



生物多样性 ›› 2025, Vol. 33 ›› Issue (3): 24101. DOI: 10.17520/biods.2024101 cstr: 32101.14.biods.2024101
• 研究报告:微生物多样性 • 下一篇
仝淼( ), 王欢(
), 王欢( ), 张文双, 王超(
), 张文双, 王超( ), 宋建潇*(
), 宋建潇*( )(
)( )
)
收稿日期:2024-03-17
接受日期:2024-09-17
出版日期:2025-03-20
发布日期:2024-11-29
通讯作者:
*E-mail: sjx@nwpu.edu.cn
基金资助:
Tong Miao( ), Wang Huan(
), Wang Huan( ), Zhang Wenshuang, Wang Chao(
), Zhang Wenshuang, Wang Chao( ), Song Jianxiao*(
), Song Jianxiao*( )(
)( )
)
Received:2024-03-17
Accepted:2024-09-17
Online:2025-03-20
Published:2024-11-29
Contact:
*E-mail: sjx@nwpu.edu.cn
Supported by:摘要:
重金属作为常见的土壤污染物, 可通过共选择作用诱导土壤微生物群落的抗生素抗性基因(antibiotic resistance genes, ARGs)的产生和扩散, 并促进ARGs在环境中持久存在。本研究建立了不同铜(Cu)浓度和镉(Cd)浓度的土壤微宇宙实验, 基于高通量测序和实时荧光定量PCR技术分析不同重金属浓度和复合污染条件下ARGs的分布特征以及土壤细菌群落对不同浓度Cu、Cd污染的响应, 旨在解析ARGs与土壤细菌群落在重金属污染土壤中的分布特征。此外, 通过探究影响ARGs变化的关键环境因子, 以期找出有效减缓环境中抗生素抗性的传播和扩散现状的途径。研究表明: 高浓度Cu (400 mg/kg)与Cd (1 mg/kg和5 mg/kg)的复合污染显著(P < 0.05)提高了土壤中抗生素抗性基因sul1、intl1、blaVIM的相对丰度, 抗生素抗性基因tetX、tetG对Cd响应最敏感, 低浓度Cd (1 mg/kg)可明显提高tetX、tetG的丰度。此外, Cu、Cd显著(P < 0.05)改变了土壤细菌群落结构, 并且使细菌群落对Cu的响应更加明显。相关性分析和网络分析表明, sul1、tetX、tetM02、blaVIM广泛分布在多个细菌门, 而ARGs的变化与细菌群落(如链霉菌属Streptomyces、慢生根瘤菌属Bradyrhizobium、BIrii41属(Polyangiales)、海无柄孢囊黏细菌属Haliangium等)的变化密切相关, 推测这些细菌可能是携带ARGs的主要宿主。
仝淼, 王欢, 张文双, 王超, 宋建潇 (2025) 重金属污染土壤中细菌抗生素抗性基因分布特征. 生物多样性, 33, 24101. DOI: 10.17520/biods.2024101.
Tong Miao, Wang Huan, Zhang Wenshuang, Wang Chao, Song Jianxiao (2025) Distribution characteristics of antibiotic resistance genes in soil bacterial communities exposed to heavy metal pollution. Biodiversity Science, 33, 24101. DOI: 10.17520/biods.2024101.
| 处理名称 Treatment | Cu (mg/kg) | Cd (mg/kg) |
|---|---|---|
| CK | 0 | 0 |
| Cu200 | 200 | 0 |
| Cu400 | 400 | 0 |
| Cd1 | 0 | 1 |
| Cd5 | 0 | 5 |
| Cu200 + Cd1 | 200 | 1 |
| Cu200 + Cd5 | 200 | 5 |
| Cu400 + Cd1 | 400 | 1 |
| Cu400 + Cd5 | 400 | 5 |
表1 本研究中重金属浓度设置
Table 1 Concentrations of heavy metals used in this study
| 处理名称 Treatment | Cu (mg/kg) | Cd (mg/kg) |
|---|---|---|
| CK | 0 | 0 |
| Cu200 | 200 | 0 |
| Cu400 | 400 | 0 |
| Cd1 | 0 | 1 |
| Cd5 | 0 | 5 |
| Cu200 + Cd1 | 200 | 1 |
| Cu200 + Cd5 | 200 | 5 |
| Cu400 + Cd1 | 400 | 1 |
| Cu400 + Cd5 | 400 | 5 |
| 目标基因 Target genes | 引物(5°-3°) Primers (5°-3°) | 片段长度 Fragment length (bp) | 退火温度 Annealing temperature (℃) | 参考文献 References |
|---|---|---|---|---|
| sul1 | F: CGGCGTGGGCTACCTGAACG R: GCCGATCGCGTGAAGTTCCG | 433 | 60 | Frank et al, |
| tetG | F: GCAGAGCAGGTCGCTGG R: CCYGCAAGAGAAGCCAGAAG | 134 | 54 | Aminov et al, |
| tetM02 | F: ACAGAAAGCTTATTATATAAC R: TGGCGTGTCTATGATGTTCAC | 171 | 52 | Aminov et al, |
| blaVIM | F: CAGATTGCCGATGGTGTTTGG R: AGGTGGGCCATTCAGCCAGA | 523 | 55 | Ktari et al, |
| tetX | F: CAATAATTGGTGGTGGACCC R: TTCTTACCTTGGACATCCCG | 468 | 55 | Ng et al, |
| intl1 | F: CTGGATTTCGATCACGGCACG R: ACATGCGTGTAAATCATCGTCG | 473 | 60 | Frank et al, |
| 16S V3 | F: CCTACGGGAGGCAGCAG R: ATTACCGCGGCTGCTGG | 193 | 55 | Aminov et al, |
| 16S V3-V4 | F: ACTCCTACGGGAGGCAGCA R: GGACTACHVGGGTWTCTAAT | 480 | 53 | Claesson et al, |
表2 引物序列及扩增条件信息
Table 2 Primer sequences and amplification conditions
| 目标基因 Target genes | 引物(5°-3°) Primers (5°-3°) | 片段长度 Fragment length (bp) | 退火温度 Annealing temperature (℃) | 参考文献 References |
|---|---|---|---|---|
| sul1 | F: CGGCGTGGGCTACCTGAACG R: GCCGATCGCGTGAAGTTCCG | 433 | 60 | Frank et al, |
| tetG | F: GCAGAGCAGGTCGCTGG R: CCYGCAAGAGAAGCCAGAAG | 134 | 54 | Aminov et al, |
| tetM02 | F: ACAGAAAGCTTATTATATAAC R: TGGCGTGTCTATGATGTTCAC | 171 | 52 | Aminov et al, |
| blaVIM | F: CAGATTGCCGATGGTGTTTGG R: AGGTGGGCCATTCAGCCAGA | 523 | 55 | Ktari et al, |
| tetX | F: CAATAATTGGTGGTGGACCC R: TTCTTACCTTGGACATCCCG | 468 | 55 | Ng et al, |
| intl1 | F: CTGGATTTCGATCACGGCACG R: ACATGCGTGTAAATCATCGTCG | 473 | 60 | Frank et al, |
| 16S V3 | F: CCTACGGGAGGCAGCAG R: ATTACCGCGGCTGCTGG | 193 | 55 | Aminov et al, |
| 16S V3-V4 | F: ACTCCTACGGGAGGCAGCA R: GGACTACHVGGGTWTCTAAT | 480 | 53 | Claesson et al, |
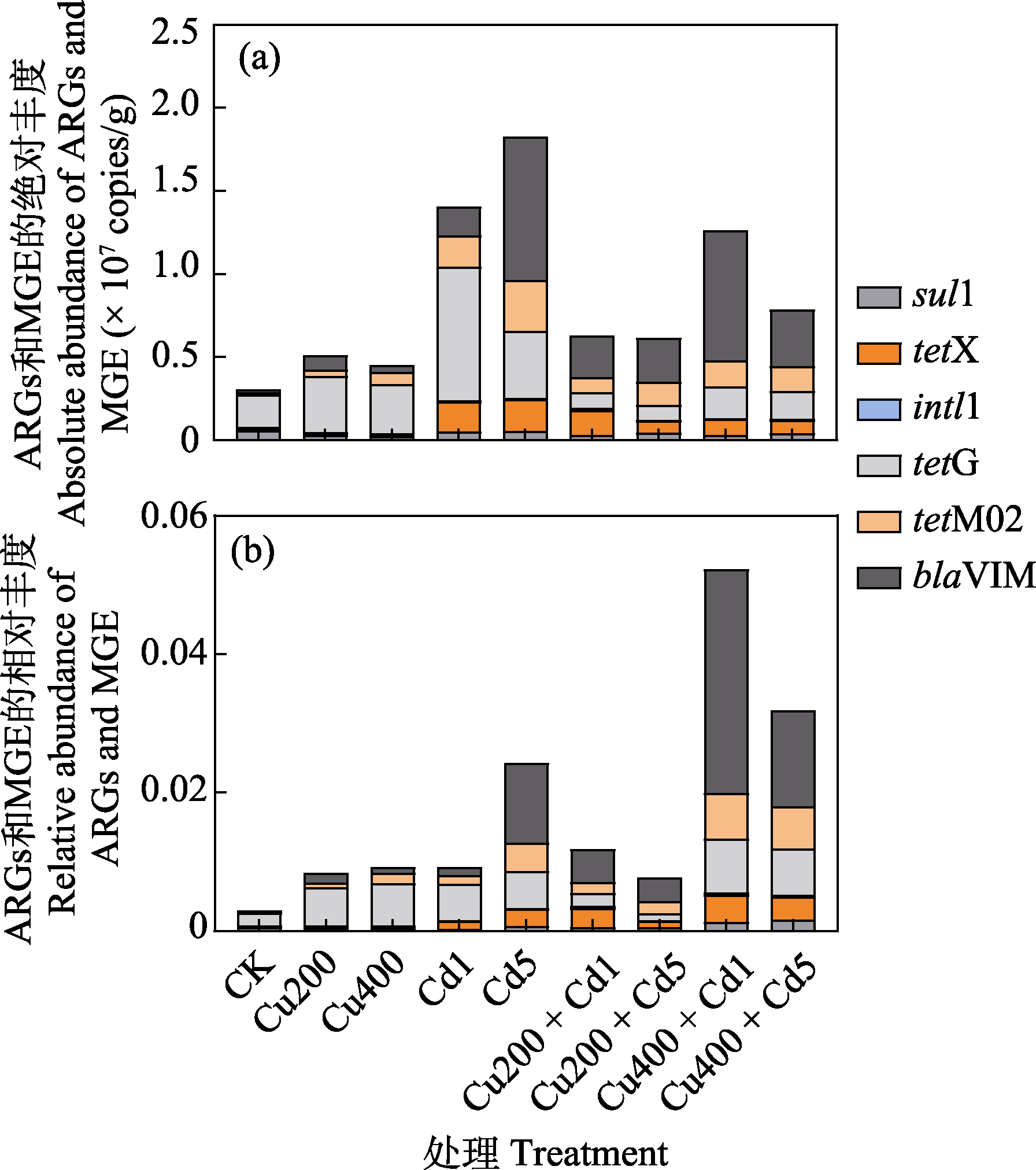
图1 不同浓度Cu和Cd污染土壤抗生素抗性基因(ARGs)和移动基因元件(MGE)绝对丰度(a)与相对丰度(b)。各处理全称见表1。Sul1、tetG、tetM02、blaVIM、tetX和intl1为ARGs和MGE。
Fig. 1 The absolute and relative abundance of antibiotic resistance genes (ARGs) and mobile genetic element (MGE) in different concentration of Cu and Cd polluted soils. The full names of all treatments are listed in Table 1. Sul1, tetG, tetM02, blaVIM, tetX, and intl1 are ARGs and MGE.
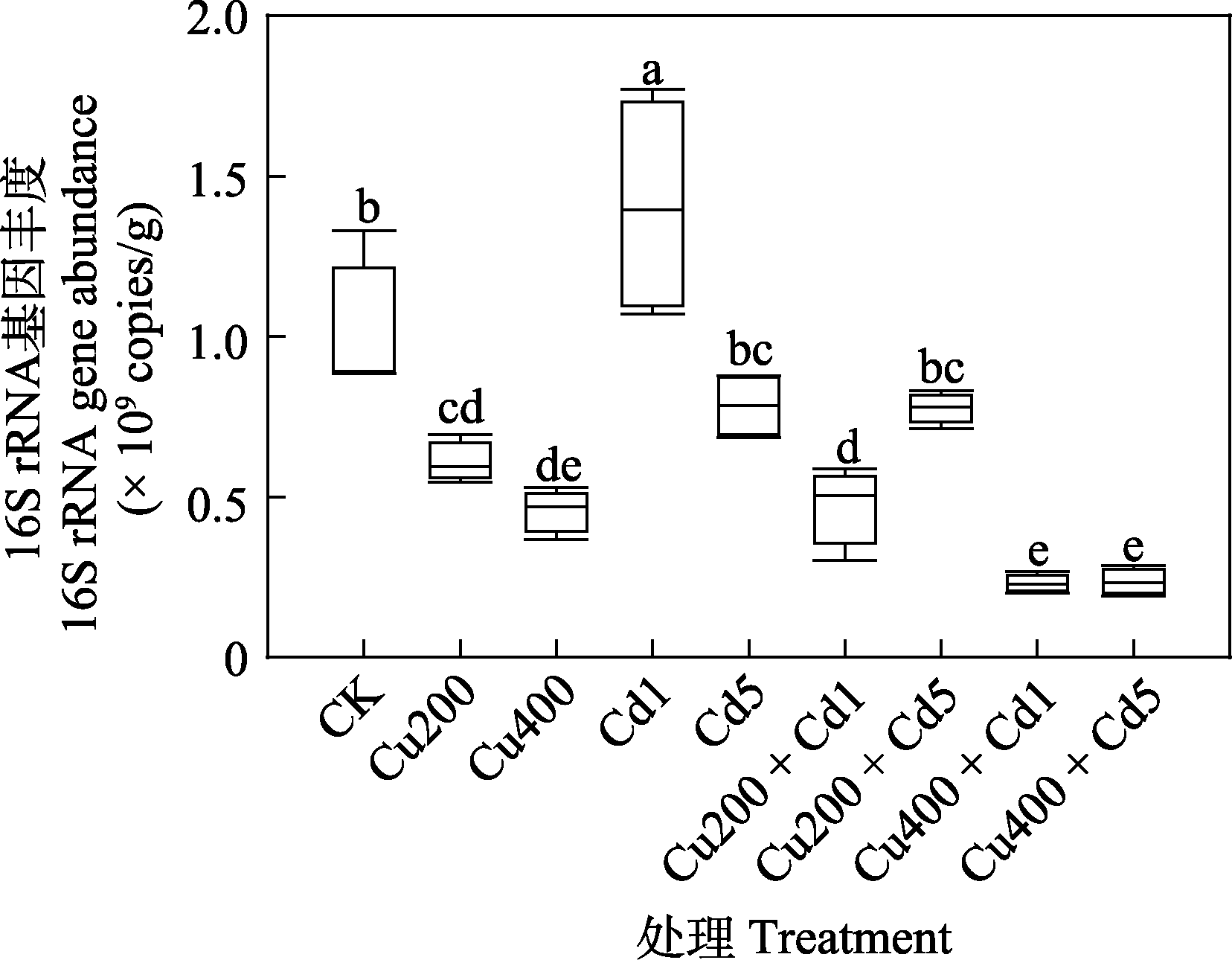
图2 不同浓度Cu和Cd污染的土壤细菌16S rRNA基因绝对丰度。各处理全称见表1。不同小写字母表示不同处理间差异显著(P < 0.05)。
Fig. 2 Absolute abundance of soil bacteria (16S rRNA gene) in different concentration of Cu and Cd polluted soils. The full names of all treatments are listed in Table 1. Different lowercase letters indicate a significant difference between treatments (P < 0.05).
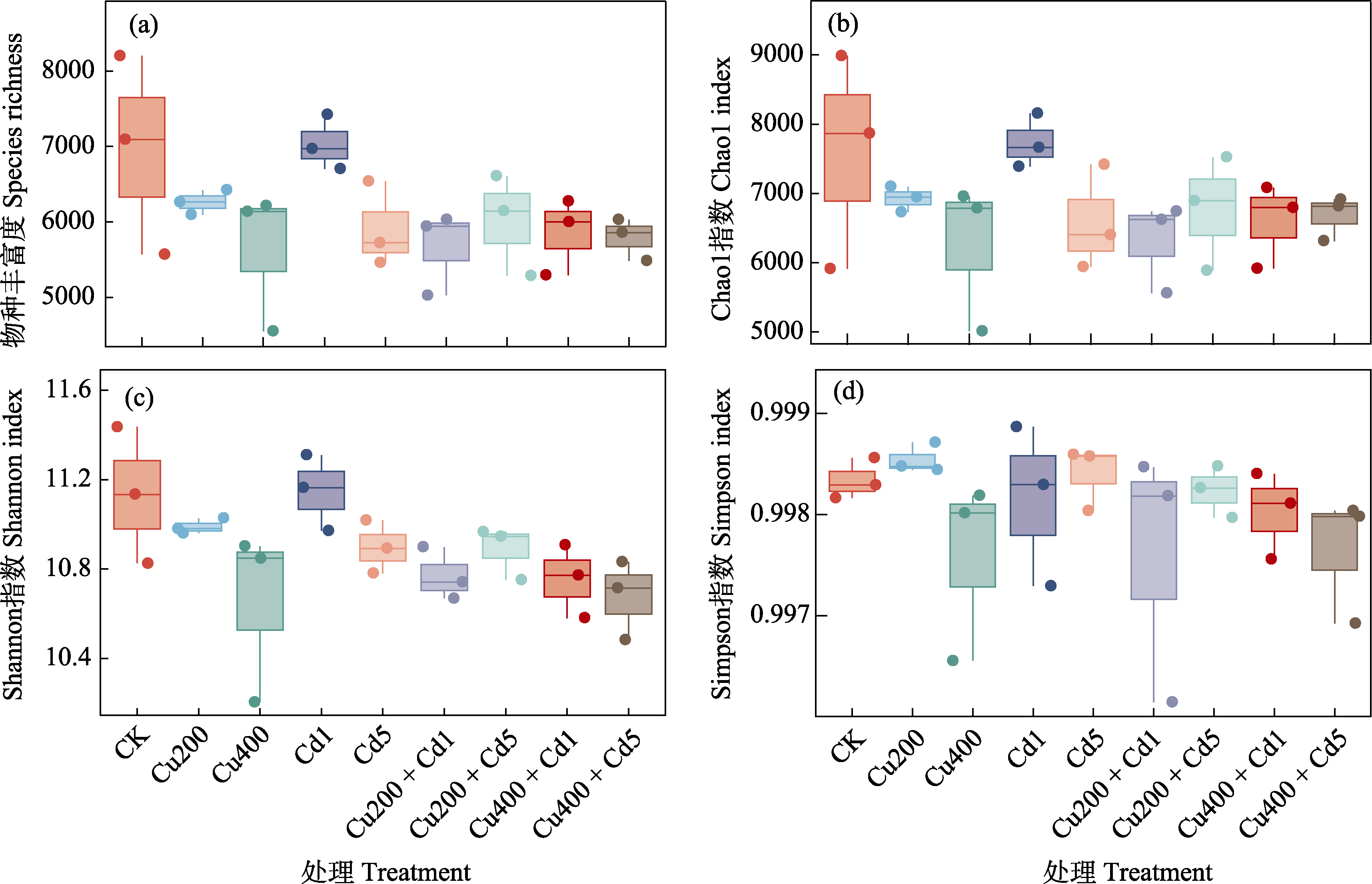
图3 不同浓度Cu和Cd重金属污染土壤细菌群落α多样性。各处理全称见表1。
Fig. 3 Bacterial α diversity in different concentration of Cu and Cd polluted soils. The full names of all treatments are listed in Table 1.
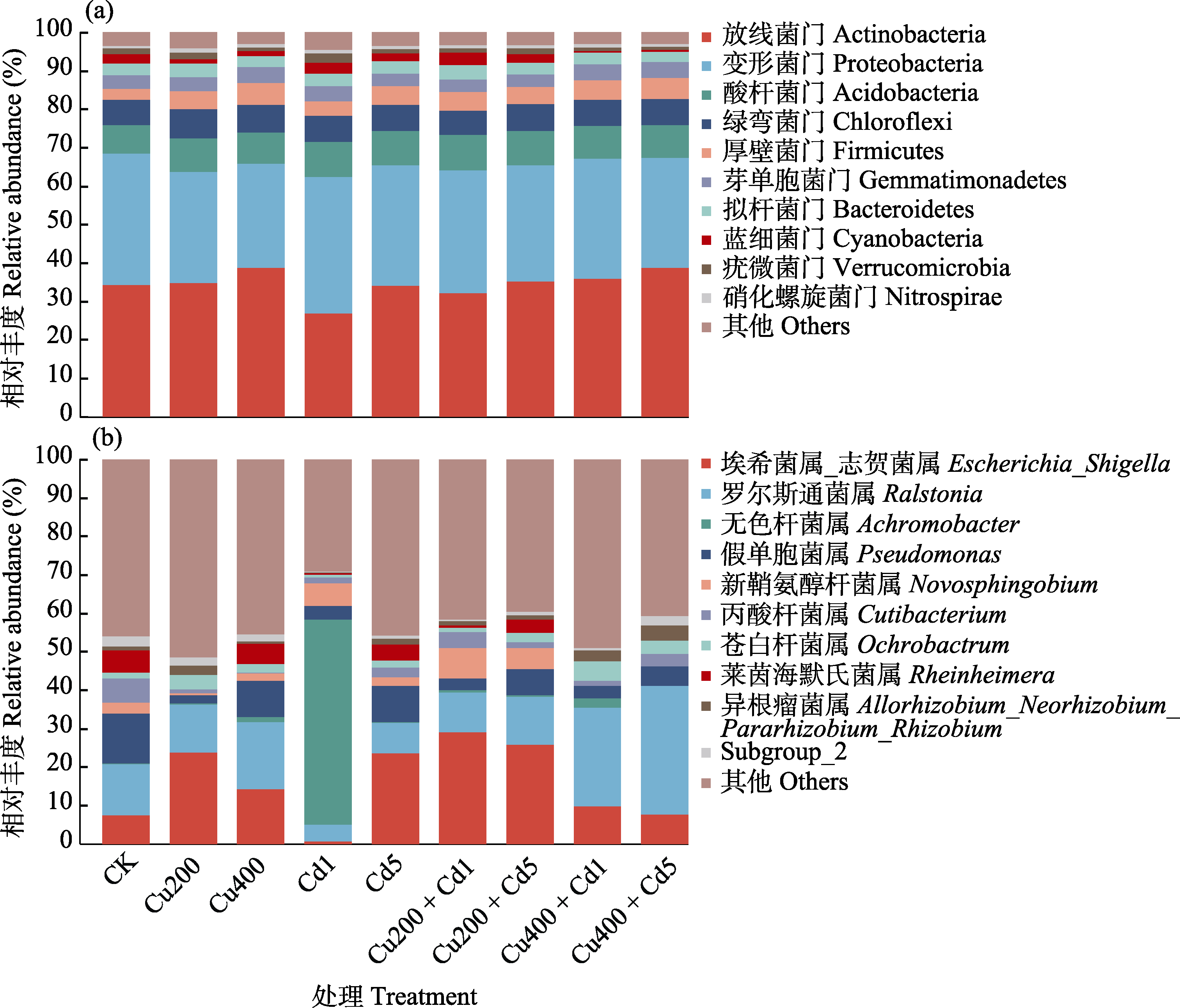
图4 不同浓度Cu和Cd重金属污染土壤细菌前10门水平(a)及属水平(b)群落结构。各处理全称见表1。
Fig. 4 Community structure of top 10 soil bacteria in different concentration of Cu and Cd polluted soils at the phylum (a) and genus level (b). The full names of all treatments are listed in Table 1.
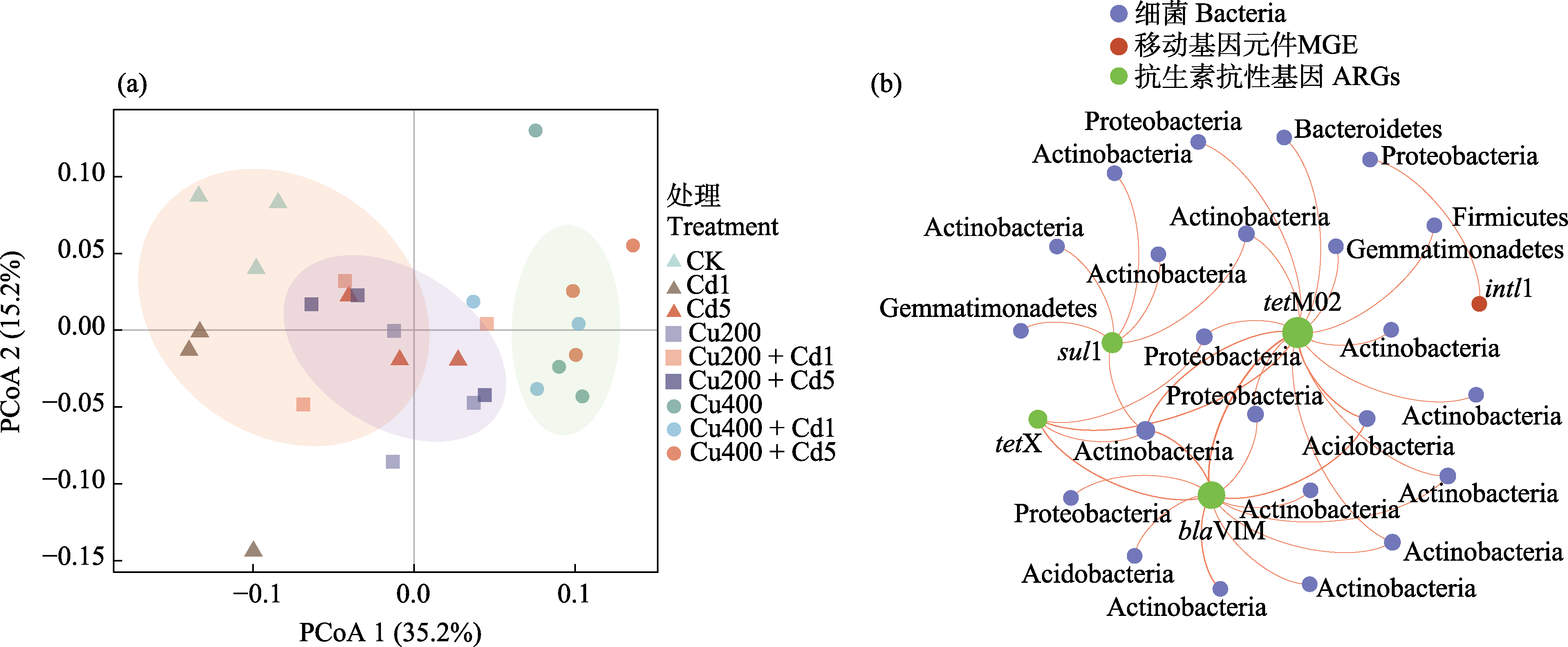
图5 土壤细菌群落结构的PCoA分析(a)与土壤抗生素抗性基因(ARGs)、移动基因元件(MGE)和细菌群落(ASVs)网络分析(b)。各处理全称见表1。
Fig. 5 PCoA analysis of soil bacterial community structure (a) and network analysis of soil antibiotic resistance genes (ARGs), mobile genetic elements (MGE) and bacterial communities at ASV level (b).The full names of all treatments are listed in Table 1.
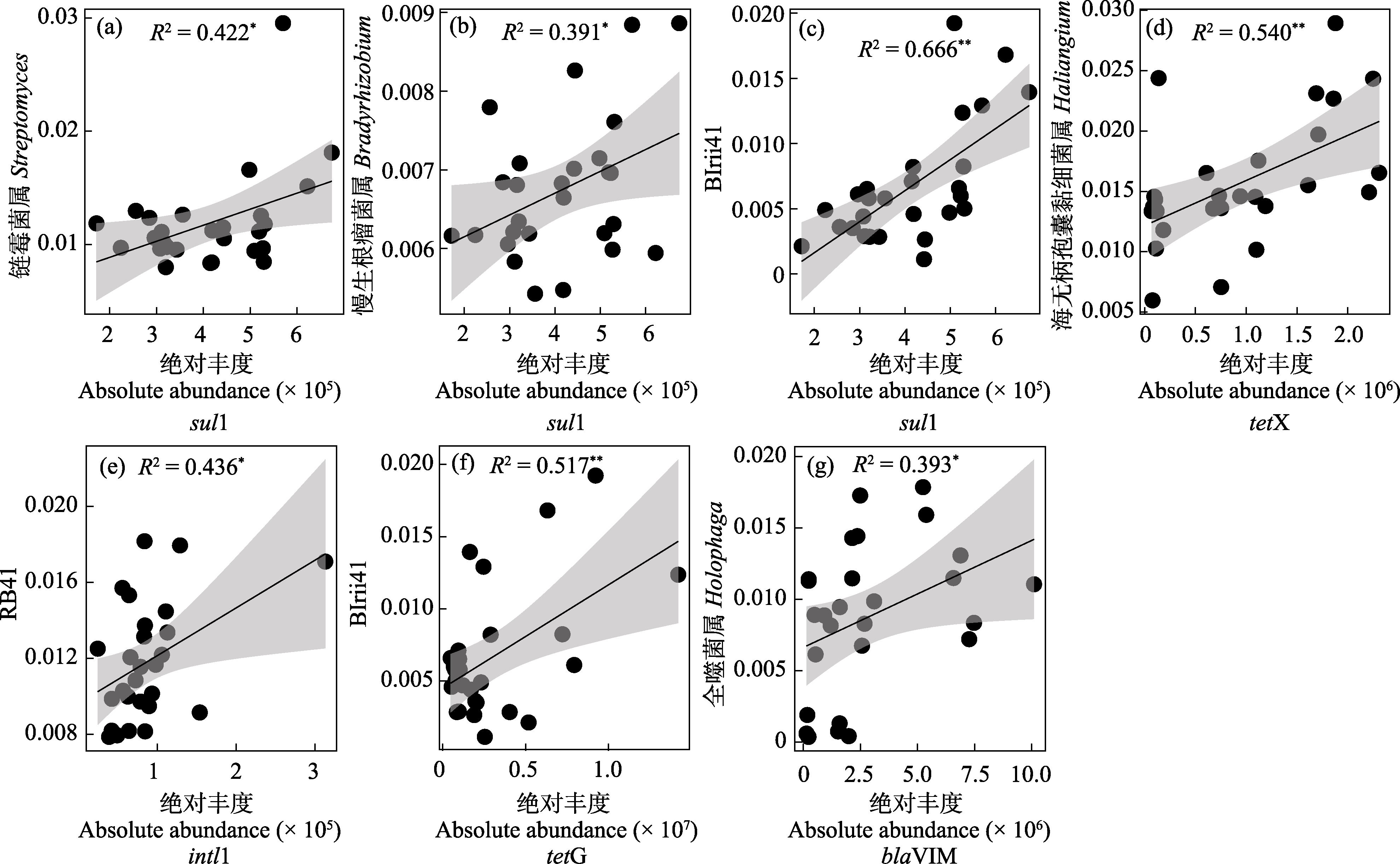
图6 属水平前30细菌与ARGs相关性分析(R2表示相关系数, *P < 0.05, **P < 0.01)。Sul1、tetX、blaVIM和intl1为ARGs和MGE。
Fig. 6 Correlation analysis between the top 30 bacteria genera and ARGs (R2 represents the correlation coefficient. * P < 0.05, ** P < 0.01). Sul1, tetX, blaVIM, and intl1 are ARGs and MGE.
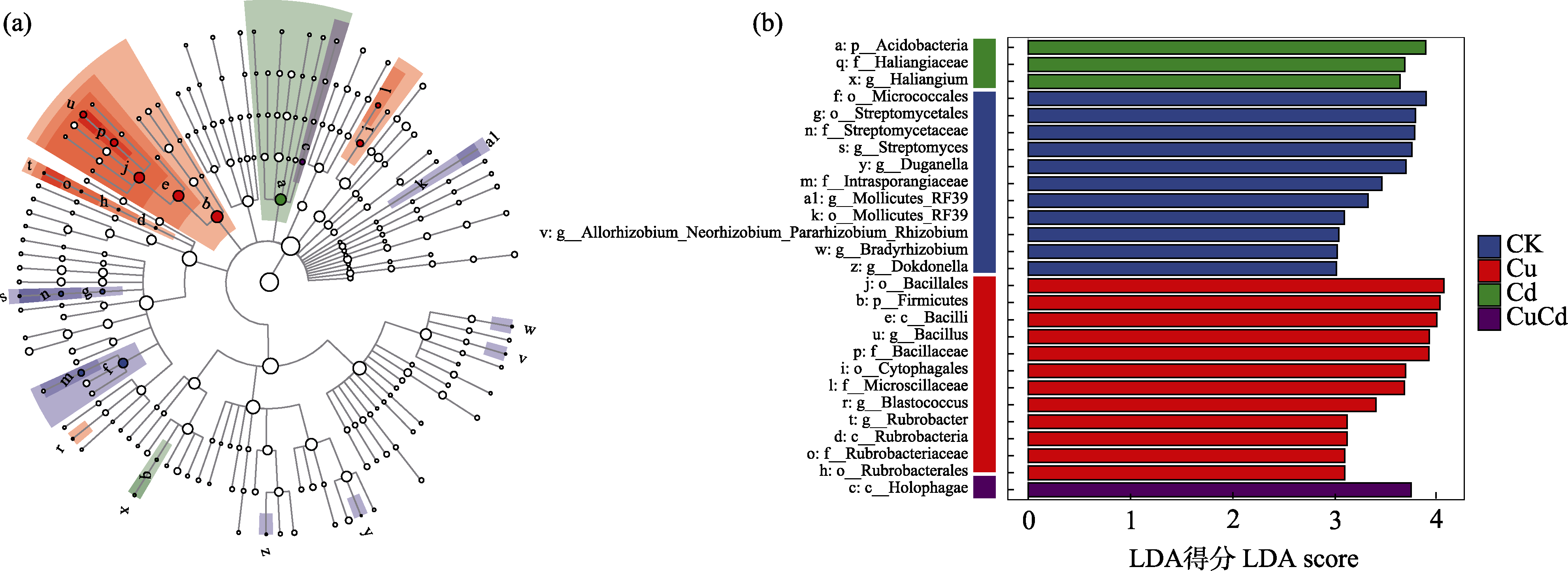
图7 不同浓度Cu和Cd重金属污染土壤中显著富集的细菌LEfSe分析(a: 进化分支图; b: 线性判别分析(LDA)柱状图)。CK代表对照组, Cu代表单独施加Cu的处理(Cu200、Cu400), Cd代表单独施加Cd的处理(Cd1、Cd5), CuCd代表二者复合污染(Cu200 + Cd1、Cu200 + Cd5、Cu400 + Cd1、Cu400 + Cd5)。当前LDA临界值为3。
Fig. 7 LEfSe analysis of significantly enriched bacteria in soil under different concentrations of Cu and Cd (a, Cladogram; b, Histogram of linear discriminant analysis (LDA)). CK, Control group; Cu, Treatment with Cu alone (Cu200, Cu400); Cd, Treatment with Cd alone (Cd1, Cd5); CuCd, Combined pollution of Cu and Cd (Cu200 + Cd1, Cu200 + Cd5, Cu400 + Cd1, Cu400 + Cd5). The current LDA threshold is 3.
| 相对丰度 Relative abundance | Cu | Cd | sul1 | tetX | intl1 | tetG | tetM02 | blaVIM |
|---|---|---|---|---|---|---|---|---|
| ARGs | 0.265 | 0.714** | ‒0.156 | 0.497** | ‒0.356 | ‒0.164 | 0.714** | 0.875** |
| Cu | 0.106 | ‒0.653** | ‒0.557** | ‒0.621** | 0 | ‒0.228 | 0.026 | |
| Cd | 0.182 | 0.499** | ‒0.522** | ‒0.358 | 0.659** | 0.77** | ||
| sul1 | 0.42* | 0.255 | 0.004 | 0.232 | ‒0.013 | |||
| tetX | 0.337 | ‒0.179 | 0.733** | 0.632** | ||||
| intl1 | 0.185 | ‒0.086 | ‒0.213 | |||||
| tetG | ‒0.009 | ‒0.291 | ||||||
| tetM02 | 0.819** |
表3 重金属Cu、Cd、抗性基因相对丰度相关性分析
Table 3 Correlation analysis of relative abundance between Cu, Cd, and ARGs
| 相对丰度 Relative abundance | Cu | Cd | sul1 | tetX | intl1 | tetG | tetM02 | blaVIM |
|---|---|---|---|---|---|---|---|---|
| ARGs | 0.265 | 0.714** | ‒0.156 | 0.497** | ‒0.356 | ‒0.164 | 0.714** | 0.875** |
| Cu | 0.106 | ‒0.653** | ‒0.557** | ‒0.621** | 0 | ‒0.228 | 0.026 | |
| Cd | 0.182 | 0.499** | ‒0.522** | ‒0.358 | 0.659** | 0.77** | ||
| sul1 | 0.42* | 0.255 | 0.004 | 0.232 | ‒0.013 | |||
| tetX | 0.337 | ‒0.179 | 0.733** | 0.632** | ||||
| intl1 | 0.185 | ‒0.086 | ‒0.213 | |||||
| tetG | ‒0.009 | ‒0.291 | ||||||
| tetM02 | 0.819** |
| [1] | Akiba T, Koyama K, Ishiki Y, Kimura S, Fukushima T(1960) On the mechanism of the development of multiple-drug- resistant clones of Shigella. Japanese Journal of Microbiology, 4, 219-227. |
| [2] |
Almakki A, Jumas-Bilak E, Marchandin H, Licznar-Fajardo P(2019) Antibiotic resistance in urban runoff. Science of the Total Environment, 667, 64-76.
DOI |
| [3] |
Aminov RI, Garrigues-Jeanjean N, Mackie RI(2001) Molecular ecology of tetracycline resistance: Development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Applied and Environmental Microbiology, 67, 22-32.
DOI PMID |
| [4] |
Awasthi MK, Liu T, Chen HY, Verma S, Duan YM, Awasthi SK, Wang Q, Ren XN, Zhao JC, Zhang ZQ(2019) The behavior of antibiotic resistance genes and their associations with bacterial community during poultry manure composting. Bioresource Technology, 280, 70-78.
DOI PMID |
| [5] | Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV(2006) Co-selection of antibiotic and metal resistance. Trends in Microbiology, 14, 176-182. |
| [6] | Bao YM, Choct M, Iji PA, Bruerton K(2010) The digestibility of organic trace minerals along the small intestine in broiler chickens. Asian-Australasian Journal of Animal Sciences, 23, 90-97. |
| [7] |
Bartha NA, Sóki J, Urbán E, Nagy E(2011) Investigation of the prevalence of tetQ, tetX and tetX1 genes in Bacteroides strains with elevated tigecycline minimum inhibitory concentrations. International Journal of Antimicrobial Agents, 38, 522-525.
DOI PMID |
| [8] |
Bassetti M, Peghin M, Vena A, Giacobbe DR(2019) Treatment of infections due to MDR gram-negative bacteria. Frontiers in Medicine, 6, 74.
DOI PMID |
| [9] | Cheng JH, Tang XY, Liu C(2019) Characteristics of antibiotic resistance genes in various livestock feedlot soils of the hilly purple soil region. Environmental Science, 40, 3257-3262. (in Chinese with English abstract) |
| [ 程建华, 唐翔宇, 刘琛 (2019) 紫色土丘陵区畜禽养殖场土壤中抗生素抗性基因分布特征. 环境科学, 40, 3257-3262.] | |
| [10] | Claesson MJ, O’Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O’Toole PW(2009) Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE, 4, e6669. |
| [11] | Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, Dantas G(2014) Bacterial phylogeny structures soil resistomes across habitats. Nature, 509, 612-616. |
| [12] |
Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G(2012) The shared antibiotic resistome of soil bacteria and human pathogens. Science, 337, 1107-1111.
DOI PMID |
| [13] |
Frank T, Gautier V, Talarmin A, Bercion R, Arlet G(2007) Characterization of sulphonamide resistance genes and class 1 integron gene cassettes in Enterobacteriaceae, Central African Republic (CAR). Journal of Antimicrobial Chemotherapy, 59, 742-745.
DOI PMID |
| [14] | Ghosh D, Veeraraghavan B, Elangovan R, Vivekanandan P(2020) Antibiotic resistance and epigenetics: More to it than meets the eye. Antimicrobial Agents and Chemotherapy, 64, e02225-19. |
| [15] | Gillings MR, Gaze WH, Pruden A, Smalla K, Tiedje JM, Zhu YG(2015) Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. The ISME Journal, 9, 1269-1279. |
| [16] |
Goldstein C, Lee MD, Sanchez S, Hudson C, Phillips B, Register B, Grady M, Liebert C, Summers AO, White DG, Maurer JJ(2001) Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrobial Agents and Chemotherapy, 45, 723-726.
DOI PMID |
| [17] | Gupta S, Graham DW, Sreekrishnan TR, Ahammad SZ(2022) Effects of heavy metals pollution on the co-selection of metal and antibiotic resistance in urban rivers in UK and India. Environmental Pollution, 306, 119326. |
| [18] |
Hou SN, Zheng N, Tang L, Ji XF(2018) Effects of cadmium and copper mixtures to carrot and pakchoi under greenhouse cultivation condition. Ecotoxicology and Environmental Safety, 159, 172-181.
DOI PMID |
| [19] |
Huang B, Yuan Z, Li DQ, Nie XD, Xie ZY, Chen JY, Liang C, Liao YS, Liu T(2019) Loss characteristics of Cd in soil aggregates under simulated rainfall conditions. Science of the Total Environment, 650, 313-320.
DOI |
| [20] | Jain R, Rivera MC, Lake JA(1999) Horizontal gene transfer among genomes: The complexity hypothesis. Proceedings of the National Academy of Sciences, USA, 96, 3801-3806. |
| [21] |
Knapp CW, Callan AC, Aitken B, Shearn R, Koenders A, Hinwood A(2017) Relationship between antibiotic resistance genes and metals in residential soil samples from Western Australia. Environmental Science and Pollution Research International, 24, 2484-2494.
DOI PMID |
| [22] | Ktari S, Arlet G, Mnif B, Gautier V, Mahjoubi F, Ben Jmeaa M, Bouaziz M, Hammami A(2006) Emergence of multidrug-resistant Klebsiella pneumoniae isolates producing VIM-4 metallo-β-lactamase, CTX-M-15 extended- spectrum β-lactamase, and CMY-4 AmpC β-lactamase in a Tunisian university hospital. Antimicrobial Agents and Chemotherapy, 50, 4198-4201. |
| [23] | Leski TA, Bangura U, Jimmy DH, Ansumana R, Lizewski SE, Stenger DA, Taitt CR, Vora GJ(2013) Multidrug-resistant tet(X)-containing hospital isolates in Sierra Leone. International Journal of Antimicrobial Agents, 42, 83-86. |
| [24] | Levy SB, Marshall B(2004) Antibacterial resistance worldwide: Causes, challenges and responses. Nature Medicine,10, S122-S129. |
| [25] | Li Y, Wang XJ, Li J, Wang Y, Song JK, Xia SQ, Jing HP, Zhao JF(2019) Effects of struvite-humic acid loaded biochar/bentonite composite amendment on Zn(II) and antibiotic resistance genes in manure-soil. Chemical Engineering Journal, 375, 122013. |
| [26] | Li ZK, Han M, Qin C, Hu XJ, Gao YZ(2024) Research progress and hot spot analysis on antibiotic resistance gene pollution in soil environment. Journal of Ecology and Rural Environment, 40, 11-22. (in Chinese with English abstract) |
| [ 李泽楷, 韩淼, 秦超, 胡小婕, 高彦征 (2024) 土壤环境中抗生素抗性基因污染研究进展和热点分析. 生态与农村环境学报, 40, 11-22.] | |
| [27] |
Liebert CA, Hall RM, Summers AO(1999) Transposon Tn21, flagship of the floating genome. Microbiology and Molecular Biology Reviews, 63, 507-522.
DOI PMID |
| [28] |
Liu H, Sun HF, Zhang M, Liu Y(2019) Dynamics of microbial community and tetracycline resistance genes in biological nutrient removal process. Journal of Environmental Management, 238, 84-91.
DOI PMID |
| [29] |
Ng LK, Martin I, Alfa M, Mulvey M(2001) Multiplex PCR for the detection of tetracycline resistant genes. Molecular and Cellular Probes, 15, 209-215.
DOI PMID |
| [30] | Pal C, Asiani K, Arya S, Rensing C, Stekel DJ, Larsson DJ, Hobman JL(2017) Metal resistance and its association with antibiotic resistance. Microbiology of Metal Ions, 70, 261-313. |
| [31] |
Partridge SR, Brown HJ, Stokes HW, Hall RM(2001) Transposons Tn1696 and Tn21 and their integrons In4 and In 2 have independent origins. Antimicrobial Agents and Chemotherapy, 45, 1263-1270.
PMID |
| [32] | Peng S, Zhang HY, Song D, Chen H, Lin XG, Wang YM, Ji LD(2022) Distribution of antibiotic, heavy metals and antibiotic resistance genes in livestock and poultry feces from different scale of farms in Ningxia, China. Journal of Hazardous Materials, 440, 129719. |
| [33] | Pruden A, Pei RT, Storteboom H, Carlson KH(2006) Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environmental Science & Technology, 40, 7445-7450. |
| [34] | Qiao LK, Liu XX, Zhang S, Zhang LY, Li XH, Hu XS, Zhao QC, Wang QY, Yu CH(2021) Distribution of the microbial community and antibiotic resistance genes in farmland surrounding gold tailings: A metagenomics approach. Science of the Total Environment, 779, 146502. |
| [35] | Shi LM, Zhang JY, Lu TD, Zhang KC(2022) Metagenomics revealed the mobility and hosts of antibiotic resistance genes in typical pesticide wastewater treatment plants. Science of the Total Environment, 817, 153033. |
| [36] | Song JX, Rensing C, Holm PE, Virta M, Brandt KK(2017) Comparison of metals and tetracycline as selective agents for development of tetracycline resistant bacterial communities in agricultural soil. Environmental Science & Technology, 51, 3040-3047. |
| [37] | Stepanauskas R, Glenn TC, Jagoe CH, Tuckfield RC, Lindell AH, McArthur JV(2005) Elevated microbial tolerance to metals and antibiotics in metal-contaminated industrial environments. Environmental Science & Technology, 39, 3671-3678. |
| [38] |
Stokes HW, Gillings MR(2011) Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into gram-negative pathogens. FEMS Microbiology Reviews, 35, 790-819.
DOI PMID |
| [39] |
Su JQ, Huang FY, Zhu YG(2013) Antibiotic resistance genes in the environment. Biodiversity Science, 21, 481-487. (in Chinese with English abstract)
DOI |
|
[ 苏建强, 黄福义, 朱永官 (2013) 环境抗生素抗性基因研究进展. 生物多样性, 21, 481-487.]
DOI |
|
| [40] | Sun J, Chen C, Cui CY, Zhang Y, Liu X, Cui ZH, Ma XY, Feng YJ, Fang LX, Lian XL, Zhang RM, Tang YZ, Zhang KX, Liu HM, Zhuang ZH, Zhou SD, Lv JN, Du H, Huang B, Yu FY, Mathema B, Kreiswirth BN, Liao XP, Chen L, Liu YH(2019) Plasmid-encoded tet (X) genes that confer high-level tigecycline resistance in Escherichia coli. Nature Microbiology, 4, 1457-1464. |
| [41] | Tian QF, He QS, Lu AX, Wang HO(2020) Relationship between antibiotic resistance genes and microbial communities in farmland soil. Environment Chemistry, 39, 1346-1355. (in Chinese with English abstract) |
| [ 田其凡, 何玘霜, 陆安祥, 王海鸥 (2020) 农田土壤抗生素抗性基因与微生物群落的关系. 环境化学, 39, 1346-1355.] | |
| [42] | Wales AD, Davies RH(2015) Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics, 4, 567-604. |
| [43] |
Wang H, Dong YH, Yang YY, Toor GS, Zhang XM(2013) Changes in heavy metal contents in animal feeds and manures in an intensive animal production region of China. Journal of Environmental Sciences, 25, 2435-2442.
PMID |
| [44] | Wang J, Zhou JT(2021) Petroleum exploitation enriches the sulfonamide resistance gene sul2 in offshore sediments. Journal of Oceanology and Limnology, 39, 946-954. |
| [45] |
Wen X, Wang Y, Zou YD, Ma BH, Wu YB(2018) No evidential correlation between veterinary antibiotic degradation ability and resistance genes in microorganisms during the biodegradation of doxycycline. Ecotoxicology and Environmental Safety, 147, 759-766.
DOI PMID |
| [46] | Wu J, Wang JY, Li ZT, Guo SM, Li KJ, Xu PS, Ok YS, Jones DL, Zou JW(2023) Antibiotics and antibiotic resistance genes in agricultural soils: A systematic analysis. Critical Reviews in Environmental Science and Technology, 53, 847-864. |
| [47] | Xia YH, Tan L, Xu M, Lei BY, Gao CX, Zhu Y, Chen Q, Hu P, Xiong W, Tang F(2019) Antibiotic resistance gene contamination status in air conditioning filter dust of a hospital in Wuhan City. Journal of Hygiene Research, 48, 583-593. (in Chinese with English abstract) |
| [ 夏雨荷, 谭莉, 徐敏, 雷博阳, 高彩霞, 朱源, 陈琪, 胡平, 熊薇, 唐非 (2019) 武汉市某医院空调过滤网积尘中抗生素抗性基因污染状况. 卫生研究, 48, 583-593.] | |
| [48] |
Yang XP, Li Q, Tang Z, Zhang WW, Yu GH, Shen QR, Zhao FJ(2017) Heavy metal concentrations and arsenic speciation in animal manure composts in China. Waste Management, 64, 333-339.
DOI PMID |
| [49] | Yazdankhah S, Skjerve E, Wasteson Y(2018) Antimicrobial resistance due to the content of potentially toxic metals in soil and fertilizing products. Microbial Ecology in Health and Disease, 29, 1548248. |
| [50] | Yuan XH, Xue ND, Han ZG(2021) A meta-analysis of heavy metals pollution in farmland and urban soils in China over the past 20 years. Journal of Environmental Sciences, 101, 217-226. |
| [51] | Zhang B, Zhao L, Chen T(2023) Research progress of antibiotic resistance genes in wastewater treatment plants. Journal of Environmental Engineering Technology, 13, 1384-1394. |
| [52] |
Zhang FS, Li YX, Yang M, Li W(2012) Content of heavy metals in animal feeds and manures from farms of different scales in Northeast China. International Journal of Environmental Research and Public Health, 9, 2658-2668.
DOI PMID |
| [53] | Zhang HC, Chang FY, Shi P, Ye L, Zhou Q, Pan Y, Li AM(2019) Antibiotic resistome alteration by different disinfection strategies in a full-scale drinking water treatment plant deciphered by metagenomic assembly. Environmental Science & Technology, 53, 2141-2150. |
| [54] | Zhang XR, Gong ZQ, Allinson G, Xiao M, Li XJ, Jia CY, Ni ZJ(2022) Environmental risks caused by livestock and poultry farms to the soils: Comparison of swine, chicken, and cattle farms. Journal of Environmental Management, 317, 115320. |
| [55] |
Zhang Y, Gu AZ, Cen TY, Li XY, He M, Li D, Chen JM(2018) Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environmental Pollution, 237, 74-82.
DOI PMID |
| [56] | Zhang YA, Wang TT, Hu XJ, Qin C, Gao YZ(2024) Research progress and hotspots analysis of soil antibiotic resistance genes. Acta Pedologica Sinica, 61, 607-618. (in Chinese with English abstract) |
| [ 张友爱, 王婷婷, 胡小婕, 秦超, 高彦征 (2024) 土壤抗生素抗性基因研究进展及热点分析. 土壤学报, 61, 607-618.] | |
| [57] | Zhang YJ, Hu HW, Gou M, Wang JT, Chen DL, He JZ(2017) Temporal succession of soil antibiotic resistance genes following application of swine, cattle and poultry manures spiked with or without antibiotics. Environmental Pollution, 231, 1621-1632. |
| [58] |
Zhang ZY, Zhang Q, Wang TZ, Xu NH, Lu T, Hong WJ, Penuelas J, Gillings M, Wang MX, Gao WW, Qian HF(2022) Assessment of global health risk of antibiotic resistance genes. Nature Communications, 13, 1553.
DOI PMID |
| [59] | Zhao RX, Yu K, Zhang JY, Zhang GJ, Huang J, Ma LP, Deng CF, Li XY, Li B(2020) Deciphering the mobility and bacterial hosts of antibiotic resistance genes under antibiotic selection pressure by metagenomic assembly and binning approaches. Water Research, 186, 116318. |
| [60] | Zou JH, Lin Q, Shao MY, Xu SH(2023) Leaching behavior of Cu, Cd, and Zn in combined pollution soil as affected by biochar. Acta Scientiae Circumstantiae, 43(9), 333-345. (in Chinese with English abstract) |
| [ 邹佳慧, 林青, 邵明艳, 徐绍辉 (2023) 生物炭影响下土壤中铜镉锌复合污染物的淋溶迁移. 环境科学学报, 43(9), 333-345.] |
| [1] | 魏诗雨, 宋天骄, 罗佳宜, 张燕, 赵子萱, 茹靖雯, 易华, 林雁冰. 秦岭火地塘针叶林土壤细菌群落的海拔分布格局[J]. 生物多样性, 2024, 32(9): 24180-. |
| [2] | 毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌. 毛竹扩张对杉木林土壤细菌群落的影响[J]. 生物多样性, 2023, 31(6): 22659-. |
| [3] | 刘厶瑶, 李柱, 柯欣, 孙丽娜, 吴龙华, 赵杰杰. 贵州省典型汞铊矿区周边农田土壤跳虫群落特征[J]. 生物多样性, 2022, 30(12): 22265-. |
| [4] | 张全国, 张大勇. 生产力、可靠度与物种多样性:微宇宙实验研究[J]. 生物多样性, 2002, 10(2): 135-142. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn