



生物多样性 ›› 2015, Vol. 23 ›› Issue (1): 61-67. DOI: 10.17520/biods.2014189 cstr: 32101.14.biods.2014189
王盼盼, 武永秀, 宋彤彤, 马春玲, 赵文, 王瑛, 孙磊*( )
)
收稿日期:2014-09-15
接受日期:2014-12-30
出版日期:2015-01-20
发布日期:2015-05-04
通讯作者:
孙磊
作者简介:E-mail: sunlei1018@126.com基金资助:
Panpan Wang, Yongxiu Wu, Tongtong Song, Chunling Ma, Wen Zhao, Ying Wang, Lei Sun*( )
)
Received:2014-09-15
Accepted:2014-12-30
Online:2015-01-20
Published:2015-05-04
Contact:
Lei Sun
摘要:
惠兰(Cymbidium faberi)是中国兰属代表种之一, 具有很高的观赏价值和经济价值, 对其内生细菌进行研究不仅可以丰富植物内生细菌资源, 还可以为探讨兰花与微生物之间的相互作用关系提供基础数据。本研究采用分离培养方法及16S rRNA基因序列测定对天目山野生蕙兰、在温室培养1年后的蕙兰根内生细菌遗传多样性进行了研究。结果表明: 从野生蕙兰根内分离得到的97株细菌分属于变形菌门的α-变形菌纲、β-变形菌纲、γ-变形菌纲及厚壁菌门的13个属, 其最优势类群为γ-变形菌纲(86.60%), Lelliottia (26.80%)为最优势菌属。从温室盆栽蕙兰根内分离得到的52株细菌分属于变形菌门的α-变形菌纲、β-变形菌纲、γ-变形菌纲及放线菌门的9个属, 优势类群为β-变形菌纲(48.08%), 优势菌属为草螺菌属(Herbaspirillum) (34.62%), 其中菌株ehR17为潜在的新种。这些结果表明天目山野生蕙兰可培养根内生细菌多样性较其在温室培养1年后更为丰富, 同时也说明植物内生细菌的群落结构与生长环境密切相关。
王盼盼, 武永秀, 宋彤彤, 马春玲, 赵文, 王瑛, 孙磊 (2015) 野生及温室盆栽蕙兰可培养根内生细菌的遗传多样性. 生物多样性, 23, 61-67. DOI: 10.17520/biods.2014189.
Panpan Wang, Yongxiu Wu, Tongtong Song, Chunling Ma, Wen Zhao, Ying Wang, Lei Sun (2015) Genetic diversity of culturable endophytic bacteria in the roots of wild and greenhouse Cymbidium faberi. Biodiversity Science, 23, 61-67. DOI: 10.17520/biods.2014189.
| 类群 Group | 菌株数 Strain numbers | 所占比例 % | 代表菌株 Representative strains | 最相近菌种(登录号) Nearest strain (accession no.) | 相似度 Similarity (%) |
|---|---|---|---|---|---|
| 野生蕙兰 Wild C. faberi | |||||
| α-变形菌纲 Alphaproteobacteria (2.06%) | 2 | ||||
| 1 | 1.03 | 6hN10 | Agrobacterium tumefaciens (AJ389904) | 100.00 | |
| 1 | 1.03 | 6hR78 | Devosia riboflavin (AJ549086) | 99.85 | |
| β-变形菌纲 Betaproteobacteria (4.12%) | 4 | ||||
| 4 | 4.12 | 6hR5 | Burkholderia stabilis (AF148554) | 99.67 | |
| γ-变形菌纲 Gammaproteobacteria (86.60%) | 84 | ||||
| 13 | 13.40 | 5hR3 | Cedecea neteri (AB086230) | 98.58 | |
| 1 | 1.03 | 6hR30 | Enterobacter asburiae LF7a (CP003026) | 99.09 | |
| 4 | 4.12 | 5hR6 | Erwinia rhapontici (ERU80206) | 98.51 | |
| 9 | 9.28 | 6hN2 | Klebsiella michiganensis (JQ070300) | 99.30 | |
| 21 | 21.65 | 6hR10 | Kluyvera cryocrescens (AF310218) | 99.34 | |
| 26 | 26.80 | 6hR36 | Lelliottia amnigena (AB004749) | 99.10 | |
| 2 | 2.06 | 6hR56 | Pantoea septic (EU216734) | 98.86 | |
| 8 | 8.25 | 6hN21 | Serratia plymuthica (AJ233433) | 98.49 | |
| 厚壁菌门 Firmicutes (7.22%) | 7 | ||||
| 1 | 1.03 | 6hR3 | Bacillus luciferensis (AJ419629) | 98.31 | |
| 6 | 6.19 | 6hR75 | Paenibacillus kribbensis (AF391123) | 97.62 | |
| 温室盆栽蕙兰 C. faberi in the greenhouse | |||||
| α-变形菌纲 Alphaproteobacteria (34.62%) | 18 | ||||
| 2 | 3.85 | ehN1 | Novosphingobium rosa (D13945) | 98.26 | |
| 6 | 11.54 | ehN5 | Rhizobium mayense (JX855172) | 100.00 | |
| 9 | 17.31 | ehR13 | Rhizobium multihospitium (EF035074) | 100.00 | |
| 1 | 1.92 | ehR17 | Sphingomonas polyaromaticivorans (EF467848) | 96.52 | |
| β-变形菌纲 Betaproteobacteria (48.08%) | 25 | ||||
| 6 | 11.54 | ehR33 | Burkholderia lata (CP000150) | 99.71 | |
| 18 | 34.62 | ehK14 | Herbaspirillum chlorophenolicum (AB094401) | 99.18 | |
| 1 | 1.92 | ehR14 | Variovorax paradoxus (D88006) | 99.56 | |
| γ-变形菌纲 Gammaproteobacteria (15.38%) | 8 | ||||
| 7 | 13.46 | ehR5 | Dyella koreensis (AY884571) | 99.43 | |
| 1 | 1.92 | ehK21 | Moraxella osloensis (X74897) | 99.86 | |
| 放线菌门 Actinobacteria (1.92%) | 1 | ||||
| 1 | 1.92 | ehT15 | Microbacterium oxydans (Y17227) | 100.00 | |
表1 野生及温室盆栽蕙兰根内生细菌16S rRNA基因序列相似性分析
Table 1 Similarity analysis of the 16S rRNA gene partial sequences of endophytic bacteria from the roots of wild Cymbidium faberi and C. faberi in the greenhouse
| 类群 Group | 菌株数 Strain numbers | 所占比例 % | 代表菌株 Representative strains | 最相近菌种(登录号) Nearest strain (accession no.) | 相似度 Similarity (%) |
|---|---|---|---|---|---|
| 野生蕙兰 Wild C. faberi | |||||
| α-变形菌纲 Alphaproteobacteria (2.06%) | 2 | ||||
| 1 | 1.03 | 6hN10 | Agrobacterium tumefaciens (AJ389904) | 100.00 | |
| 1 | 1.03 | 6hR78 | Devosia riboflavin (AJ549086) | 99.85 | |
| β-变形菌纲 Betaproteobacteria (4.12%) | 4 | ||||
| 4 | 4.12 | 6hR5 | Burkholderia stabilis (AF148554) | 99.67 | |
| γ-变形菌纲 Gammaproteobacteria (86.60%) | 84 | ||||
| 13 | 13.40 | 5hR3 | Cedecea neteri (AB086230) | 98.58 | |
| 1 | 1.03 | 6hR30 | Enterobacter asburiae LF7a (CP003026) | 99.09 | |
| 4 | 4.12 | 5hR6 | Erwinia rhapontici (ERU80206) | 98.51 | |
| 9 | 9.28 | 6hN2 | Klebsiella michiganensis (JQ070300) | 99.30 | |
| 21 | 21.65 | 6hR10 | Kluyvera cryocrescens (AF310218) | 99.34 | |
| 26 | 26.80 | 6hR36 | Lelliottia amnigena (AB004749) | 99.10 | |
| 2 | 2.06 | 6hR56 | Pantoea septic (EU216734) | 98.86 | |
| 8 | 8.25 | 6hN21 | Serratia plymuthica (AJ233433) | 98.49 | |
| 厚壁菌门 Firmicutes (7.22%) | 7 | ||||
| 1 | 1.03 | 6hR3 | Bacillus luciferensis (AJ419629) | 98.31 | |
| 6 | 6.19 | 6hR75 | Paenibacillus kribbensis (AF391123) | 97.62 | |
| 温室盆栽蕙兰 C. faberi in the greenhouse | |||||
| α-变形菌纲 Alphaproteobacteria (34.62%) | 18 | ||||
| 2 | 3.85 | ehN1 | Novosphingobium rosa (D13945) | 98.26 | |
| 6 | 11.54 | ehN5 | Rhizobium mayense (JX855172) | 100.00 | |
| 9 | 17.31 | ehR13 | Rhizobium multihospitium (EF035074) | 100.00 | |
| 1 | 1.92 | ehR17 | Sphingomonas polyaromaticivorans (EF467848) | 96.52 | |
| β-变形菌纲 Betaproteobacteria (48.08%) | 25 | ||||
| 6 | 11.54 | ehR33 | Burkholderia lata (CP000150) | 99.71 | |
| 18 | 34.62 | ehK14 | Herbaspirillum chlorophenolicum (AB094401) | 99.18 | |
| 1 | 1.92 | ehR14 | Variovorax paradoxus (D88006) | 99.56 | |
| γ-变形菌纲 Gammaproteobacteria (15.38%) | 8 | ||||
| 7 | 13.46 | ehR5 | Dyella koreensis (AY884571) | 99.43 | |
| 1 | 1.92 | ehK21 | Moraxella osloensis (X74897) | 99.86 | |
| 放线菌门 Actinobacteria (1.92%) | 1 | ||||
| 1 | 1.92 | ehT15 | Microbacterium oxydans (Y17227) | 100.00 | |
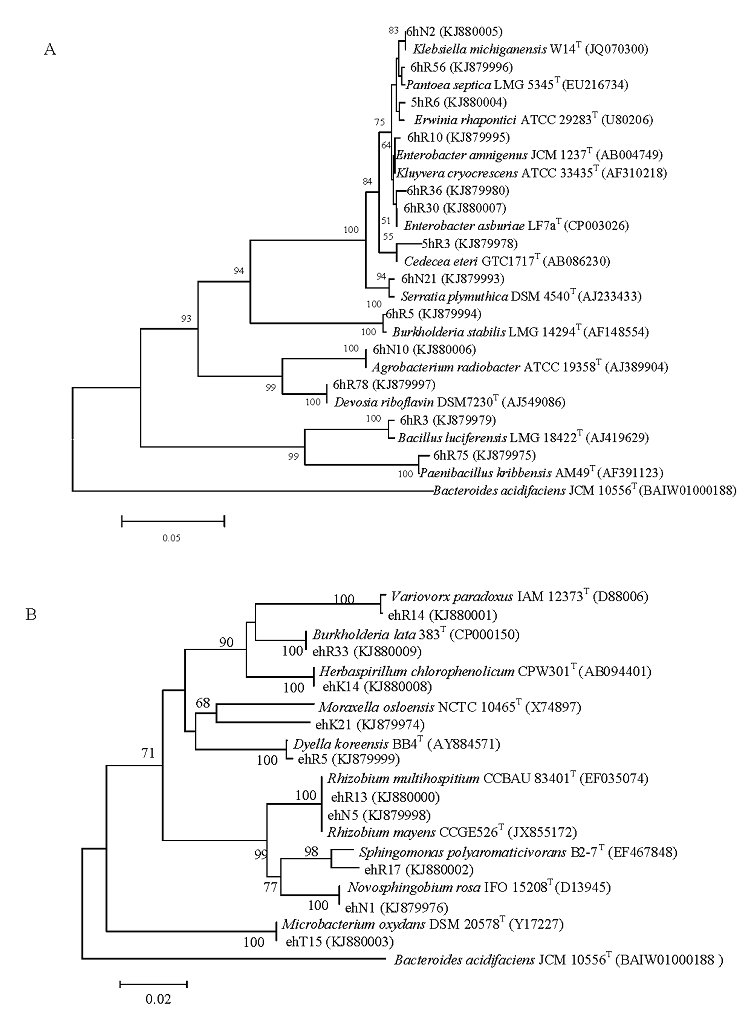
图1 蕙兰根内生细菌16S rRNA基因序列系统发育树。(A)野生蕙兰; (B)温室盆栽蕙兰。
Fig. 1 A dendrogram based on the 16S rRNA gene partial sequences of endophytic bacteria. GenBank accession numbers are given in parentheses. Numbers at the nodes indicate the bootstrap values (>50%) based on 1,000 replicates. (A) A dendrogram of roots endophytic bacteria of wilid Cymbidium faberi. (B) A dendrogram of roots endophytic bacteria of C. faberi in the greenhouse.
| 类群 Group | 属 Genera | |
|---|---|---|
| 野生蕙兰 Wild | 温室盆栽蕙兰 In the greenhouse | |
| Alphaproteobacteria | Agrobacterium | Novosphingobium |
| Devosia | Rhizobium | |
| - | Sphingomonas | |
| Betaproteobacteria | Burkholderia | Burkholderia |
| - | Herbaspirillum | |
| - | Variovorax | |
| Gammaproteobacteria | Cedecea | Dyella |
| Enterobacter | Moraxella | |
| Erwinia | - | |
| Klebsiella | - | |
| Kluyvera | - | |
| Lelliottia | - | |
| Pantoea | - | |
| Serratia | - | |
| Firmicutes | Bacillus | - |
| Paenibacillus | - | |
| Actinobacteria | - | Microbacterium |
表2 野生蕙兰和温室盆栽蕙兰根内生细菌群落结构比较
Table 2 Comparison of endophytic bacteria community struc- tures in the roots from wild Cymbidium faberi and C. faberi in the greenhouse
| 类群 Group | 属 Genera | |
|---|---|---|
| 野生蕙兰 Wild | 温室盆栽蕙兰 In the greenhouse | |
| Alphaproteobacteria | Agrobacterium | Novosphingobium |
| Devosia | Rhizobium | |
| - | Sphingomonas | |
| Betaproteobacteria | Burkholderia | Burkholderia |
| - | Herbaspirillum | |
| - | Variovorax | |
| Gammaproteobacteria | Cedecea | Dyella |
| Enterobacter | Moraxella | |
| Erwinia | - | |
| Klebsiella | - | |
| Kluyvera | - | |
| Lelliottia | - | |
| Pantoea | - | |
| Serratia | - | |
| Firmicutes | Bacillus | - |
| Paenibacillus | - | |
| Actinobacteria | - | Microbacterium |
| 1 |
Ardanov P, Ovcharenko L, Zaets I, Kozyrovska N, Pirttilä AM ( 2011) Endophytic bacteria enhancing growth and disease resistance of potato (Solanum tuberosum L.). Biological Control, 56, 43-49.
DOI URL |
| 2 |
Bulgari D, Bozkurt AI, Casati P, Çağlayan K, Quaglino F, Bianco PA ( 2012) Endophytic bacterial community living in roots of healthy and ‘Candidatus Phytoplasma mali’-infected apple (Malus domestica, Borkh.) trees. Antonie van Leeuwenhoek, 102, 677-687.
DOI URL PMID |
| 3 |
Catalán AI, Ferreira F, Gill PR, Batista S ( 2007) Production of polyhydroxyalkanoates by Herbaspirillum seropedicae grown with different sole carbon sources and on lactose when engineered to express the lacZlacY genes. Enzyme and Microbial Technology, 40, 1352-1367.
DOI URL |
| 4 | Chu XL ( 褚晓玲), Yang B ( 杨波), Gao L ( 高丽), Li HL ( 李洪林), Hu L ( 胡磊), Yang N ( 杨娜 ) ( 2010) Species diversity of cultivable bacteria isolated from the roots of Cymbidium faberi Rolfe. Journal of Wuhan Botanical Research (武汉植物学研究), 28, 199-205. (in Chinese with English abstract) |
| 5 |
Compant S, Clément C, Sessitsch A ( 2010) Colonization of plant growth-promoting bacteria in the rhizo- and endosphere of plants: importance, mechanisms involved and future prospects. Soil Biology and Biochemistry, 42, 669-678.
DOI URL |
| 6 |
Edwards U, Rogall T, Blöker H, Emde M, Böttger EC ( 1989) Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Research, 17, 7843-7853.
DOI URL PMID |
| 7 |
Germaine KJ, Liu X, Cabellos GG, Hogan JP, Ryan D, Dowling DN ( 2006) Bacterial endophyte-enhanced phytoremediation of the organochlorine herbicide 2,4-dichloro- phenoxyacetic acid. FEMS Microbiology Ecology, 57, 302-310.
DOI URL PMID |
| 8 | Huang G ( 黄盖), Gao H ( 高焓), Wang C ( 王琛), Zhao Q ( 赵倩), Zhang WF ( 张文峰), Dang J ( 党娟), Ma XT ( 马晓棠), Yan X ( 颜霞), Gao JP ( 高建平 ) ( 2013) ACC 30 strain with ACC deaminase activity: its isolation identification and growth-promoting effect. Microbiology China (微生物学通报), 40, 812-821. (in Chinese with English abstract) |
| 9 | Kado CI ( 1991) Plant pathogenic bacteria. In: The Prokaryotes (eds Balows A, Truper HG, Dworkin M, Schleifer KH), pp. 659-674. Springer-Verlag, New York. |
| 10 | Lane DJ ( 1991) 16S/23S rRNA sequencing. In: Nucleic Acid Techniques in Bacterial Systematics (eds Stackebrandt E, Goodfellow M), pp. 115-175. John Wiley & Sons, Chichester, UK. |
| 11 | Liu L ( 刘琳), Sun L ( 孙磊), Zhang RY ( 张瑞英), Yao N ( 姚娜), Li LB ( 李潞滨 ) ( 2010) Diversity of IAA-producing endophytic bacteria isolated from the roots of Cymbidium goeringii. Biodiversity Science (生物多样性), 18, 182-187. (in Chinese with English abstract) |
| 12 |
Luo HF, Qi HY, Zhang HX ( 2004) Assessment of the bacterial diversity in fenvalerate-treated soil. World Journal of Microbiology and biotechnology, 20, 509-515.
DOI URL |
| 13 |
Mehnaz S, Baig DN, Lazarovits G ( 2010) Genetic and phenotypic diversity of plant growth promoting rhizobacteria isolated from sugarcane plants growing in Pakistan. Journal of Microbiology and Biotechnology, 20, 1614-1623.
DOI URL PMID |
| 14 |
Rincón-Rosales R, Villalobos-Escobedo JM, Rogel MA, Mart- inez J, Ormeño-Orrillo E, Martínez-Romero E ( 2013) Rhizobium calliandrae sp. nov., Rhizobium mayense sp. nov. and Rhizobium jaguaris sp. nov., rhizobial species nodulating the medicinal legume Calliandra grandiflora. International Journal of Systematic and Evolutionary Microbiology, 63, 3423-3429.
DOI URL PMID |
| 15 |
Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN ( 2008) Bacterial endophytes: recent developments and applications. FEMS Microbiology Letters, 278, 1-9.
DOI URL PMID |
| 16 |
Serrato RV, Sassaki GL, Cruz LM, Carlson RW, Muszyński A, Monteiro RA, Pedrosa FO, Souza EM, Iacomini M ( 2010) Chemical composition of lipopolysaccharides isolated from various endophytic nitrogen-fixing bacteria of the genus Herbaspirillum. Canadian Journal of Microbiology, 56, 342-347.
DOI URL PMID |
| 17 |
Stępniewska Z, Kuźniar A ( 2013) Endophytic microorganisms—promising applications in bioremediation of greenhouse gases. Applied Microbiology Biotechnology, 97, 9589-9596.
DOI URL PMID |
| 18 |
Sun L ( 孙磊), Shao H ( 邵红), Liu L ( 刘琳), Zhang RY ( 张瑞英), Zhao LH ( 赵立华), Li LB ( 李潞滨), Yao N ( 姚娜 ) ( 2011) Diversity of siderophore-producing endophytic bacteria of Cymbidium goeringii roots. Acta Microbiologica Sinica (微生物学报), 51, 189-195. (in Chinese with English abstract)
URL PMID |
| 19 | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S ( 2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 10, 2731-2739. |
| 20 |
Tamura K, Dudley J, Nei M, Kumar S ( 2011) Study on mechanisms of colonization of nitrogen-fixing PGPB, Klebsiella pneumoniae NG14 on the root surface of rice and the formation of biofilm. Current Microbiology, 62, 1113-1122.
DOI URL PMID |
| 21 |
Von Bulow JFW, Döbereiner J ( 1975) Potential for nitrogen-fixation in maize genotypes in Brazil. Proceedings of the National Academy of Sciences, USA, 72, 2389-2393.
DOI URL |
| 22 |
Wang MX, Liu J, Chen SL, Yan SZ ( 2012) Isolation, characterization and colonization of 1-aminocyclopropane-1- carboxylate deaminase-producing bacteria XG32 and DP24. World Journal of Microbiology and Biotechnology, 28, 1155-1162.
DOI URL PMID |
| 23 |
Wilkinson HH, Siegel MR, Blankenship JD, Mallory AC, Bush LP, Schardl CL ( 2000) Contribution of fungal loline alkaloids to protection from aphids in a grass-endophyte mutualism. Molecular Plant-Microbe Interactions, 13, 1027-1033.
DOI URL PMID |
| 24 | Wu YX ( 武永秀), Song TT ( 宋彤彤), Zhang RY ( 张瑞英), Liu L ( 刘亮), Shao H ( 邵红), Li LB ( 李潞滨), Sun L ( 孙磊 ) ( 2014) Diversity of culturable endophytic bacteria in the roots of Cymbidium goeringii from Tianmu Mountain. Biotechnology Bulletin (生物技术通报), 5, 170-173. (in Chinese with English abstract) |
| 25 | Yang N ( 杨娜), Yang B ( 杨波 ) ( 2011) Diversity and seasonal fluctuations of endophytic bacteria isolated from the root tissues of Cymbidum faberi. Plant Science Journal (植物科学学报), 29, 156-163. (in Chinese with English abstract) |
| [1] | 吴晓晴 张美惠 葛苏婷 李漫淑 宋坤 沈国春 达良俊 张健. 上海近自然林重建过程中木本植物物种多样性与地上生物量的时空动态——以闵行区生态岛为例[J]. 生物多样性, 2025, 33(5): 24444-. |
| [2] | 干靓 刘巷序 鲁雪茗 岳星. 全球生物多样性热点地区大城市的保护政策与优化方向[J]. 生物多样性, 2025, 33(5): 24529-. |
| [3] | 曾子轩 杨锐 黄越 陈路遥. 清华大学校园鸟类多样性特征与环境关联[J]. 生物多样性, 2025, 33(5): 24373-. |
| [4] | 周昊, 王茗毅, 张楚格, 肖治术, 欧阳芳. 昆虫旅馆在独栖蜂多样性保护中的现状与挑战[J]. 生物多样性, 2025, 33(5): 24472-. |
| [5] | 臧明月, 刘立, 马月, 徐徐, 胡飞龙, 卢晓强, 李佳琦, 于赐刚, 刘燕. 《昆明-蒙特利尔全球生物多样性框架》下的中国城市生物多样性保护[J]. 生物多样性, 2025, 33(5): 24482-. |
| [6] | 祝晓雨, 王晨灏, 王忠君, 张玉钧. 城市绿地生物多样性研究进展与展望[J]. 生物多样性, 2025, 33(5): 25027-. |
| [7] | 袁琳, 王思琦, 侯静轩. 大都市地区的自然留野:趋势与展望[J]. 生物多样性, 2025, 33(5): 24481-. |
| [8] | 胡敏, 李彬彬, Coraline Goron. 只绿是不够的: 一个生物多样性友好的城市公园管理框架[J]. 生物多样性, 2025, 33(5): 24483-. |
| [9] | 王欣, 鲍风宇. 基于鸟类多样性提升的南滇池国家湿地公园生态修复效果分析[J]. 生物多样性, 2025, 33(5): 24531-. |
| [10] | 明玥, 郝培尧, 谭铃千, 郑曦. 基于城市绿色高质量发展理念的中国城市生物多样性保护与提升研究[J]. 生物多样性, 2025, 33(5): 24524-. |
| [11] | 徐欢, 辛凤飞, 施宏亮, 袁琳, 薄顺奇, 赵欣怡, 邓帅涛, 潘婷婷, 余婧, 孙赛赛, 薛程. 生态修复技术集成应用对长江口北支生境与鸟类多样性提升效果评估[J]. 生物多样性, 2025, 33(5): 24478-. |
| [12] | 谢淦, 宣晶, 付其迪, 魏泽, 薛凯, 雒海瑞, 高吉喜, 李敏. 草地植物多样性无人机调查的物种智能识别模型构建[J]. 生物多样性, 2025, 33(4): 24236-. |
| [13] | 王太, 宋福俊, 张永胜, 娄忠玉, 张艳萍, 杜岩岩. 河西走廊内陆河水系鱼类多样性及资源现状[J]. 生物多样性, 2025, 33(4): 24387-. |
| [14] | 褚晓琳, 张全国. 演化速率假说的实验验证研究进展[J]. 生物多样性, 2025, 33(4): 25019-. |
| [15] | 张浩斌, 肖路, 刘艳杰. 夜间灯光对外来入侵植物和本地植物群落多样性和生长的影响[J]. 生物多样性, 2025, 33(4): 24553-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn