



生物多样性 ›› 2010, Vol. 18 ›› Issue (5): 509-515. DOI: 10.3724/SP.J.1003.2010.509 cstr: 32101.14.SP.J.1003.2010.509
陈碧云1, 胡琼1, Christina Dixelius2, 李国庆3, 伍晓明1,*( )
)
收稿日期:2010-02-25
接受日期:2010-07-27
出版日期:2010-09-20
发布日期:2010-09-20
通讯作者:
伍晓明
作者简介:* E-mail: wuxm@oilcrops.cn基金资助:
Biyun Chen1, Qiong Hu1, Christina Dixelius2, Guoqing Li3, Xiaoming Wu1,*( )
)
Received:2010-02-25
Accepted:2010-07-27
Online:2010-09-20
Published:2010-09-20
Contact:
Xiaoming Wu
摘要:
核盘菌(Sclerotinia sclerotiorum)是危害油菜等多种经济作物的重要病原真菌。研究不同地区、相同或不同寄主核盘菌的遗传多样性对了解核盘菌的遗传演化过程和指导病害防控具有重要意义。我们采用序列相关扩增多态性(sequence-related amplified polymorphism, SRAP)标记对不同地理来源、不同寄主来源的76个核盘菌菌株的遗传多样性进行了分析。7对SRAP引物共获得260个位点, 其中114个为多态位点, 占43.85%。UPGMA聚类结果显示, 在相似性系数为0.64时, 76个核盘菌菌株分为4个组, 每组包含的菌株数分别为54、18、2和2。聚类及主成分分析结果显示, 来源于春油菜生态区和冬油菜生态区油菜上的核盘菌菌株可以明显分为两簇, 而油菜、大豆、莴苣等不同寄主植物上的核盘菌菌株没有明显的遗传分化。分子变异分析(AMOVA)结果显示, 不同地理来源、不同油菜生态区和不同寄主来源的核盘菌群体内的变异率分别为75.2%、81.2%和97.6%, 均达到极显著水平(P<0.001); 不同地理来源和不同油菜生态区的核盘菌群体间的变异率分别为24.8%和18.8%, 也达到极显著水平(P<0.001); 不同寄主来源的核盘菌群体间的变异率仅为2.4%, 变异不显著(P = 0.8673)。研究结果表明, 来源于春油菜生态区的核盘菌的遗传多样性高于冬油菜生态区。
陈碧云, 胡琼, Christina Dixelius, 李国庆, 伍晓明 (2010) 利用SRAP分析核盘菌遗传多样性. 生物多样性, 18, 509-515. DOI: 10.3724/SP.J.1003.2010.509.
Biyun Chen, Qiong Hu, Christina Dixelius, Guoqing Li, Xiaoming Wu (2010) Genetic diversity in Sclerotinia sclerotiorum assessed with SRAP markers. Biodiversity Science, 18, 509-515. DOI: 10.3724/SP.J.1003.2010.509.
| 编号 No. | 菌株名称 Isolate | 寄主 Host | 来源 Origin | 编号 No. | 菌株名称 Isolate | 寄主 Host | 来源 Origin | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RSC-1 | 油菜 Rape | 陕西勉县 Mianxian, Shaanxi | 31 | F5-219-S2 clone 39 | 大豆 Soybean | 加拿大 Canada | ||||||
| 2 | RSC-2 | 油菜 Rape | 陕西咸阳 Xianyang, Shaanxi | 32 | F5-138-L4 clone 321 | 大豆 Soybean | 加拿大 Canada | ||||||
| 3 | RSC-3 | 油菜 Rape | 陕西大荔 Dali, Shaanxi | 33 | LMK211 clone 2 | 油菜 Rape | 加拿大 Canada | ||||||
| 4 | RSC-4 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 34 | CW312 clone 1 | 大豆 Soybean | 加拿大 Canada | ||||||
| 5 | RSC-5 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 35 | CW317 clone 2 | 大豆 Soybean | 加拿大 Canada | ||||||
| 6 | RSC-6 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 36 | SC-13 Sweden | 油菜 Rape | 瑞典 Sweden | ||||||
| 7 | RSC-7 | 油菜 Rape | 湖北襄樊 Xiangfan, Hubei | 37 | IC16 | 油菜 Rape | 挪威 Norway | ||||||
| 8 | RSC-8 | 油菜 Rape | 河南确山 Queshan, Henan | 38 | IC6 | 油菜 Rape | 挪威 Norway | ||||||
| 9 | RSC-9 | 油菜 Rape | 湖南汉寿 Hanshou, Hunan | 39 | 824 clone 1 | 油菜 Rape | 加拿大 Canada | ||||||
| 10 | RSC-10 | 油菜 Rape | 河南遂平 Suiping, Henan | 40 | Xbz-4 | 小白菜 Pakchoi | 湖北罗田 Luotian, Hubei | ||||||
| 11 | RSC-11 | 油菜 Rape | 河南信阳 Xinyang, Henan | 41 | Xbz-5 | 小白菜 Pakchoi | 湖北十堰 Shiyan, Hubei | ||||||
| 12 | RSC-12 | 油菜 Rape | 河南郑州 Zhengzhou, Henan | 42 | Xbz-9 | 小白菜 Pakchoi | 湖北洪湖 Honghu, Hubei | ||||||
| 13 | RSC-13 | 油菜 Rape | 河南郑州 Zhengzhou, Henan | 43 | Sb-2 | 大豆 Soybean | 黑龙江 Heilongjiang | ||||||
| 14 | RSC-14 | 油菜 Rape | 江西宜春 Yichun, Jiangxi | 44 | Sb-4 | 大豆 Soybean | 美国 USA | ||||||
| 15 | RSC-15 | 油菜 Rape | 浙江杭州 Hangzhou, Zhejiang | 45 | Sb-6 | 大豆 Soybean | 加拿大 Canada | ||||||
| 16 | RSC-16 | 油菜 Rape | 浙江杭州 Hangzhou, Zhejiang | 46 | Cor-3 | 芫荽 Coriander | 湖北神农架 Shennongjia, Hubei | ||||||
| 17 | RSC-17 | 油菜 Rape | 贵州思南 Sinan, Guizhou | 47 | Cor-6 | 芫荽 Coriander | 湖北通城 Tongcheng, Hubei | ||||||
| 18 | RSC-18 | 油菜 Rape | 湖北罗田 Luotian, Hubei | 48 | Brb-1 | 蚕豆 Horsebean | 湖北武汉 Wuhan, Hubei | ||||||
| 19 | RSC-19 | 油菜 Rape | 湖北英山 Yingshan, Hubei | 49 | Brb-9 | 蚕豆 Horsebean | 湖北通城 Tongcheng, Hubei | ||||||
| 20 | RSC-20 | 油菜 Rape | 江苏南京 Nanjing, Jiangsu | 50 | Rap-1 | 油菜 Rape | 湖北云梦 Yunmeng, Hubei | ||||||
| 21 | RSC-21 | 油菜 Rape | 江苏南京 Nanjing, Jiangsu | 51 | Rap-2 | 油菜 Rape | 湖北沙市 Shashi, Hubei | ||||||
| 22 | RSC-22 | 油菜 Rape | 江西南昌 Nanchang, Jiangxi | 52 | Rap-6 | 油菜 Rape | 四川雅安 Yaan, Sichuan | ||||||
| 23 | RSC-23 | 油菜 Rape | 四川绵阳 Mianyang, Sichuan | 53 | Rap-20 | 油菜 Rape | 湖北罗田 Luotian, Hubei | ||||||
| 24 | RSC-24 | 油菜 Rape | 陕西杨凌 Yangling, Shaanxi | 54 | Rap-21 | 油菜 Rape | 湖北宣恩 Xuanen, Hubei | ||||||
| 25 | RSC-25 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 55 | Rap-31 | 油菜 Rape | 湖北宜昌 Yichang, Hubei | ||||||
| 26 | RSC-26 | 油菜 Rape | 黑龙江佳木斯 Jiamusi, Heilongjiang | 56 | Rap-35 | 油菜 Rape | 湖北五峰 Wufeng, Hubei | ||||||
| 27 | RSC-27 | 油菜 Rape | 青海西宁 Xining, Qinghai | 57 | Rap-36 | 油菜 Rape | 英国 Britain | ||||||
| 28 | RSC-28 | 油菜 Rape | 青海西宁 Xining, Qinghai | 58 | Rap-37 | 油菜 Rape | 加拿大 Canada | ||||||
| 29 | RSC-29 | 油菜 Rape | 青海西宁 Xining, Qinghai | 59 | Hzd-3 | 小白菜 Pakchoi | 湖北仙桃 Xiantao, Hubei | ||||||
| 30 | RSC-30 | 油菜 Rape | 青海西宁 Xining, Qinghai | 60 | Hzd-4 | 小白菜 Pakchoi | 湖北黄梅 Huangmei, Hubei | ||||||
| 编号 No. | 菌株名称 Isolate | 寄主 Host | 来源 Origin | 编号 No. | 菌株名称 Isolate | 寄主 Host | 来源 Origin | ||||||
| 61 | Nic-1 | 烟草 Tobacco | 湖北咸丰 Xianfeng, Hubei | 69 | Let-46 | 莴苣 Lettuce | 湖北神农架 Shennongjia, Hubei | ||||||
| 62 | Zxd-1 | 红豆 Red bean | 内蒙古扎兰屯 Zalanten, Inner Monglia | 70 | Brb-8 | 蚕豆 Horsebean | 湖北通山 Tongshan, Hubei | ||||||
| 63 | Rad-1 | 萝卜 Radish | 湖北竹山 Zhushan, Hubei | 71 | Let-9 | 莴苣 Lettuce | 湖北五峰 Wufeng, Hubei | ||||||
| 64 | clone-22 | 油菜 Rape | 加拿大 Canada | 72 | Let-3 | 莴苣 Lettuce | 湖北武汉 Wuhan, Hubei | ||||||
| 65 | UK-2 | 不清 Unknown | 湖北武汉 Wuhan, Hubei | 73 | Let-27 | 莴苣 Lettuce | 台湾 Taiwan | ||||||
| 66 | Can02-6 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 74 | Let-11 | 莴苣 Lettuce | 湖北宜昌 Yichang, Hubei | ||||||
| 67 | Cor-10 | 芫荽 Coriander | 湖北宜昌 Yichang, Hubei | 75 | Can02-22 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | ||||||
| 68 | Rad-30-2 | 萝卜 Radish | 湖北神农架 Shennongjia, Hubei | 76 | Let-26 | 莴苣 Lettuce | 加拿大 Canada | ||||||
附录Ⅰ 76个核盘菌菌株、寄主及来源
Appendix I Name, host and origin of 76 Sclerotinia sclerotiorum isolates in this study
| 编号 No. | 菌株名称 Isolate | 寄主 Host | 来源 Origin | 编号 No. | 菌株名称 Isolate | 寄主 Host | 来源 Origin | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RSC-1 | 油菜 Rape | 陕西勉县 Mianxian, Shaanxi | 31 | F5-219-S2 clone 39 | 大豆 Soybean | 加拿大 Canada | ||||||
| 2 | RSC-2 | 油菜 Rape | 陕西咸阳 Xianyang, Shaanxi | 32 | F5-138-L4 clone 321 | 大豆 Soybean | 加拿大 Canada | ||||||
| 3 | RSC-3 | 油菜 Rape | 陕西大荔 Dali, Shaanxi | 33 | LMK211 clone 2 | 油菜 Rape | 加拿大 Canada | ||||||
| 4 | RSC-4 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 34 | CW312 clone 1 | 大豆 Soybean | 加拿大 Canada | ||||||
| 5 | RSC-5 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 35 | CW317 clone 2 | 大豆 Soybean | 加拿大 Canada | ||||||
| 6 | RSC-6 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 36 | SC-13 Sweden | 油菜 Rape | 瑞典 Sweden | ||||||
| 7 | RSC-7 | 油菜 Rape | 湖北襄樊 Xiangfan, Hubei | 37 | IC16 | 油菜 Rape | 挪威 Norway | ||||||
| 8 | RSC-8 | 油菜 Rape | 河南确山 Queshan, Henan | 38 | IC6 | 油菜 Rape | 挪威 Norway | ||||||
| 9 | RSC-9 | 油菜 Rape | 湖南汉寿 Hanshou, Hunan | 39 | 824 clone 1 | 油菜 Rape | 加拿大 Canada | ||||||
| 10 | RSC-10 | 油菜 Rape | 河南遂平 Suiping, Henan | 40 | Xbz-4 | 小白菜 Pakchoi | 湖北罗田 Luotian, Hubei | ||||||
| 11 | RSC-11 | 油菜 Rape | 河南信阳 Xinyang, Henan | 41 | Xbz-5 | 小白菜 Pakchoi | 湖北十堰 Shiyan, Hubei | ||||||
| 12 | RSC-12 | 油菜 Rape | 河南郑州 Zhengzhou, Henan | 42 | Xbz-9 | 小白菜 Pakchoi | 湖北洪湖 Honghu, Hubei | ||||||
| 13 | RSC-13 | 油菜 Rape | 河南郑州 Zhengzhou, Henan | 43 | Sb-2 | 大豆 Soybean | 黑龙江 Heilongjiang | ||||||
| 14 | RSC-14 | 油菜 Rape | 江西宜春 Yichun, Jiangxi | 44 | Sb-4 | 大豆 Soybean | 美国 USA | ||||||
| 15 | RSC-15 | 油菜 Rape | 浙江杭州 Hangzhou, Zhejiang | 45 | Sb-6 | 大豆 Soybean | 加拿大 Canada | ||||||
| 16 | RSC-16 | 油菜 Rape | 浙江杭州 Hangzhou, Zhejiang | 46 | Cor-3 | 芫荽 Coriander | 湖北神农架 Shennongjia, Hubei | ||||||
| 17 | RSC-17 | 油菜 Rape | 贵州思南 Sinan, Guizhou | 47 | Cor-6 | 芫荽 Coriander | 湖北通城 Tongcheng, Hubei | ||||||
| 18 | RSC-18 | 油菜 Rape | 湖北罗田 Luotian, Hubei | 48 | Brb-1 | 蚕豆 Horsebean | 湖北武汉 Wuhan, Hubei | ||||||
| 19 | RSC-19 | 油菜 Rape | 湖北英山 Yingshan, Hubei | 49 | Brb-9 | 蚕豆 Horsebean | 湖北通城 Tongcheng, Hubei | ||||||
| 20 | RSC-20 | 油菜 Rape | 江苏南京 Nanjing, Jiangsu | 50 | Rap-1 | 油菜 Rape | 湖北云梦 Yunmeng, Hubei | ||||||
| 21 | RSC-21 | 油菜 Rape | 江苏南京 Nanjing, Jiangsu | 51 | Rap-2 | 油菜 Rape | 湖北沙市 Shashi, Hubei | ||||||
| 22 | RSC-22 | 油菜 Rape | 江西南昌 Nanchang, Jiangxi | 52 | Rap-6 | 油菜 Rape | 四川雅安 Yaan, Sichuan | ||||||
| 23 | RSC-23 | 油菜 Rape | 四川绵阳 Mianyang, Sichuan | 53 | Rap-20 | 油菜 Rape | 湖北罗田 Luotian, Hubei | ||||||
| 24 | RSC-24 | 油菜 Rape | 陕西杨凌 Yangling, Shaanxi | 54 | Rap-21 | 油菜 Rape | 湖北宣恩 Xuanen, Hubei | ||||||
| 25 | RSC-25 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 55 | Rap-31 | 油菜 Rape | 湖北宜昌 Yichang, Hubei | ||||||
| 26 | RSC-26 | 油菜 Rape | 黑龙江佳木斯 Jiamusi, Heilongjiang | 56 | Rap-35 | 油菜 Rape | 湖北五峰 Wufeng, Hubei | ||||||
| 27 | RSC-27 | 油菜 Rape | 青海西宁 Xining, Qinghai | 57 | Rap-36 | 油菜 Rape | 英国 Britain | ||||||
| 28 | RSC-28 | 油菜 Rape | 青海西宁 Xining, Qinghai | 58 | Rap-37 | 油菜 Rape | 加拿大 Canada | ||||||
| 29 | RSC-29 | 油菜 Rape | 青海西宁 Xining, Qinghai | 59 | Hzd-3 | 小白菜 Pakchoi | 湖北仙桃 Xiantao, Hubei | ||||||
| 30 | RSC-30 | 油菜 Rape | 青海西宁 Xining, Qinghai | 60 | Hzd-4 | 小白菜 Pakchoi | 湖北黄梅 Huangmei, Hubei | ||||||
| 编号 No. | 菌株名称 Isolate | 寄主 Host | 来源 Origin | 编号 No. | 菌株名称 Isolate | 寄主 Host | 来源 Origin | ||||||
| 61 | Nic-1 | 烟草 Tobacco | 湖北咸丰 Xianfeng, Hubei | 69 | Let-46 | 莴苣 Lettuce | 湖北神农架 Shennongjia, Hubei | ||||||
| 62 | Zxd-1 | 红豆 Red bean | 内蒙古扎兰屯 Zalanten, Inner Monglia | 70 | Brb-8 | 蚕豆 Horsebean | 湖北通山 Tongshan, Hubei | ||||||
| 63 | Rad-1 | 萝卜 Radish | 湖北竹山 Zhushan, Hubei | 71 | Let-9 | 莴苣 Lettuce | 湖北五峰 Wufeng, Hubei | ||||||
| 64 | clone-22 | 油菜 Rape | 加拿大 Canada | 72 | Let-3 | 莴苣 Lettuce | 湖北武汉 Wuhan, Hubei | ||||||
| 65 | UK-2 | 不清 Unknown | 湖北武汉 Wuhan, Hubei | 73 | Let-27 | 莴苣 Lettuce | 台湾 Taiwan | ||||||
| 66 | Can02-6 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | 74 | Let-11 | 莴苣 Lettuce | 湖北宜昌 Yichang, Hubei | ||||||
| 67 | Cor-10 | 芫荽 Coriander | 湖北宜昌 Yichang, Hubei | 75 | Can02-22 | 油菜 Rape | 湖北武汉 Wuhan, Hubei | ||||||
| 68 | Rad-30-2 | 萝卜 Radish | 湖北神农架 Shennongjia, Hubei | 76 | Let-26 | 莴苣 Lettuce | 加拿大 Canada | ||||||
| 上游引物 Forward primers | 下游引物 Reverse primers | |||
|---|---|---|---|---|
| em1 | 5′-GACTGCGTACGAATTAAT-3′ | me1 | 5′-TGAGTCCAAACCGGATA-3′ | |
| em2 | 5′-GACTGCGTACGAATTTGC-3′ | me2 | 5′-TGAGTCCAAACCGGAGC-3′ | |
| em3 | 5′-GACTGCGTACGAATTGAC-3′ | me3 | 5′-TGAGTCCAAACCGGAAT-3′ | |
| em4 | 5′-GACTGCGTACGAATTTGA-3′ | me4 | 5′-TGAGTCCAAACCGGACC-3′ | |
| em5 | 5′-GACTGCGTACGAATTAAC-3′ | me5 | 5′-TGAGTCCAAACCGGAAG-3′ | |
| em6 | 5′-GACTGCGTACGAATTGCA-3′ | |||
表1 用于核盘菌SRAP分析的引物信息
Table 1 SRAP primers used in genetic diversity analysis of Sclerotinia sclerotiorum
| 上游引物 Forward primers | 下游引物 Reverse primers | |||
|---|---|---|---|---|
| em1 | 5′-GACTGCGTACGAATTAAT-3′ | me1 | 5′-TGAGTCCAAACCGGATA-3′ | |
| em2 | 5′-GACTGCGTACGAATTTGC-3′ | me2 | 5′-TGAGTCCAAACCGGAGC-3′ | |
| em3 | 5′-GACTGCGTACGAATTGAC-3′ | me3 | 5′-TGAGTCCAAACCGGAAT-3′ | |
| em4 | 5′-GACTGCGTACGAATTTGA-3′ | me4 | 5′-TGAGTCCAAACCGGACC-3′ | |
| em5 | 5′-GACTGCGTACGAATTAAC-3′ | me5 | 5′-TGAGTCCAAACCGGAAG-3′ | |
| em6 | 5′-GACTGCGTACGAATTGCA-3′ | |||
| 引物组合 Primer combination | 位点总数 Total fragments | 多态位点数 Number of polymorphic loci | 多态位点比例 Percentage of polymorphic loci (%) |
|---|---|---|---|
| em1-me3 | 43 | 16 | 37.21 |
| em2-me2 | 35 | 12 | 34.29 |
| em2-me3 | 38 | 14 | 36.84 |
| em1-me4 | 51 | 26 | 50.98 |
| em6-me5 | 32 | 18 | 56.25 |
| em4-me1 | 28 | 15 | 53.57 |
| em1-me2 | 33 | 13 | 39.39 |
| 总计 Total | 260 | 114 | 43.85 |
| 平均 Mean | 37.1 | 16.3 | 43.85 |
表2 7对SRAP引物在76份材料中的多态性信息
Table 2 Polymorphic information of seven selected SRAP primers on 76 isolates
| 引物组合 Primer combination | 位点总数 Total fragments | 多态位点数 Number of polymorphic loci | 多态位点比例 Percentage of polymorphic loci (%) |
|---|---|---|---|
| em1-me3 | 43 | 16 | 37.21 |
| em2-me2 | 35 | 12 | 34.29 |
| em2-me3 | 38 | 14 | 36.84 |
| em1-me4 | 51 | 26 | 50.98 |
| em6-me5 | 32 | 18 | 56.25 |
| em4-me1 | 28 | 15 | 53.57 |
| em1-me2 | 33 | 13 | 39.39 |
| 总计 Total | 260 | 114 | 43.85 |
| 平均 Mean | 37.1 | 16.3 | 43.85 |
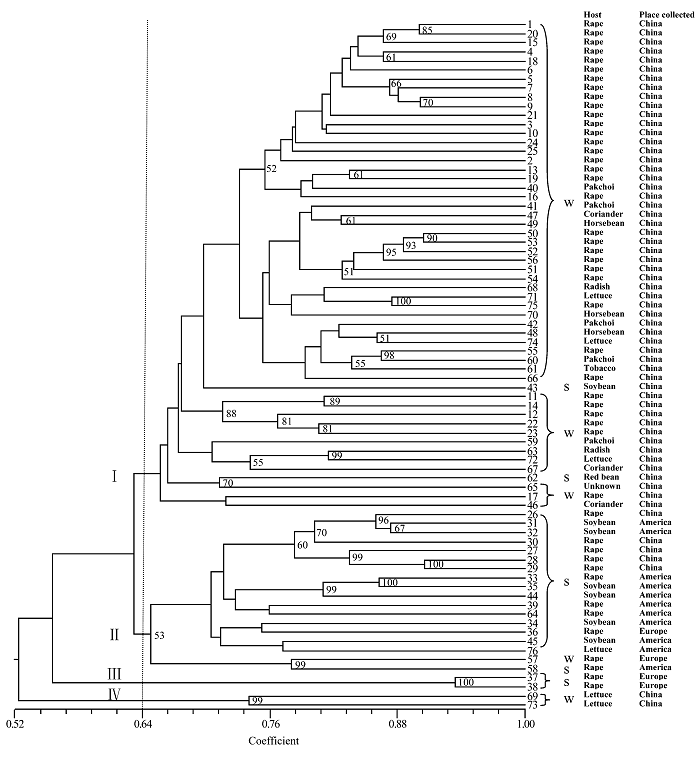
图1 76个核盘菌菌株的UPGMA聚类图(基于500次重复运算的大于50%的Bootstrap值在图中列出。W: 来源于冬油菜生态区; S: 来源于春油菜生态区)
Fig. 1 UPGMA dendrogram of 76 Sclerotinia sclerotiorum isolates. Bootstrap values over 50% are shown in the branches based on 500 resamplings of the data set. W, Winter rapeseed ecological region; S, Spring rapeseed ecological region.
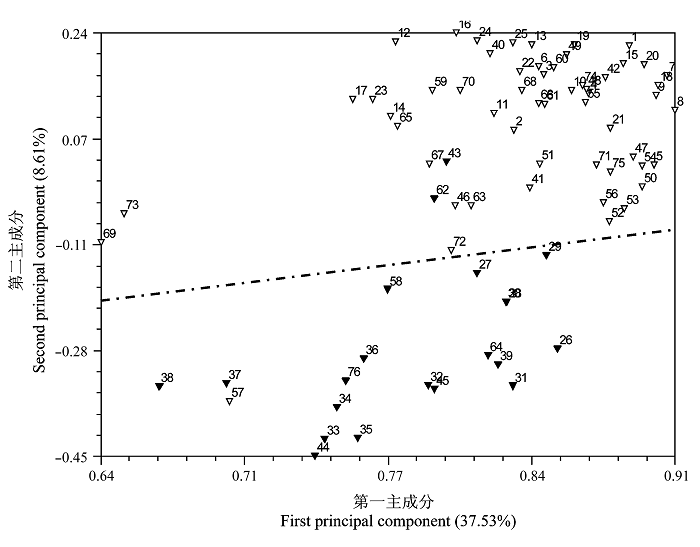
图2 基于SRAP标记的76个核盘菌菌株的主成分分析二维散点图(▽来源于冬油菜生态区, ▼来源于春油菜生态区)
Fig. 2 Two dimensional principal component plot of 76 Sclerotinia sclerotiorum isolates for two principal components estimated with 114 SRAP markers.▽ Winter rapeseed ecological region; ▼ Spring rapeseed ecological region.
| 组 Group | 份数 No. | Shannon指数 Shannon index | Simpson指数 Simpson index |
|---|---|---|---|
| 寄主 Host plant | |||
| 油菜 Rape | 47 | 0.3822 | 0.2417 |
| 大豆 Soybean | 7 | 0.3083 | 0.2034 |
| 莴苣 Lettuce | 6 | 0.2870 | 0.1901 |
| 小白菜 Pakchoi | 5 | 0.2340 | 0.1614 |
| 芫荽 Coriander | 3 | 0.1898 | 0.1326 |
| 蚕豆 Horsebean | 3 | 0.1731 | 0.1236 |
| 萝卜 Radish | 2 | 0.1702 | 0.1228 |
| 地区 Geographical region | |||
| 中国 China | 61 | 0.3516 | 0.2239 |
| 美洲 America | 11 | 0.3278 | 0.2175 |
| 欧洲 Europe | 4 | 0.3368 | 0.2216 |
| 油菜生态区 Rape ecological region | |||
| 春 Spring | 21 | 0.4050 | 0.2628 |
| 冬 Winter | 55 | 0.3401 | 0.2175 |
| 总计 Total | 76 | 0.4002 | 0.2550 |
表3 不同地理来源、不同油菜生态区和不同寄主来源的核盘菌各群体的遗传多样性指数
Table 3 Genetic diversity indices of Sclerotinia sclerotiorum populations from different geographical regions, ecological regions and hosts
| 组 Group | 份数 No. | Shannon指数 Shannon index | Simpson指数 Simpson index |
|---|---|---|---|
| 寄主 Host plant | |||
| 油菜 Rape | 47 | 0.3822 | 0.2417 |
| 大豆 Soybean | 7 | 0.3083 | 0.2034 |
| 莴苣 Lettuce | 6 | 0.2870 | 0.1901 |
| 小白菜 Pakchoi | 5 | 0.2340 | 0.1614 |
| 芫荽 Coriander | 3 | 0.1898 | 0.1326 |
| 蚕豆 Horsebean | 3 | 0.1731 | 0.1236 |
| 萝卜 Radish | 2 | 0.1702 | 0.1228 |
| 地区 Geographical region | |||
| 中国 China | 61 | 0.3516 | 0.2239 |
| 美洲 America | 11 | 0.3278 | 0.2175 |
| 欧洲 Europe | 4 | 0.3368 | 0.2216 |
| 油菜生态区 Rape ecological region | |||
| 春 Spring | 21 | 0.4050 | 0.2628 |
| 冬 Winter | 55 | 0.3401 | 0.2175 |
| 总计 Total | 76 | 0.4002 | 0.2550 |
| 变异来源 Source of variation | 自由度 df | 均方 Mean square | 方差分量 Variance component | 方差分量比率 Percentage of variation (%) | P |
|---|---|---|---|---|---|
| 2个油菜生态区 Two rape ecological regions | |||||
| 群体间 Among populations | 1 | 108.034 | 3.111 | 18.8 | <0.001 |
| 群体内 Within population | 74 | 13.467 | 13.467 | 81.2 | <0.001 |
| 7个寄主来源 Seven plant hosts | |||||
| 群体间 Among populations | 6 | 26.784 | 0.334 | 2.40 | 0.8673 |
| 群体内 Within population | 66 | 13.590 | 13.590 | 97.6 | <0.001 |
| 3个地理来源 Three geographical regions | |||||
| 群体间 Among populations | 2 | 68.315 | 4.363 | 24.8 | <0.001 |
| 群体内 Within population | 73 | 13.260 | 13.260 | 75.2 | <0.001 |
表4 不同地理来源、不同油菜生态区和不同寄主来源的核盘菌各群体基于SRAP标记的遗传变异分析(AMOVA)
Table 4 AMOVA analysis of genetic variation in Sclerotinia sclerotiorum populations from different geographical regions, ecological regions and hosts based on SRAP markers
| 变异来源 Source of variation | 自由度 df | 均方 Mean square | 方差分量 Variance component | 方差分量比率 Percentage of variation (%) | P |
|---|---|---|---|---|---|
| 2个油菜生态区 Two rape ecological regions | |||||
| 群体间 Among populations | 1 | 108.034 | 3.111 | 18.8 | <0.001 |
| 群体内 Within population | 74 | 13.467 | 13.467 | 81.2 | <0.001 |
| 7个寄主来源 Seven plant hosts | |||||
| 群体间 Among populations | 6 | 26.784 | 0.334 | 2.40 | 0.8673 |
| 群体内 Within population | 66 | 13.590 | 13.590 | 97.6 | <0.001 |
| 3个地理来源 Three geographical regions | |||||
| 群体间 Among populations | 2 | 68.315 | 4.363 | 24.8 | <0.001 |
| 群体内 Within population | 73 | 13.260 | 13.260 | 75.2 | <0.001 |
| [1] |
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. The American Journal of Human Genetics, 32, 314-331.
URL PMID |
| [2] | Chen BY (陈碧云), Wu XM (伍晓明) , Lu GY (陆光远), Gao GZ (高桂珍), Xu K (许鲲), Li XZ (李响枝) (2006) Molecular mapping of the gene(s) controlling petal-loss trait in Brassica napus L. Hereditas (Beijing) (遗传), 28, 707-712. (in Chinese with English abstract) |
| [3] | Dong YC (董玉琛), Cao YS (曹永生), Zhang XY (张学勇), Liu SC (刘三才), Wang LF (王兰芬), You GX (游光霞), Pang BS (庞斌双), Li LH (李立会), Jia JZ (贾继增) (2003) Establishment of candidate core collections in Chinese common wheat germplasm. Journal of Plant Genetic Resources (植物遗传资源学报), 4, 1-8. (in Chinese with English abstract) |
| [4] | Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 179-191. |
| [5] | Felsenstein J (1993) PHYLIP (Phylogeny Inference Package), Version 3.6. Department of Genetics, University of Washington, Seattle, USA. |
| [6] | Fu HT (傅洪拓), Qiao H (乔慧), Yao JH (姚建华), Gong YS (龚永生), Wu Y (吴滟), Jiang SF (蒋速飞), Xiong YW (熊贻伟) (2010) Genetic diversity in five Macrobrachium hainanense populations using SRAP markers. Biodiversity Science (生物多样性), 18, 145-149. (in Chinese with English abstract) |
| [7] | Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theoretical and Applied Genetics, 103, 455-461. |
| [8] | Li GQ (李国庆) (1996) Study on Diversity of Crop Sclerotinia Disease Pathogeny: Sclerotinia sclerotiorum (作物菌核病病原——核盘菌的多样性研究). PhD dissertation, Huazhong Agricultural University, Wuhan. (in Chinese with English abstract) |
| [9] | Li GQ (李国庆), Wang DB (王道本), Zhou Q (周启), Huang HZ (黄鸿章) (1997) Study on diversity of Sclerotinia sclerotiorum in sclerotial germinations. Journal of Plant Protection (植物保护学报), 24, 59-64. (in Chinese with English abstract) |
| [10] | Li GQ (李国庆), Wang DB (王道本), Jiang DH (姜道宏), Huang HZ (黄鸿章), Erickson RS, Yi XH (易先宏), Zhou Q (周启) (2000) RAPD assay of strains of Sclerotinia sclerotiorum and its related species. Acta Phytopathologica Sinica (植物病理学报), 30, 166-170. (in Chinese with English abstract) |
| [11] | Li J (李佳), Shen BZ (沈斌章), Han JX (韩继祥), Gan L (甘莉) (1994) An effective procedure for extracting total DNA in rape. Journal of Huazhong Agricultural University (华中农业大学学报), 13, 521-523. (in Chinese with English abstract) |
| [12] | Li LL (李丽丽) (1994) Advances in study of the world rape disease. Chinese Journal of Oil Crop Sciences (中国油料), 16, 79-81. (in Chinese with English abstract) |
| [13] | Liu XH (刘晓红), Zhou H (周航), Ji HL (姬红丽), Peng LN (彭丽年), Wen CJ (文成敬), Peng YL (彭云良) (2004) Primary studies on population structure of Sclerotinia sclerotiorum on vegetables in Sichuan Province. Southwest China Journal of Agricultural Sciences (西南农业学报), 17, 618-623. (in Chinese with English abstract) |
| [14] | Lu GY (陆光远), Yang GS (杨光圣), Fu TD (傅廷栋) (2003) An effective SSR detection system in rapeseed. Chinese Journal of Oil Crop Sciences (中国油料作物学报), 25, 79-81. (in Chinese with English abstract) |
| [15] | Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences, USA, 76, 569-573. |
| [16] | Rolhf FJ (1997) NTSYSpc Version 2.1. Numerical Taxonomy and Multivariate Analysis System. Exeter Software, Setauket, New York. |
| [17] | Sexton AC, Howlett BJ (2004) Microsatellite markers reveal genetic differentiation among populations of Sclerotinia sclerotiorum from Australian canola fields. Current Genetics, 46, 357-365. |
| [18] | Sun JM, Witold I, Malgorzata J, Han FX (2005) Analysis of the genetic structure of Sclerotinia sclerotiorum (Lib.) de Bary populations from different regions and host plants by random amplified polymorphic DNA markers. Journal of Integrative Plant Biology, 47, 385-395. |
| [19] | Wen YC (文雁成), Wang HZ (王汉中), Shen JX (沈金雄), Liu GH (刘贵华), Zhang SF (张书芬) (2006) Analysis of genetic diversity and genetic basis of Chinese rapeseed cultivars (Brassica napus L.) by sequence-related amplified polymorphism markers. Scientia Agricultura Sinica (中国农业科学), 39, 246-256. (in Chinese with English abstract) |
| [1] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [2] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [3] | 翁茁先, 黄佳琼, 张仕豪, 余锴纯, 钟福生, 黄勋和, 张彬. 利用线粒体COI基因揭示中国乌骨鸡遗传多样性和群体遗传结构[J]. 生物多样性, 2019, 27(6): 667-676. |
| [4] | 武星彤, 陈璐, 王敏求, 张原, 林雪莹, 李鑫玉, 周宏, 文亚峰. 丹霞梧桐群体遗传结构及其遗传分化[J]. 生物多样性, 2018, 26(11): 1168-1179. |
| [5] | 刘青青, 董志军. 基于线粒体COI基因分析钩手水母的群体遗传结构[J]. 生物多样性, 2018, 26(11): 1204-1211. |
| [6] | 盛芳, 陈淑英, 田嘉, 李鹏, 秦雪, 罗淑萍, 李疆. 新疆准噶尔山楂不同居群的遗传多样性[J]. 生物多样性, 2017, 25(5): 518-530. |
| [7] | 刘若愚, 孙忠民, 姚建亭, 胡自民, 段德麟. 中国近海重要生态建群红藻真江蓠的群体遗传多样性[J]. 生物多样性, 2016, 24(7): 781-790. |
| [8] | 于少帅, 徐启聪, 林彩丽, 王圣洁, 田国忠. 植原体遗传多样性研究现状与展望[J]. 生物多样性, 2016, 24(2): 205-215. |
| [9] | 向妮, 肖炎农, 段灿星, 王晓鸣, 朱振东. 利用SSR标记分析茄镰孢豌豆专化型的遗传多样性[J]. 生物多样性, 2012, 20(6): 693-702. |
| [10] | 周蓉, 李佳琦, 李铀, 刘迺发, 房峰杰, 施丽敏, 王莹. 基于线粒体DNA的大石鸡种群遗传变异[J]. 生物多样性, 2012, 20(4): 451-459. |
| [11] | 张俊红, 黄华宏, 童再康, 程龙军, 梁跃龙, 陈奕良. 光皮桦6个南方天然群体的遗传多样性[J]. 生物多样性, 2010, 18(3): 233-240. |
| [12] | 张根, 席贻龙, 薛颖昊, 胡忻, 项贤领, 温新利. 基于rDNA ITS序列探讨粉煤灰污染对萼花臂尾轮虫种群遗传多样性的影响[J]. 生物多样性, 2010, 18(3): 241-250. |
| [13] | 傅洪拓, 乔慧, 姚建华, 龚永生, 吴滟, 蒋速飞, 熊贻伟. 基于SRAP分子标记的海南沼虾种群遗传多样性[J]. 生物多样性, 2010, 18(2): 145-149. |
| [14] | 王长忠, 李忠, 梁宏伟, 呼光富, 吴勤超, 邹桂伟, 罗相忠. 长江下游地区4个克氏原螯虾群体的遗传多样性分析[J]. 生物多样性, 2009, 17(5): 518-523. |
| [15] | 戴李川, 张明龙, 刘继业, 李小白, 崔海瑞. 用EST-SSR标记检测我国甘蓝型油菜品种的遗传多样性[J]. 生物多样性, 2009, 17(5): 482-489. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2026 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()