 (2011) 北京云峰山自然保护区生物多样性及保护研究. 中国大地出版社, 北京.] | |
| [24] | Masuoka H, Shimada K, Kiyosue-Yasuda T, Kiyosue M, Oishi Y, Kimura S, Ohashi Y, Fujisawa T, Hotta K, Yamada A, Hirayama K (2017) Transition of the intestinal microbiota of cats with age. PLoS ONE, 12, e0181739. |
| [25] |
Mittal P, Saxena R, Gupta A, Mahajan S, Sharma VK (2020) The gene catalog and comparative analysis of gut microbiome of big cats provide new insights on Panthera species. Frontiers in Microbiology, 11, 1012.
DOI PMID |
| [26] | Mozejko-Ciesielska J, Pokoj T, Ciesielski S (2018) Transcriptome remodeling of Pseudomonas putida KT 2440 during mcl-PHAs synthesis: Effect of different carbon sources and response to nitrogen stress. Journal of Industrial Microbiology & Biotechnology, 45, 433-446. |
| [27] |
Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI (2011) Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science, 332, 970-974.
DOI PMID |
| [28] | Ning Y (2020) Study on Amur Tiger Dispersal, Inbreeding and Gut Microbiota Based on DNA Analysis. PhD dissertation, Northeast Forestry University, Harbin. (in Chinese with English abstract) |
| [宁瑶 (2020) 基于DNA分析的东北虎扩散状况、近交以及肠道菌群的研究. 博士学位论文, 东北林业大学, 哈尔滨.] | |
| [29] |
Phillips CD, Phelan G, Dowd SE, McDonough MM, Ferguson AW, Hanson JD, Siles L, Ordonez-Garza N, San Francisco M, Baker RJ (2012) Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Molecular Ecology, 21, 2617-2627.
DOI PMID |
| [30] |
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: A versatile open source tool for metagenomics. PeerJ, 4, e2584.
DOI URL |
| [31] |
Saka T, Nishita Y, Masuda R (2018) Low genetic variation in the MHC class II DRB gene and MHC-linked microsatellites in endangered island populations of the leopard cat (Prionailurus bengalensis) in Japan. Immunogenetics, 70, 115-124.
DOI URL |
| [32] |
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60.
DOI URL |
| [33] |
Sheppard SK, Harwood JD (2005) Advances in molecular ecology: Tracking trophic links through predator prey food-webs. Functional Ecology, 19, 751-762.
DOI URL |
| [34] |
Sicuro FL, Oliveira LFB (2015) Variations in leopard cat (Prionailurus bengalensis) skull morphology and body size: Sexual and geographic influences. PeerJ, 3, e1309.
DOI URL |
| [35] |
Sinha T, Vich VA, Garmaeva S, Jankipersadsing SA, Imhann F, Collij V, Bonder MJ, Jiang XF, Gurry T, Alm EJ, D’Amato M, Weersma RK, Scherjon S, Wijmenga C, Fu JY, Kurilshikov A, Zhernakova A (2019) Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles. Gut Microbes, 10, 358-366.
DOI PMID |
| [36] |
Sommer F, Bäckhed F (2013) The gut microbiota—Masters of host development and physiology. Nature Reviews Microbiology, 11, 227-238.
DOI PMID |
| [37] |
Sun BH, Wang X, Bernstein S, Huffman MA, Xia DP, Gu ZY, Chen R, Sheeran LK, Wagner RS, Li JH (2016) Marked variation between winter and spring gut microbiota in free-ranging Tibetan macaques (Macaca thibetana). Scientific Reports, 6, 26035.
DOI URL |
| [38] |
Tamada T, Siriaroonrat B, Subramaniam V, Hamachi M, Lin LK, Oshida T, Rerkamnuaychoke W, Masuda R (2008) Molecular diversity and phylogeography of the Asian leopard cat, Felis bengalensis, inferred from mitochondrial and Y-Chromosomal DNA sequences. Zoological Science, 25, 154-163.
DOI URL |
| [39] |
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI (2009) A core gut microbiome in obese and lean twins. Nature, 457, 480-484.
DOI URL |
| [40] |
Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M (2018) Glycan utilization and cross-feeding activities by bifidobacteria. Trends in Microbiology, 26, 339-350.
DOI PMID |
| [41] | Wang Y, Sheng HF, He Y, Wu JY, Jiang YX, Tam NFY, Zhou HW (2012) Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of Illumina tags. Applied & Environmental Microbiology, 78, 8264-8271. |
| [42] |
Wasimuddin, Menke S, Melzheimer J, Thalwitzer S, Heinrich S, Wachter B, Sommer S (2017) Gut microbiomes of free-ranging and captive Namibian cheetahs: Diversity, putative functions and occurrence of potential pathogens. Molecular Ecology, 26, 5515-5527.
DOI PMID |
| [43] |
Wei FW, Wang X, Wu Q (2015) The giant panda gut microbiome. Trends in Microbiology, 23, 450-452.
DOI PMID |
| [44] | Xu XM, Zhang ZB (2021) Sex- and age-specific variation of gut microbiota in Brandt’s voles. PeerJ, 9, e11434. |
| [45] |
Zhang HH, Liu GS, Chen L, Sha WL (2015) Composition and diversity of the bacterial community in snow leopard (Uncia uncia) distal gut. Annals of Microbiology, 65, 703-711.
DOI URL |
| [46] |
Zhang TH, Yang Y, Liang Y, Jiao X, Zhao CH (2018) Beneficial effect of intestinal fermentation of natural polysaccharides. Nutrients, 10, 1055.
DOI URL |
| [47] | Zhao B (2005) Studies on Plant Diversity in Mountain Areas in Beijing. PhD dissertation, Beijing Forestry University, Beijing. (in Chinese with English abstract) |
| [赵勃 (2005) 北京山区植物多样性研究. 博士学位论文, 北京林业大学, 北京.] |
| [1] | 刘志禹, 吉鑫, 隋国辉, 杨定, 李轩昆. 北京首都国际机场野牛草与杂草草坪无脊椎动物多样性[J]. 生物多样性, 2025, 33(4): 24456-. |
| [2] | 韩思成, 陆道炜, 韩宇辰, 栗若寒, 杨晶, 孙戈, 杨陆, 钱俊伟, 方翔, 罗述金. 北京近郊浅山地区的野生豹猫分布及环境影响因素[J]. 生物多样性, 2024, 32(8): 24138-. |
| [3] | 郝操, 吴东辉, 莫凌梓, 徐国良. 越冬动物肠道微生物多样性及功能研究进展[J]. 生物多样性, 2024, 32(3): 23407-. |
| [4] | 罗正明, 刘晋仙, 张变华, 周妍英, 郝爱华, 杨凯, 柴宝峰. 不同退化阶段亚高山草甸土壤原生生物群落多样性特征及驱动因素[J]. 生物多样性, 2023, 31(8): 23136-. |
| [5] | 毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌. 毛竹扩张对杉木林土壤细菌群落的影响[J]. 生物多样性, 2023, 31(6): 22659-. |
| [6] | 殷鲁秦, 王成, 韩文静. 基于取食行为探究北京居民区鸟类的食源特征及多样性[J]. 生物多样性, 2023, 31(5): 22473-. |
| [7] | 赵也茜, 张家语, 李子涵, 解秦米佳, 邓歆, 王楠. 北京城市鸟类夜栖时对本土和外来植物的利用[J]. 生物多样性, 2023, 31(3): 22399-. |
| [8] | 赵雯, 王丹丹, 热依拉·木民, 黄开钏, 刘顺, 崔宝凯. 阿尔山地区兴安落叶松林土壤微生物群落结构[J]. 生物多样性, 2023, 31(2): 22258-. |
| [9] | 邓雪琴, 刘统, 刘天时, 徐恺, 姚松, 黄小群, 肖治术. 河南内乡宝天曼国家级自然保护区豹猫及其潜在猎物之间日活动节律的季节性[J]. 生物多样性, 2022, 30(9): 22263-. |
| [10] | 崔夏, 刘全儒, 吴超然, 何宇飞, 马金双. 京津冀外来入侵植物[J]. 生物多样性, 2022, 30(8): 21497-. |
| [11] | 林秦文, 吴超然, 崔夏, 马金双. 北京维管植物三十年之变化[J]. 生物多样性, 2022, 30(6): 22107-. |
| [12] | 肖翠, 刘冰, 吴超然, 马金双, 叶建飞, 夏晓飞, 林秦文. 北京维管植物编目和分布数据集[J]. 生物多样性, 2022, 30(6): 22064-. |
| [13] | 孙翌昕, 李英滨, 李玉辉, 李冰, 杜晓芳, 李琪. 高通量测序技术在线虫多样性研究中的应用[J]. 生物多样性, 2022, 30(12): 22266-. |
| [14] | 高程, 郭良栋. 微生物物种多样性、群落构建与功能性状研究进展[J]. 生物多样性, 2022, 30(10): 22429-. |
| [15] | 夏呈强, 李毅, 党延茹, 察倩倩, 贺晓艳, 秦启龙. 中印度洋与南海西部表层海水细菌多样性[J]. 生物多样性, 2022, 30(1): 21407-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn