



生物多样性 ›› 2023, Vol. 31 ›› Issue (2): 22258. DOI: 10.17520/biods.2022258 cstr: 32101.14.biods.2022258
所属专题: 土壤生物与土壤健康
赵雯, 王丹丹, 热依拉·木民, 黄开钏, 刘顺, 崔宝凯*( )
)
收稿日期:2022-05-11
接受日期:2022-09-13
出版日期:2023-02-20
发布日期:2022-10-28
通讯作者:
*崔宝凯, E-mail: cuibaokai@bjfu.edu.cn
基金资助:
Wen Zhao, Dandan Wang, Mumin Reyila, Kaichuan Huang, Shun Liu, Baokai Cui*( )
)
Received:2022-05-11
Accepted:2022-09-13
Online:2023-02-20
Published:2022-10-28
Contact:
*Baokai Cui, E-mail: cuibaokai@bjfu.edu.cn
摘要:
土壤微生物在森林生态系统能量流动和物质循环过程中发挥着不可替代的作用。兴安落叶松(Larix gmelinii)是我国大兴安岭地区的优势树种, 而阿尔山地区是其在我国分布的最南端, 受气候变暖影响较为显著, 因此探索兴安落叶松林下土壤微生物群落结构对了解和维护我国东北地区森林生态系统的稳定有重要意义。在本研究中, 我们从阿尔山白狼镇、天池镇采集兴安落叶松林下土样, 通过高通量测序技术, 依据97%相似性原则, 将细菌序列划分为5,163个可操作分类单位(operational taxonomic units, OTUs), 真菌序列划分为2,439个OTUs, 其中门水平优势细菌为放线菌门、变形菌门、疣微菌门、绿弯菌门和酸杆菌门, 优势真菌为担子菌门和子囊菌门; 属水平上, 优势细菌为Candidatus_Udaeobacter, 优势真菌属为丝盖伞属(Inocybe)、蜡壳耳属(Sebacina)、Piloderma和棉革菌属(Tomentella)。天池镇的细菌多样性显著高于白狼镇, 而真菌多样性在两地间无显著差异。土壤pH和阳离子交换量是驱动土壤细菌多样性的主要因子, 而土壤全氮和有机碳是驱动真菌多样性的主要因子。土壤全氮、有机碳、速效磷和阳离子交换量是影响土壤细菌群落组成的重要理化因子, 而土壤理化性质对真菌群落组成的影响不显著。绿弯菌门的相对多度与pH显著正相关; 担子菌门与土壤全氮和有机碳显著负相关, 而子囊菌门与土壤全氮、有机碳和阳离子交换量显著正相关。
赵雯, 王丹丹, 热依拉·木民, 黄开钏, 刘顺, 崔宝凯 (2023) 阿尔山地区兴安落叶松林土壤微生物群落结构. 生物多样性, 31, 22258. DOI: 10.17520/biods.2022258.
Wen Zhao, Dandan Wang, Mumin Reyila, Kaichuan Huang, Shun Liu, Baokai Cui (2023) Soil microbial community structure of Larix gmelinii forest in the Aershan area. Biodiversity Science, 31, 22258. DOI: 10.17520/biods.2022258.
| 土壤性质 Soil property | 白狼镇 Bailang Town | 天池镇 Tianchi Town | P值 P value |
|---|---|---|---|
| 全氮 Total nitrogen (%) | 0.54 ± 0.03 | 0.56 ± 0.01 | 0.188 |
| 土壤有机碳 Soil organic carbon (%) | 6.29 ± 0.32 | 6.77 ± 0.16 | 0.242 |
| 速效磷 Available phosphorus (mg/kg) | 10.22 ± 0.79 | 9.21 ± 0.50 | 0.176 |
| pH | 5.50 ± 0.01 | 5.61 ± 0.03 | 0.002 |
| 阳离子交换量 Cation exchange capacity (cmol(+)/kg) | 43.57 ± 0.96 | 47.47 ± 0.44 | 0.039 |
表1 土壤理化性质(平均值 ± 标准误差)
Table 1 Soil physicochemical properties (mean ± SE)
| 土壤性质 Soil property | 白狼镇 Bailang Town | 天池镇 Tianchi Town | P值 P value |
|---|---|---|---|
| 全氮 Total nitrogen (%) | 0.54 ± 0.03 | 0.56 ± 0.01 | 0.188 |
| 土壤有机碳 Soil organic carbon (%) | 6.29 ± 0.32 | 6.77 ± 0.16 | 0.242 |
| 速效磷 Available phosphorus (mg/kg) | 10.22 ± 0.79 | 9.21 ± 0.50 | 0.176 |
| pH | 5.50 ± 0.01 | 5.61 ± 0.03 | 0.002 |
| 阳离子交换量 Cation exchange capacity (cmol(+)/kg) | 43.57 ± 0.96 | 47.47 ± 0.44 | 0.039 |
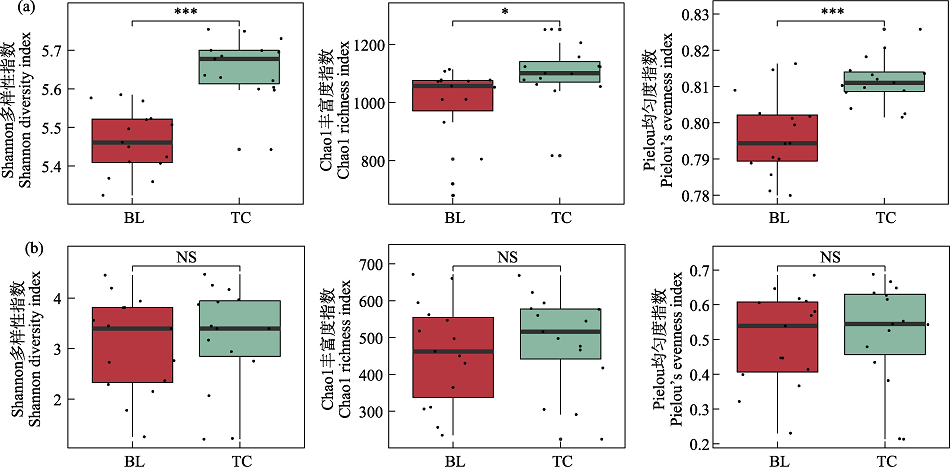
图1 两地的细菌(a)和真菌(b) alpha多样性。BL: 白狼镇; TC: 天池镇; NS: 不显著。* P < 0.05; *** P < 0.001。
Fig. 1 Alpha diversity of bacteria (a) and fungi (b) in two sites. BL, Bailang Town; TC, Tianchi Town; NS, Not significant. * P < 0.05; *** P < 0.001.
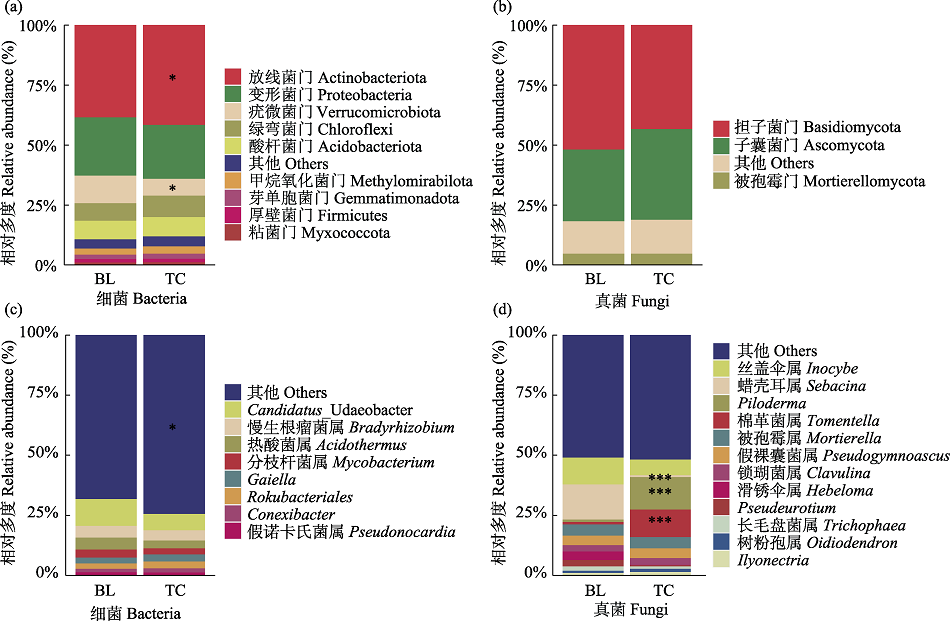
图2 两个兴安落叶松林分细菌和真菌的门水平(a、b)和属水平(c、d)相对多度。BL: 白狼镇; TC: 天池镇。* P < 0.05; *** P < 0.001。
Fig. 2 Relative abundance of bacteria and fungi at the phylum level (a, b) and genus level (c, d) in two Larix gmelinii stand. BL, Bailang Town; TC, Tianchi Town. * P < 0.05; *** P < 0.001.
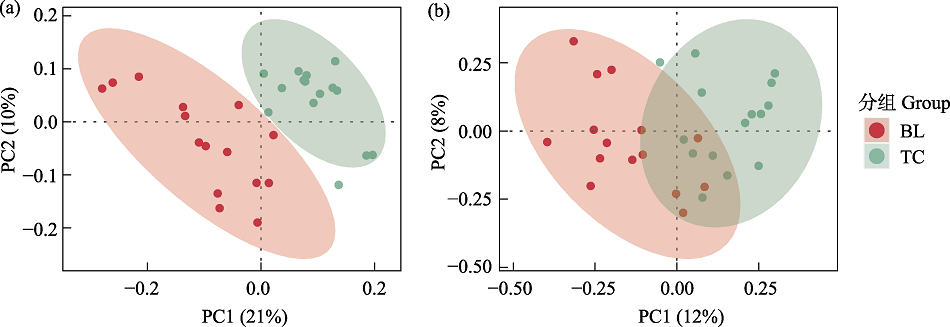
图3 基于OTU水平的Bray-Curtis距离的主坐标分析比较两个样地间的细菌(a)和真菌(b)群落相似性。BL: 白狼镇; TC: 天池镇。
Fig. 3 Principal coordinate analysis based on Bray-Curtis distance at OTU level to compare the similarity of bacterial (a) and fungal (b) communities between two sites. BL, Bailang Town; TC, Tianchi Town.
| 土壤性质 Soil properties | Shannon多样性指数 Shannon diversity index | Chao1丰富度指数 Chao1 richness index | Pielou均匀度指数 Pielou’s evenness index | |
|---|---|---|---|---|
| 细菌 Bacteria | 全氮 Total nitrogen | 0.208 | 0.210 | -0.110 |
| 有机碳 Soil organic carbon | 0.223 | 0.132 | 0.273 | |
| 速效磷 Available phosphorus | -0.256 | -0.231 | 0.156 | |
| pH | 0.436* | 0.361* | 0.203 | |
| 阳离子交换量 Cation exchange capacity | 0.430* | 0.247 | 0.453* | |
| 真菌 Fungi | 全氮 Total nitrogen | 0.561*** | 0.523** | 0.582*** |
| 有机碳 Soil organic carbon | 0.369* | 0.451* | 0.372* | |
| 速效磷 Available phosphorus | -0.336 | -0.238 | -0.385* | |
| pH | 0.026 | -0.039 | 0.052 | |
| 阳离子交换量 Cation exchange capacity | 0.306 | 0.346 | 0.323 |
表2 细菌和真菌多样性与土壤理化性质相关性分析
Table 2 Spearman correlation analyses between the bacterial and fungal diversities and soil physicochemical properties
| 土壤性质 Soil properties | Shannon多样性指数 Shannon diversity index | Chao1丰富度指数 Chao1 richness index | Pielou均匀度指数 Pielou’s evenness index | |
|---|---|---|---|---|
| 细菌 Bacteria | 全氮 Total nitrogen | 0.208 | 0.210 | -0.110 |
| 有机碳 Soil organic carbon | 0.223 | 0.132 | 0.273 | |
| 速效磷 Available phosphorus | -0.256 | -0.231 | 0.156 | |
| pH | 0.436* | 0.361* | 0.203 | |
| 阳离子交换量 Cation exchange capacity | 0.430* | 0.247 | 0.453* | |
| 真菌 Fungi | 全氮 Total nitrogen | 0.561*** | 0.523** | 0.582*** |
| 有机碳 Soil organic carbon | 0.369* | 0.451* | 0.372* | |
| 速效磷 Available phosphorus | -0.336 | -0.238 | -0.385* | |
| pH | 0.026 | -0.039 | 0.052 | |
| 阳离子交换量 Cation exchange capacity | 0.306 | 0.346 | 0.323 |
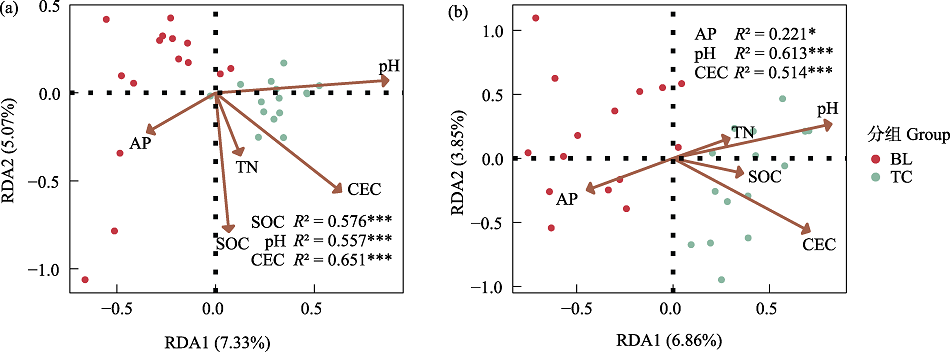
图4 细菌(a)和真菌(b)群落结构与土壤理化性质的冗余分析。BL: 白狼镇; TC: 天池镇。R 2: envfit函数得出的相关系数。* P < 0.05; *** P < 0.001。TN: 全氮; SOC: 有机碳; AP: 速效磷; CEC: 阳离子交换量。
Fig. 4 Redundancy analysis of bacterial (a) and fungal (b) community structures and soil physicochemical properties. BL, Bailang Town; TC, Tianchi Town. R2, The correlation coefficient based on the envfit function. * P < 0.05; *** P < 0.001. TN, Total nitrogen; SOC, Soil organic carbon; AP, Available phosphorus; CEC, Cation exchange capacity.
| 全氮 Total nitrogen | 有机碳 Soil organic carbon | 速效磷 Available phosphorus | pH | 阳离子交换量 Cation exchange capacity | 土壤性质 Soil property | ||
|---|---|---|---|---|---|---|---|
| 细菌 Bacteria | R | 0.289 | 0.231 | 0.194 | 0.130 | 0.263 | 0.478 |
| P | 0.012* | 0.025* | 0.037* | 0.079 | 0.008** | 0.001*** | |
| 真菌 Fungi | R | -0.056 | -0.004 | 0.169 | 0.083 | 0.130 | 0.168 |
| P | 0.655 | 0.475 | 0.073 | 0.173 | 0.120 | 0.104 |
表3 细菌和真菌群落组成与土壤理化性质的Mantel检验
Table 3 Mantel test between the dominant bacterial and fungal community compositions and soil physicochemical properties
| 全氮 Total nitrogen | 有机碳 Soil organic carbon | 速效磷 Available phosphorus | pH | 阳离子交换量 Cation exchange capacity | 土壤性质 Soil property | ||
|---|---|---|---|---|---|---|---|
| 细菌 Bacteria | R | 0.289 | 0.231 | 0.194 | 0.130 | 0.263 | 0.478 |
| P | 0.012* | 0.025* | 0.037* | 0.079 | 0.008** | 0.001*** | |
| 真菌 Fungi | R | -0.056 | -0.004 | 0.169 | 0.083 | 0.130 | 0.168 |
| P | 0.655 | 0.475 | 0.073 | 0.173 | 0.120 | 0.104 |
| 全氮 Total nitrogen | 有机碳 Soil organic carbon | 速效磷 Available phosphorus | pH | 阳离子交换量 Cation exchange capacity | ||
|---|---|---|---|---|---|---|
| 细菌 Bacteria | 放线菌门 Actinobacteriota | 0.016 | 0.011 | 0.069 | 0.302 | 0.161 |
| 变形菌门 Proteobacteria | -0.172 | -0.152 | 0.307 | -0.143 | -0.316 | |
| 疣微菌门 Verrucomicrobiota | 0.180 | 0.087 | -0.098 | -0.316 | -0.091 | |
| 绿弯菌门 Chloroflexi | -0.260 | -0.233 | -0.211 | 0.498** | 0.051 | |
| 酸杆菌门 Acidobacteriota | -0.225 | -0.046 | -0.019 | 0.127 | 0.012 | |
| 真菌 Fungi | 担子菌门 Basidiomycota | -0.475** | -0.370* | 0.226 | -0.272 | -0.250 |
| 子囊菌门 Ascomycota | 0.582*** | 0.517** | -0.267 | 0.265 | 0.366* |
表4 细菌和真菌优势菌门的相对多度与土壤理化性质相关性
Table 4 Spearman correlation analyses between the relative abundance of the dominant bacterial and fungal phyla and soil physicochemical properties
| 全氮 Total nitrogen | 有机碳 Soil organic carbon | 速效磷 Available phosphorus | pH | 阳离子交换量 Cation exchange capacity | ||
|---|---|---|---|---|---|---|
| 细菌 Bacteria | 放线菌门 Actinobacteriota | 0.016 | 0.011 | 0.069 | 0.302 | 0.161 |
| 变形菌门 Proteobacteria | -0.172 | -0.152 | 0.307 | -0.143 | -0.316 | |
| 疣微菌门 Verrucomicrobiota | 0.180 | 0.087 | -0.098 | -0.316 | -0.091 | |
| 绿弯菌门 Chloroflexi | -0.260 | -0.233 | -0.211 | 0.498** | 0.051 | |
| 酸杆菌门 Acidobacteriota | -0.225 | -0.046 | -0.019 | 0.127 | 0.012 | |
| 真菌 Fungi | 担子菌门 Basidiomycota | -0.475** | -0.370* | 0.226 | -0.272 | -0.250 |
| 子囊菌门 Ascomycota | 0.582*** | 0.517** | -0.267 | 0.265 | 0.366* |
| [1] |
Adomako MO, Xue W, Du DL, Yu FH (2022) Soil microbe-mediated N∶P stoichiometric effects on Solidago canadensis performance depend on nutrient levels. Microbial Ecology, 83, 960-970.
DOI |
| [2] |
Baker KL, Langenheder S, Nicol GW, Ricketts D, Killham K, Campbell CD, Prosser JI (2009) Environmental and spatial characterisation of bacterial community composition in soil to inform sampling strategies. Soil Biology and Biochemistry, 41, 2292-2298.
DOI URL |
| [3] | Bao SD (2000) Agriculture Chemical Analysis of Soil. China Agriculture Press, Beijing. (in Chinese) |
| [鲍士旦 (2000) 土壤农化分析. 中国农业出版社, 北京.] | |
| [4] |
Bayranvand M, Akbarinia M, Jouzani GS, Gharechahi J, Kooch Y, Baldrian P (2021) Composition of soil bacterial and fungal communities in relation to vegetation composition and soil characteristics along an altitudinal gradient. FEMS Microbiology Ecology, 97, fiaa201.
DOI URL |
| [5] |
Björnsson L, Hugenholtz P, Tyson GW, Blackall LL (2002) Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology, 148, 2309-2318.
DOI URL |
| [6] |
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo JR, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang LJ, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan YH, Wang MX, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang YL, Zhu QY, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37, 852-857.
DOI PMID |
| [7] | Bueno de Mesquita CP, Sartwell SA, Schmidt SK, Suding KN (2020) Growing-season length and soil microbes influence the performance of a generalist bunchgrass beyond its current range. Ecology, 101, e03095. |
| [8] |
Costello EK, Schmidt SK (2006) Microbial diversity in alpine tundra wet meadow soil: Novel Chloroflexi from a cold, water-saturated environment. Environmental Microbiology, 8, 1471-1486.
PMID |
| [9] |
Dedysh SN, Yilmaz P (2018) Refining the taxonomic structure of the phylum Acidobacteria. International Journal of Systematic and Evolutionary Microbiology, 68, 3796-3806.
DOI PMID |
| [10] | Ding LZ, Man XL, Xiao RH (2019) Characteristics of nitrogen content in rhizosphere soil of main tree species in northern part of Daxinganling during growing seasons. Journal of Central South University of Forestry & Technology, 39, 65-71, 92. (in Chinese with English abstract) |
| [丁令智, 满秀玲, 肖瑞晗 (2019) 大兴安岭北部主要树种生长季根际土壤氮素含量特征. 中南林业科技大学学报, 39, 65-71, 92.] | |
| [11] |
Eldridge DJ, Travers SK, Val J, Ding JY, Wang JT, Singh BK, Delgado-Baquerizo M (2021) Experimental evidence of strong relationships between soil microbial communities and plant germination. Journal of Ecology, 109, 2488-2498.
DOI URL |
| [12] |
Escalas A, Paula FS, Guilhaumon F, Yuan MT, Yang YF, Wu LW, Liu FF, Feng J, Zhang YG, Zhou JZ (2022) Macroecological distributions of gene variants highlight the functional organization of soil microbial systems. The ISME Journal, 16, 726-737.
DOI |
| [13] |
Falkowski PG, Fenchel T, Delong EF (2008) The microbial engines that drive earth’s biogeochemical cycles. Science, 320, 1034-1039.
DOI PMID |
| [14] |
Fierer N (2017) Embracing the unknown: Disentangling the complexities of the soil microbiome. Nature Reviews Microbiology, 15, 579-590.
DOI PMID |
| [15] |
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology, 88, 1354-1364.
DOI PMID |
| [16] | Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proceedings of the National Academy of Sciences, USA, 103, 626-631. |
| [17] |
Fitzpatrick CR, Mustafa Z, Viliunas J (2019) Soil microbes alter plant fitness under competition and drought. Journal of Evolutionary Biology, 32, 438-450.
DOI PMID |
| [18] |
George PBL, Lallias D, Creer S, Seaton FM, Kenny JG, Eccles RM, Griffiths RI, Lebron I, Emmett BA, Robinson DA, Jones DL (2019) Divergent national-scale trends of microbial and animal biodiversity revealed across diverse temperate soil ecosystems. Nature Communications, 10, 1107.
DOI PMID |
| [19] | Gou YN, Nan ZB (2015) The impacts of grazing on the soil microorganism population of grassland. Acta Prataculturae Sinica, 24, 194-205. (in Chinese with English abstract) |
| [苟燕妮, 南志标 (2015) 放牧对草地土壤微生物的影响. 草业学报, 24, 194-205.] | |
| [20] | Gu XN, He HS, Tao Y, Jin YH, Zhang XY, Xu ZW, Wang YT, Song XX (2017) Soil microbial community structure, enzyme activities, and their influencing factors along different altitudes of Changbai Mountain. Acta Ecologica Sinica, 37, 8374-8384. (in Chinese with English abstract) |
| [谷晓楠, 贺红士, 陶岩, 靳英华, 张心昱, 徐志伟, 王钰婷, 宋祥霞 (2017) 长白山土壤微生物群落结构及酶活性随海拔的分布特征与影响因子. 生态学报, 37, 8374-8384.] | |
| [21] |
Guo LD (2012) Progress of microbial species diversity research in China. Biodiversity Science, 20, 572-580. (in Chinese with English abstract)
DOI |
|
[郭良栋 (2012) 中国微生物物种多样性研究进展. 生物多样性, 20, 572-580.]
DOI |
|
| [22] |
Guo QX, Yan LJ, Korpelainen H, Niinemets Ü, Li CY (2019) Plant-plant interactions and N fertilization shape soil bacterial and fungal communities. Soil Biology and Biochemistry, 128, 127-138.
DOI |
| [23] |
Hannula SE, Heinen R, Huberty M, Steinauer K, de Long JR, Jongen R, Bezemer TM (2021) Persistence of plant-mediated microbial soil legacy effects in soil and inside roots. Nature Communications, 12, 5686.
DOI PMID |
| [24] | Hannula SE, Kielak AM, Steinauer K, Huberty M, Jongen R, de Long JR, Heinen R, Bezemer TM (2019) Time after time: Temporal variation in the effects of grass and forb species on soil bacterial and fungal communities. mBio, 10, e02635-19. |
| [25] |
Harris J (2009) Soil microbial communities and restoration ecology: Facilitators or followers? Science, 325, 573-574.
DOI PMID |
| [26] | He J, Tedersoo L, Hu A, Han C, He D, Wei H, Jiao M, Anslan S, Nie Y, Jia Y, Zhang G, Yu G, Liu S, Shen W (2017). Greater diversity of soil fungal communities and distinguishable seasonal variation in temperate deciduous forests compared with subtropical evergreen forests of eastern China. FEMS Microbiology Ecology, 93, fix069. |
| [27] |
Hu L, Cao LX, Zhang RD (2014) Bacterial and fungal taxon changes in soil microbial community composition induced by short-term biochar amendment in red oxidized loam soil. World Journal of Microbiology and Biotechnology, 30, 1085-1092.
DOI PMID |
| [28] |
Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR, Williams KH, Tringe SG, Banfield JF (2013) Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi Phylum and indicate roles in sediment carbon cycling. Microbiome, 1, 22.
DOI PMID |
| [29] |
Jiang S, Xing YJ, Liu GC, Hu CY, Wang XC, Yan GY, Wang QG (2021) Changes in soil bacterial and fungal community composition and functional groups during the succession of boreal forests. Soil Biology and Biochemistry, 161, 108393.
DOI URL |
| [30] | Jiang XW, Ma DL, Zang SY, Zhang DY, Sun HZ (2021) Characteristics of soil bacterial and fungal community of typical forest in the Greater Khingan Mountains based on high-throughput sequencing. Microbiology China, 48, 1093-1105. (in Chinese with English abstract) |
| [姜雪薇, 马大龙, 臧淑英, 张冬有, 孙弘哲 (2021) 高通量测序分析大兴安岭典型森林土壤细菌和真菌群落特征. 微生物学通报, 48, 1093-1105.] | |
| [31] |
Jiao S, Chen WM, Wang JL, Du NN, Li QP, Wei GH (2018) Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome, 6, 146.
DOI PMID |
| [32] |
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. The ISME Journal, 3, 442-453.
DOI |
| [33] |
Kajimoto T, Matsuura Y, Osawa A, Prokushkin AS, Sofronov MA, Abaimov AP (2003) Root system development of Larix gmelinii trees affected by micro-scale conditions of permafrost soils in central Siberia. Plant and Soil, 255, 281-292.
DOI URL |
| [34] |
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology, 75, 5111-5120.
DOI PMID |
| [35] | Li F, Zhou GS, Cao MC (2006) Responses of Larix gmelinii geographical distribution to future climate change: A simulation study. Chinese Journal of Applied Ecology, 17, 2255-2260. (in Chinese with English abstract) |
| [李峰, 周广胜, 曹铭昌 (2006) 兴安落叶松地理分布对气候变化响应的模拟. 应用生态学报, 17, 2255-2260.] | |
| [36] |
Li J, Luo YQ, Shi J (2012) The optimum mixture ratio of larch and birch in terms of biodiversity conservation: A case study in Aershan forest area. Acta Ecologica Sinica, 32, 4943-4949. (in Chinese with English abstract)
DOI URL |
| [李菁, 骆有庆, 石娟 (2012) 基于生物多样性保护的兴安落叶松与白桦最佳混交比例——以阿尔山林区为例. 生态学报, 32, 4943-4949.] | |
| [37] | Li J, Luo YQ, Shi J, Ma LY, Chen C (2011) Niche of main understory populations of Larix gmelinii Rupr forest in A’ershan area. Forest Research, 24, 651-658. (in Chinese with English abstract) |
| [李菁, 骆有庆, 石娟, 马凌云, 陈超 (2011) 阿尔山地区兴安落叶松林下植物种群生态位. 林业科学研究, 24, 651-658.] | |
| [38] |
Liang JL, Liu J, Jia P, Yang TT, Zeng QW, Zhang SC, Liao B, Shu WS, Li JT (2020) Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining. The ISME Journal, 14, 1600-1613.
DOI |
| [39] |
Liang YT, Jiang YJ, Wang F, Wen CQ, Deng Y, Xue K, Qin YJ, Yang YF, Wu LY, Zhou JZ, Sun B (2015) Long-term soil transplant simulating climate change with latitude significantly alters microbial temporal turnover. The ISME Journal, 9, 2561-2572.
DOI |
| [40] | Lin XG, Hu JL (2008) Scientific connotation and ecological service function of soil microbial diversity. Acta Pedologica Sinica, 45, 892-900. (in Chinese with English abstract) |
| [林先贵, 胡君利 (2008) 土壤微生物多样性的科学内涵及其生态服务功能. 土壤学报, 45, 892-900.] | |
| [41] | Liu CL, Zuo WY, Zhao ZY, Qiu LH (2012) Bacterial diversity of different successional stage forest soils in Dinghushan. Acta Microbiologica Sinica, 52, 1489-1496. (in Chinese with English abstract) |
| [柳春林, 左伟英, 赵增阳, 邱礼鸿 (2012) 鼎湖山不同演替阶段森林土壤细菌多样性. 微生物学报, 52, 1489-1496.] | |
| [42] | Liu HW, Li YN, Zhang SH (2014) Isolation of ectomycorrhizal fruiting bodies of Larix gmelinii and screening of culture medium. Inner Mongolia Forestry Investigation and Design, 37(05), 96-97, 118. (in Chinese) |
| [刘宏伟, 李晔男, 张淑华 (2014) 兴安落叶松外生菌根菌子实体组织分离及培养基筛选初探. 内蒙古林业调查设计, 37(05), 96-97, 118.] | |
| [43] | Luo S, Schmid B, de Deyn GB, Yu SX (2018) Soil microbes promote complementarity effects among co‐existing trees through soil nitrogen partitioning. Functional Ecology, 32, 1879-1889. |
| [44] |
McHugh TA, Koch GW, Schwartz E (2014) Minor changes in soil bacterial and fungal community composition occur in response to monsoon precipitation in a semiarid grassland. Microbial Ecology, 68, 370-378.
DOI PMID |
| [45] | Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2020) vegan: Community Ecology Package. R package version 2.5-7. https://CRAN.R-project.org/package=vegan. (accessed on 2022-04-15) |
| [46] |
Ren CJ, Liu WC, Zhao FZ, Zhong ZK, Deng J, Han XH, Yang GH, Feng YZ, Ren GX (2019) Soil bacterial and fungal diversity and compositions respond differently to forest development. CATENA, 181, 104071.
DOI URL |
| [47] | Rinaldi AC, Comandini O, Kuyper TW (2008) Ectomycorrhizal fungal diversity: Separating the wheat from the chaff. Fungal Diversity, 33, 1-45. |
| [48] |
Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. The ISME Journal, 4, 1340-1351.
DOI |
| [49] |
Schimel J (2013) Microbes and global carbon. Nature Climate Change, 3, 867-868.
DOI |
| [50] |
Singh BK, Nunan N, Millard P (2009) Response of fungal, bacterial and ureolytic communities to synthetic sheep urine deposition in a grassland soil. FEMS Microbiology Ecology, 70, 109-117.
DOI PMID |
| [51] | Stevenson A, Hallsworth JE (2014) Water and temperature relations of soil Actinobacteria. Environmental Micro- biology Reports, 6, 744-755. |
| [52] |
Štursová M, Šnajdr J, Cajthaml T, Bárta J, Šantrůčková H, Baldrian P (2014) When the forest dies: The response of forest soil fungi to a bark beetle-induced tree dieback. The ISME Journal, 8, 1920-1931.
DOI |
| [53] |
Štursová M, Žifčáková L, Leigh MB, Burgess R, Baldrian P (2012) Cellulose utilization in forest litter and soil: Identification of bacterial and fungal decomposers. FEMS Microbiology Ecology, 80, 735-746.
DOI PMID |
| [54] |
Su Y, Hu YX, Zi HY, Chen Y, Deng XP, Hu BB, Jiang YL (2022) Contrasting assembly mechanisms and drivers of soil rare and abundant bacterial communities in 22-year continuous and non-continuous cropping systems. Scientific Reports, 12, 3264.
DOI PMID |
| [55] |
Sul WJ, Asuming-Brempong S, Wang Q, Tourlousse DM, Penton CR, Deng Y, Rodrigues JLM, Adiku SGK, Jones JW, Zhou JZ, Cole JR, Tiedje JM (2013) Tropical agricultural land management influences on soil microbial communities through its effect on soil organic carbon. Soil Biology and Biochemistry, 65, 33-38.
DOI URL |
| [56] | Sun YJ, Zhang J, Han AH, Wang XJ, Wang XJ (2007) Biomass and carbon pool of Larix gmelinii young and middle age forest in Xing’an Mountains Inner Mongolia. Acta Ecologica Sinica, 27, 1756-1762. (in Chinese with English abstract) |
| [孙玉军, 张俊, 韩爱惠, 王雪军, 王新杰 (2007) 兴安落叶松(Larix gmelinii)幼中龄林的生物量与碳汇功能. 生态学报, 27, 1756-1762.] | |
| [57] |
Urbanová M, Šnajdr J, Baldrian P (2015) Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biology and Biochemistry, 84, 53-64.
DOI URL |
| [58] |
Walker TWN, Kaiser C, Strasser F, Herbold CW, Leblans NIW, Woebken D, Janssens IA, Sigurdsson BD, Richter A (2018) Microbial temperature sensitivity and biomass change explain soil carbon loss with warming. Nature Climate Change, 8, 885-889.
DOI PMID |
| [59] | Wang K, Zhang Y, Tang Z, Shangguan Z, Chang F, Jia F, Chen Y, He X, Shi W, Deng L (2019) Effects of grassland afforestation on structure and function of soil bacterial and fungal communities. Science of the Total Environment, 2019, 676, 396-406. |
| [60] |
Xu N, Tan GC, Wang HY, Gai XP (2016) Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. European Journal of Soil Biology, 74, 1-8.
DOI URL |
| [61] | Yang LB, Sui X, Zhu GD, Cui FX, Song RQ, Ni HW (2017a) Study on fungal communities characteristics of different Larix gmelinii forest types in cold temperate zone. Journal of Central South University of Forestry & Technology, 37, 76-84. (in Chinese with English abstract) |
| [杨立宾, 隋心, 朱道光, 崔福星, 李金博, 宋瑞清, 倪红伟 (2017a) 大兴安岭兴安落叶松林土壤真菌群落特征研究. 中南林业科技大学学报, 37, 76-84.] | |
| [62] | Yang LB, Zhu DG, Cui FX, Li JB, Song RQ, Ni HW (2017b) Soil microbial community characteristics of the different forest types of Larix gmelinii forest in cold temperate zone. Journal of Northeast Forestry University, 45(09), 66-72. (in Chinese with English abstract) |
| [杨立宾, 朱道光, 崔福星, 李金博, 宋瑞清, 倪红伟 (2017b) 寒温带兴安落叶松林不同林型土壤微生物群落特征. 东北林业大学学报, 45(09), 66-72.] | |
| [63] | Yang Y, Qiu YM, Wang ZB, Qu LY (2020) Effects of different harvest methods on physicochemical properties and microbial community of Larix gmelinii rhizosphere soil. Acta Ecologica Sinica, 40, 7621-7629. (in Chinese with English abstract) |
| [杨寅, 邱钰明, 王中斌, 曲来叶 (2020) 不同主伐方式对兴安落叶松(Larix gmelinii)根际土壤理化性质及微生物群落的影响. 生态学报, 40, 7621-7629.] | |
| [64] | Yu YY, Li MS, Liu XL, Yin WP, Li GF, Mu LQ, Cui XY, Cheng ZC (2020) Soil bacterial community composition and diversity of typical permafrost in Greater Khingan Mountains. Microbiology China, 47, 2759-2770. (in Chinese with English abstract) |
| [余炎炎, 李梦莎, 刘啸林, 尹伟平, 李国富, 穆立蔷, 崔晓阳, 程智超 (2020) 大兴安岭典型永久冻土土壤细菌群落组成和多样性. 微生物学通报, 47, 2759-2770.] | |
| [65] |
Zhang KR, Cheng XL, Shu X, Liu Y, Zhang QF (2018) Linking soil bacterial and fungal communities to vegetation succession following agricultural abandonment. Plant and Soil, 431, 19-36.
DOI |
| [66] | Zhang TT, Wang Q, Du C, Zhang DN, Zhang F, Ma WG, Wang ZK, Li X, Geng ZC (2017) Diversity of ectomycorrhizal fungi associated with Betula albosinensis in Xinjiashan forest region of Qinling Mountains. mycosystema, 36, 851-860. (in Chinese with English abstract) |
| [张彤彤, 王强, 杜璨, 张丹妮, 张帆, 马武功, 王志康, 李鑫, 耿增超 (2017) 秦岭辛家山林区红桦外生菌根真菌多样性. 菌物学报, 36, 851-860.] | |
| [67] |
Zhou JZ, Deng Y, Shen LN, Wen CQ, Yan QY, Ning DL, Qin YJ, Xue K, Wu LY, He ZL, Voordeckers JW, Nostrand JDV, Buzzard V, Michaletz ST, Enquist BJ, Weiser MD, Kaspari M, Waide R, Yang YF, Brown JH (2016) Temperature mediates continental-scale diversity of microbes in forest soils. Nature Communications, 7, 12083.
DOI PMID |
| [68] | Zhu P, Chen RS, Song YX, Liu GX, Chen T, Zhang W (2015) Soil microbial community diversity under four vegetation types in the Qilian Mountains, China. Acta Prataculturae Sinica, 24, 75-84. (in Chinese with English abstract) |
| [69] | [朱平, 陈仁升, 宋耀选, 刘光琇, 陈拓, 张威 (2015) 祁连山不同植被类型土壤微生物群落多样性差异. 草业学报, 24, 75-84.] |
| [1] | 吴晓晴 张美惠 葛苏婷 李漫淑 宋坤 沈国春 达良俊 张健. 上海近自然林重建过程中木本植物物种多样性与地上生物量的时空动态——以闵行区生态岛为例[J]. 生物多样性, 2025, 33(5): 24444-. |
| [2] | 干靓 刘巷序 鲁雪茗 岳星. 全球生物多样性热点地区大城市的保护政策与优化方向[J]. 生物多样性, 2025, 33(5): 24529-. |
| [3] | 曾子轩 杨锐 黄越 陈路遥. 清华大学校园鸟类多样性特征与环境关联[J]. 生物多样性, 2025, 33(5): 24373-. |
| [4] | 周昊, 王茗毅, 张楚格, 肖治术, 欧阳芳. 昆虫旅馆在独栖蜂多样性保护中的现状与挑战[J]. 生物多样性, 2025, 33(5): 24472-. |
| [5] | 臧明月, 刘立, 马月, 徐徐, 胡飞龙, 卢晓强, 李佳琦, 于赐刚, 刘燕. 《昆明-蒙特利尔全球生物多样性框架》下的中国城市生物多样性保护[J]. 生物多样性, 2025, 33(5): 24482-. |
| [6] | 祝晓雨, 王晨灏, 王忠君, 张玉钧. 城市绿地生物多样性研究进展与展望[J]. 生物多样性, 2025, 33(5): 25027-. |
| [7] | 袁琳, 王思琦, 侯静轩. 大都市地区的自然留野:趋势与展望[J]. 生物多样性, 2025, 33(5): 24481-. |
| [8] | 胡敏, 李彬彬, Coraline Goron. 只绿是不够的: 一个生物多样性友好的城市公园管理框架[J]. 生物多样性, 2025, 33(5): 24483-. |
| [9] | 王欣, 鲍风宇. 基于鸟类多样性提升的南滇池国家湿地公园生态修复效果分析[J]. 生物多样性, 2025, 33(5): 24531-. |
| [10] | 明玥, 郝培尧, 谭铃千, 郑曦. 基于城市绿色高质量发展理念的中国城市生物多样性保护与提升研究[J]. 生物多样性, 2025, 33(5): 24524-. |
| [11] | 徐欢, 辛凤飞, 施宏亮, 袁琳, 薄顺奇, 赵欣怡, 邓帅涛, 潘婷婷, 余婧, 孙赛赛, 薛程. 生态修复技术集成应用对长江口北支生境与鸟类多样性提升效果评估[J]. 生物多样性, 2025, 33(5): 24478-. |
| [12] | 谢淦, 宣晶, 付其迪, 魏泽, 薛凯, 雒海瑞, 高吉喜, 李敏. 草地植物多样性无人机调查的物种智能识别模型构建[J]. 生物多样性, 2025, 33(4): 24236-. |
| [13] | 王太, 宋福俊, 张永胜, 娄忠玉, 张艳萍, 杜岩岩. 河西走廊内陆河水系鱼类多样性及资源现状[J]. 生物多样性, 2025, 33(4): 24387-. |
| [14] | 褚晓琳, 张全国. 演化速率假说的实验验证研究进展[J]. 生物多样性, 2025, 33(4): 25019-. |
| [15] | 张浩斌, 肖路, 刘艳杰. 夜间灯光对外来入侵植物和本地植物群落多样性和生长的影响[J]. 生物多样性, 2025, 33(4): 24553-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn