



生物多样性 ›› 2010, Vol. 18 ›› Issue (3): 233-240. DOI: 10.3724/SP.J.1003.2010.233 cstr: 32101.14.SP.J.1003.2010.233
张俊红1, 黄华宏1, 童再康1,*( ), 程龙军1, 梁跃龙2, 陈奕良3
), 程龙军1, 梁跃龙2, 陈奕良3
收稿日期:2009-11-11
接受日期:2010-04-24
出版日期:2010-05-20
发布日期:2012-02-08
通讯作者:
童再康
作者简介: E-mail: zktong@zjfc.edu.cn基金资助:
Junhong Zhang1, Huahong Huang1, Zaikang Tong1,*( ), Longjun Cheng1, Yuelong Liang2, Yiliang Chen3
), Longjun Cheng1, Yuelong Liang2, Yiliang Chen3
Received:2009-11-11
Accepted:2010-04-24
Online:2010-05-20
Published:2012-02-08
Contact:
Zaikang Tong
摘要:
为揭示中国特有珍贵用材树种光皮桦(Betula luminifera)天然群体的遗传多样性和遗传结构, 采用AFLP分子标记, 分析了采自浙江、福建、江西、广西和贵州5个省区6个天然群体的120份样品。9对引物获得了355个位点, 其中多态性位点323个。分析结果表明光皮桦天然群体具有较高的遗传多样性, 多态位点百分率(PPL)达90.99%, 各群体的PPL和Nei’s基因多样性(hj)分别为93.20-98.60%和0.3143-0.3645; 总群体遗传多样性指数(Ht)为0.3616, 群体间遗传分化系数(Fst)为0.0650, 群体间总的基因流较高(Nm= 3.5962)。AMOVA分析表明群体间的遗传变异占总变异的11.49%, 浙江临安群体和贵州修文群体间的遗传距离最大(0.0665), 江西龙南群体和广西龙胜群体间的遗传距离最小(0.0173), 且遗传结构分析显示这两个群体的部分个体可能来自同一近祖。Mantel检测发现, 群体间的遗传距离与地理距离没有显著相关性(r = 0.423, P = 0.113), 而与两两群体所在地的均温差呈显著相关(r = 0.449, P = 0.017)。结合群体实地调查, 可以得出光皮桦天然群体的遗传多样性和遗传结构的形成不仅与其广域分布、自然杂交、种子特性以及生活史有关, 而且与群体被人为砍伐、生境片断化等因素有重要关系。基于上述结果我们提出了光皮桦天然种群的保护策略。
张俊红, 黄华宏, 童再康, 程龙军, 梁跃龙, 陈奕良 (2010) 光皮桦6个南方天然群体的遗传多样性. 生物多样性, 18, 233-240. DOI: 10.3724/SP.J.1003.2010.233.
Junhong Zhang, Huahong Huang, Zaikang Tong, Longjun Cheng, Yuelong Liang, Yiliang Chen (2010) Genetic diversity in six natural populations of Betula luminifera from southern China. Biodiversity Science, 18, 233-240. DOI: 10.3724/SP.J.1003.2010.233.
| 群体 Population | 采样地 Collection site | 经纬度 Locality | 温度 Temperature (℃) | 降水量 Precipitation (mm) |
|---|---|---|---|---|
| 浙江临安 LA | 浙江临安太湖源 Taihuyuan, Lin’an, Zhejiang | 119°33′ E 30°25′ N | 15.5 | 1,375.8 |
| 浙江庆元 QY | 浙江庆元巾子峰 Jinzifeng, Qingyuan, Zhejiang | 119°08′ E 27°41′ N | 17.7 | 1,645.5 |
| 江西龙南 LN | 江西赣州九连山 Jiulianshan, Ganzhou, Jiangxi | 114°28′ E 24°33′ N | 19.5 | 1,395.3 |
| 广西龙胜 LS | 广西龙胜花坪 Huaping, Longsheng, Guangxi | 109°55′ E 25°38′ N | 18.7 | 1,577.3 |
| 贵州修文 XW | 贵州修文 Xiuwen, Guizhou | 106°34′ E 26°49′ N | 15.2 | 1,128.3 |
| 福建尤溪 YX | 福建尤溪 Youxi, Fujian | 118°10′ E 25°56′ N | 19.3 | 1,322.8 |
表1 光皮桦天然群体采样地概况
Table 1 Survey of sampling locations in natural populations of Betula luminifera
| 群体 Population | 采样地 Collection site | 经纬度 Locality | 温度 Temperature (℃) | 降水量 Precipitation (mm) |
|---|---|---|---|---|
| 浙江临安 LA | 浙江临安太湖源 Taihuyuan, Lin’an, Zhejiang | 119°33′ E 30°25′ N | 15.5 | 1,375.8 |
| 浙江庆元 QY | 浙江庆元巾子峰 Jinzifeng, Qingyuan, Zhejiang | 119°08′ E 27°41′ N | 17.7 | 1,645.5 |
| 江西龙南 LN | 江西赣州九连山 Jiulianshan, Ganzhou, Jiangxi | 114°28′ E 24°33′ N | 19.5 | 1,395.3 |
| 广西龙胜 LS | 广西龙胜花坪 Huaping, Longsheng, Guangxi | 109°55′ E 25°38′ N | 18.7 | 1,577.3 |
| 贵州修文 XW | 贵州修文 Xiuwen, Guizhou | 106°34′ E 26°49′ N | 15.2 | 1,128.3 |
| 福建尤溪 YX | 福建尤溪 Youxi, Fujian | 118°10′ E 25°56′ N | 19.3 | 1,322.8 |
| 群体 Population | 样本数量 Sample size | 多态位点数 N | 多态位点百分率 PPL(%) | Nei’s基因多样性(标准差) hj(standard error) | 个体取样的变异比例 VarI (%) | 位点导致的变异比例 VarL (%) |
|---|---|---|---|---|---|---|
| 浙江临安 LA | 20 | 337 | 94.9 | 0.3422(0.0074) | 40.3 | 59.7 |
| 浙江庆元 QY | 20 | 350 | 98.6 | 0.3645(0.0065) | 46.5 | 53.5 |
| 江西龙南 LN | 20 | 333 | 93.8 | 0.3309(0.0072) | 37.5 | 62.5 |
| 广西龙胜 LS | 20 | 332 | 93.5 | 0.3143(0.0076) | 37.6 | 62.4 |
| 贵州修文 XW | 20 | 331 | 93.2 | 0.3230(0.0076) | 35.1 | 64.9 |
| 福建尤溪 YX | 20 | 335 | 94.4 | 0.3535(0.0069) | 37.7 | 62.3 |
表2 AFLP检测的光皮桦6个群体的遗传多样性水平
Table 2 Genetic diversity of six Betula luminifera populations detected by AFLP
| 群体 Population | 样本数量 Sample size | 多态位点数 N | 多态位点百分率 PPL(%) | Nei’s基因多样性(标准差) hj(standard error) | 个体取样的变异比例 VarI (%) | 位点导致的变异比例 VarL (%) |
|---|---|---|---|---|---|---|
| 浙江临安 LA | 20 | 337 | 94.9 | 0.3422(0.0074) | 40.3 | 59.7 |
| 浙江庆元 QY | 20 | 350 | 98.6 | 0.3645(0.0065) | 46.5 | 53.5 |
| 江西龙南 LN | 20 | 333 | 93.8 | 0.3309(0.0072) | 37.5 | 62.5 |
| 广西龙胜 LS | 20 | 332 | 93.5 | 0.3143(0.0076) | 37.6 | 62.4 |
| 贵州修文 XW | 20 | 331 | 93.2 | 0.3230(0.0076) | 35.1 | 64.9 |
| 福建尤溪 YX | 20 | 335 | 94.4 | 0.3535(0.0069) | 37.7 | 62.3 |
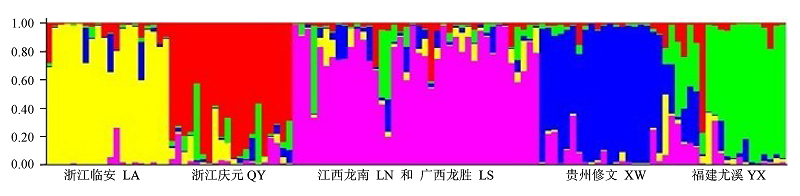
图1 基于STRUCTURE软件对6个光皮桦群体的亲缘关系分析。纵坐标表示各群体中的单株占某群体祖先成分的比例, 其中江西龙南群体和广西龙胜群体混合为一个群体; 浙江临安群体、浙江庆元群体、江西龙南与广西龙胜混合群体、贵州修文群体和福建尤溪群体分别用黄色、红色、粉红色、蓝色和绿色表示。
Fig. 1 Ancestry of Betula luminifera from six population estimated using STRUCTURE. Population LA, QY, admixture of LN and LS, XW and YX gene pools are shown in yellow, red, pink, blue and green respectively. Y-coordinate denotes the proportion of ancestry components in an individual in relation to other population.
| 变异来源 Source of variance | 自由度 df | 变异组分 Variance components | 变异贡献率(%) Contribution of variation | 遗传分化系数(фST) Genetic differentiation index | P |
|---|---|---|---|---|---|
| 群体间 Among populations | 5 | 6.38404 | 11.49% | 0.11489 | <0.0001** |
| 群体内 Within populations | 113 | 49.18305 | 88.51% | ||
| 总计 Total | 118 | 55.56709 |
表3 AFLP检测获得的6个群体的分子变异分析
Table 3 Analysis of molecular variance (AMOVA) of six populations in Betula luminifera
| 变异来源 Source of variance | 自由度 df | 变异组分 Variance components | 变异贡献率(%) Contribution of variation | 遗传分化系数(фST) Genetic differentiation index | P |
|---|---|---|---|---|---|
| 群体间 Among populations | 5 | 6.38404 | 11.49% | 0.11489 | <0.0001** |
| 群体内 Within populations | 113 | 49.18305 | 88.51% | ||
| 总计 Total | 118 | 55.56709 |
| 群体 | LA | QY | LN | LS | XW | YX |
|---|---|---|---|---|---|---|
| LA | **** | 0.0637 | 0.1115 | 0.1135 | 0.1146 | 0.0881 |
| QY | 0.0377 | **** | 0.0534 | 0.0516 | 0.0612 | 0.0358 |
| LN | 0.0657 | 0.0302 | **** | 0.0349 | 0.0513 | 0.0385 |
| LS | 0.0643 | 0.0276 | 0.0173 | **** | 0.0551 | 0.0552 |
| XW | 0.0665 | 0.0342 | 0.0266 | 0.0276 | **** | 0.0379 |
| YX | 0.0529 | 0.0210 | 0.0209 | 0.0293 | 0.0201 | **** |
表4 6个群体间的遗传距离(对角线下半部分)及遗传分化系数(对角线上半部分)
Table 4 Nei’s genetic distance (below diagonal) and genetic differentiation (pairwise фST,above diagonal) between populations in Betula luminifera
| 群体 | LA | QY | LN | LS | XW | YX |
|---|---|---|---|---|---|---|
| LA | **** | 0.0637 | 0.1115 | 0.1135 | 0.1146 | 0.0881 |
| QY | 0.0377 | **** | 0.0534 | 0.0516 | 0.0612 | 0.0358 |
| LN | 0.0657 | 0.0302 | **** | 0.0349 | 0.0513 | 0.0385 |
| LS | 0.0643 | 0.0276 | 0.0173 | **** | 0.0551 | 0.0552 |
| XW | 0.0665 | 0.0342 | 0.0266 | 0.0276 | **** | 0.0379 |
| YX | 0.0529 | 0.0210 | 0.0209 | 0.0293 | 0.0201 | **** |
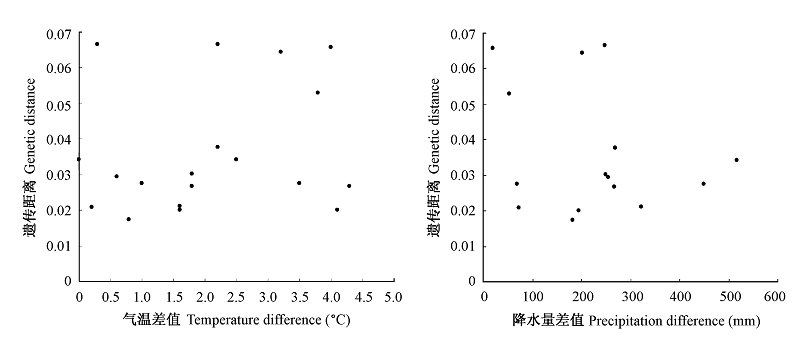
图4 光皮桦群体间的遗传距离与气温差值(A)和降水量差值(B)的Mantel相关性检测
Fig. 4 Mantel correlations between genetic distance and temperature difference (A) and precipitation difference (B)
| [1] | Chen W (陈伟) (2006) Genetic diversity of the natural populations of Betula luminifera. Journal of Beijing Forestry University (北京林业大学学报), 28 (6),28-34. (in Chinese with English abstract) |
| [2] | Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus, 12, 13-14. |
| [3] |
Evanno G, Regnaut S, Goudetd J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611-2620.
DOI URL PMID |
| [4] | Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics (Online), 1, 47-50. |
| [5] |
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics, 164, 1567-1587.
URL PMID |
| [6] | Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes, 7, 574-578. |
| [7] | Feng FJ, Han SJ, Wang HM (2006) Genetic diversity and genetic differentiation of natural Pinus koraiensis population. Journal of Forestry Research, 17, 21-24. |
| [8] | Gao YK (高亦珂), Nie SQ (聂绍荃), Zu YG (祖元刚) (1999) Genetic structure analysis by RAPD in Betula platyphylla nature population in Northeast of China. In: The Application, Method and Theory of Molecular Ecology (分子生态学理论、方法和应用), pp.196-205. China Higher Education Press, Beijing. (in Chinese) |
| [9] |
Gillies ACM, Navarro C, Lowe AJ, Newton AC, Hernandez M, Wilson J, Cornelius JP (1999) Genetic diversity in Mesoamerican populations of mahogany (Swietenia macrophylla), assessed using RAPDs. Heredity, 83, 722-732.
URL PMID |
| [10] | Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Plant Population Genetics, Breeding and Genetic Resources (eds Brown AHD, Clegg MT, Kahler AL,Weir BS), pp.43-63. Sinauer Associates, Sunderland. |
| [11] | Hamrick JL, Godt MJW, Sherman-Broyes SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forests, 6, 95-124. |
| [12] |
He JS, Chen L, Si Y, Huang B, Ban XQ, Wang YW (2008) Population structure and genetic diversity distribution in wild and cultivated populations of the traditional Chinese medicinal plant Magnolia officinalis subsp. biloba (Magnoliaceae). Genetica, 135, 233-243.
URL PMID |
| [13] | Li QM (李巧明), Zhao JL (赵建立) (2007) Genetic diversity of Phyllanthus embilica populations in dry-hot valleys in Yunnan. Biodiversity Science (生物多样性), 15, 84-91. (in Chinese with English abstract) |
| [14] | Luo JX (罗建勋), Gu WC (顾万春), Chen SY (陈少瑜) (2006) Allozyme variation in 10 natural population of Picea asperata. Acta Phytoecologica Sinica (植物生态学报), 30, 165-173. (in Chinese with English abstract) |
| [15] | Miller MP (1997) Tools for population genetic analyses (TFPGA) version 1.3: A windows program for the analysis of allozyme and molecular genetic data. Department of Biological Sciences, Northern Arizona University, Flagstaff. |
| [16] | Nei M (1972) Genetic distance between populations. The American Naturalist, 106, 283-292. |
| [17] | Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583-590. |
| [18] | Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences, USA, 76, 5269-5273. |
| [19] | Powell W, Thomas WTB, Baird E (1997) Analysis of quantitative traits in barley by the use of amplified fragment polymorphism. Heredity, 79, 48-59. |
| [20] |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
URL PMID |
| [21] | de la Torre A, López C, Yglesias E, Cornelius JP (2008) Genetic (AFLP) diversity of nine Cedrela odorata populations in Madre de Dios, southern Peruvian Amazon. Forest Ecology and Management, 255, 334-339. |
| [22] |
Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molecular Ecology, 11, 139-151.
DOI URL PMID |
| [23] | Wang ZF (王峥峰), Ge XJ (葛学军) (2009) Not only genetic diversity: advances in plant conservation genetics. Biodiversity Science (生物多样性), 17, 330-339. (in Chinese with English abstract) |
| [24] | Wu ZC (吴子诚), Wang LH (王乐辉) (1996) The characteristics of selective population and improved technology of Betula luminifera. Journal of Sichuan Forestry Science and Technology (四川林业科技), 17(4),17-28. (in Chinese with English abstract) |
| [25] | Xie YQ (谢一青), Li ZZ (李志真), Huang RZ (黄儒珠), Xiao XX (肖祥希), Huang Y (黄勇) (2008) Genetic diversity of Betula luminifera populations at different altitudes in Wuyi Mountains and its association with ecological factors. Scientia Silvae Sinicae (林业科学), 44(3),50-55. (in Chinese with English abstract) |
| [26] | Yeh FC, Chong DKX, Yang RC (1995) RAPD variation within and among natural populations of trembling aspen ( Populus tremuloides) from Alberta. Heredity, 86, 454-460. |
| [27] | You WY (尤卫艳), Huang HH (黄华宏), Tong ZK (童再康), Zhu YQ (朱玉球) (2008) Establishment of AFLP molecular labeling technique system for Betula luminifera. Biotechnology (生物技术), 18(6),42-47. (in Chinese with English abstract) |
| [28] | Zabeau M, Vos P (1993) Selective restriction fragment amplification: a general method for DNA fingerprinting. European Patent Application, EP 534858A1. |
| [29] | Zeng J (曾杰), Wang ZR (王中仁), Zhou SL (周世良), Zheng HS (郑海水), Bai JY (白嘉雨) (2003) Allozymed diversity in natural populations of Betula alnoides from Guangxi, China. Acta Phytoecologica Sinica (植物生态学报), 27, 66-72. (in Chinese with English abstract) |
| [30] | Zeng J, Zou YP, Bai JY, Zheng HS (2003) RAPD analysis of genetic variation in natural populations of Betula alnoides from Guangxi, China. Euphytica, 134, 33-41. |
| [31] | Zhang P (张萍), Zhou ZC (周志春), Jin GQ (金国庆), Fan HH (范辉华), Hu HB (胡红宝) (2006) Genetic diversity analysis and provenance zone allocation of Schima superba in China using RAPD markers. Scientia Silvae Sinicae (林业科学), 42(2),38-42. (in Chinese with English abstract) |
| [32] | Zheng WJ (郑万钧) (1985) Records of Chinese Trees (Vol.2)(中国树木志)(第二卷), pp.2124-2131. China Forestry Publishing House, Beijing. (in Chinese) |
| [33] | Zheng J (郑健), Zheng YQ (郑勇奇), Zhang CH (张川红), Zong YC (宗亦臣), Li BJ (李伯菁), Wu C (吴超) (2008) Genetic diversity in natural populations of Sorbus pohuashanensis. Biodiversity Science (生物多样性), 16, 562-569. (in Chinese with English abstract) |
| [1] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [2] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [3] | 吕燕文, 王子韵, 肖钰, 何梓晗, 吴超, 胡新生. 谱系分选理论与检测方法的研究进展[J]. 生物多样性, 2024, 32(4): 23400-. |
| [4] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [5] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [6] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [7] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [8] | 孙琼, 王嵘, 陈小勇. 物种形成过程中的分化基因组岛及其形成机制[J]. 生物多样性, 2022, 30(3): 21383-. |
| [9] | 向登高, 李跃飞, 李新辉, 陈蔚涛, 马秀慧. 多基因联合揭示海南鲌的遗传结构与遗传多样性[J]. 生物多样性, 2021, 29(11): 1505-1512. |
| [10] | 李潮,金锦锦,罗锦桢,王春晖,王俊杰,赵俊. 唐鱼养殖种群与广州附近4个野生种群的遗传关系[J]. 生物多样性, 2020, 28(4): 474-484. |
| [11] | 翁茁先, 黄佳琼, 张仕豪, 余锴纯, 钟福生, 黄勋和, 张彬. 利用线粒体COI基因揭示中国乌骨鸡遗传多样性和群体遗传结构[J]. 生物多样性, 2019, 27(6): 667-676. |
| [12] | 张亚红, 贾会霞, 王志彬, 孙佩, 曹德美, 胡建军. 滇杨种群遗传多样性与遗传结构[J]. 生物多样性, 2019, 27(4): 355-365. |
| [13] | 张剑, 董路, 张雁云. 基于基因组SNPs的南极恩克斯堡岛阿德利企鹅繁殖种群的遗传结构[J]. 生物多样性, 2019, 27(12): 1291-1297. |
| [14] | 武星彤, 陈璐, 王敏求, 张原, 林雪莹, 李鑫玉, 周宏, 文亚峰. 丹霞梧桐群体遗传结构及其遗传分化[J]. 生物多样性, 2018, 26(11): 1168-1179. |
| [15] | 刘青青, 董志军. 基于线粒体COI基因分析钩手水母的群体遗传结构[J]. 生物多样性, 2018, 26(11): 1204-1211. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn