



生物多样性 ›› 2021, Vol. 29 ›› Issue (11): 1505-1512. DOI: 10.17520/biods.2021166 cstr: 32101.14.biods.2021166
向登高1,2, 李跃飞2, 李新辉2, 陈蔚涛2,*( ), 马秀慧1,*(
), 马秀慧1,*( )
)
收稿日期:2021-04-29
接受日期:2021-07-05
出版日期:2021-11-20
发布日期:2021-07-27
通讯作者:
陈蔚涛,马秀慧
作者简介:E-mail: lovemxh@126.com基金资助:
Denggao Xiang1,2, Yuefei Li2, Xinhui Li2, Weitao Chen2,*( ), Xiuhui Ma1,*(
), Xiuhui Ma1,*( )
)
Received:2021-04-29
Accepted:2021-07-05
Online:2021-11-20
Published:2021-07-27
Contact:
Weitao Chen,Xiuhui Ma
摘要:
海南鲌(Culter recurviceps)是我国华南地区重要经济鱼类, 由于受到近些年水利开发、过度捕捞、环境污染等诸多因素的影响, 其资源量快速下降, 亟需得到更多的关注和保护。为保护和合理开发海南鲌种质资源, 本研究采集了华南地区23个地理群体207尾海南鲌样本, 测定了2个线粒体基因(Cytb和ND2)并从Barcode of Life Data System数据库获得相对应线粒体COI基因, 结合多种分析方法(系统发育分析、分化时间估算、单倍型网状图、群体遗传分析和Mantel检验)对海南鲌的遗传结构和遗传多样性展开研究。系统发育分析和单倍型网状图表明华南地区海南鲌群体被分成3个谱系(I、II和III), 其中谱系I和III由珠江的群体组成, 谱系II由海南岛的群体组成。分化时间估算发现3个谱系之间的分化时间介于0.028-0.251 Ma之间, 表明华南地区更新世气候变化可能是造成海南鲌谱系分化的重要原因。群体遗传分析发现海南鲌群体之间存在极显著的遗传分化(FST = 0.511, P < 0.001), 并且符合距离隔离模式(R = 0.348, P = 0.0010)。群体动态历史分析表明, 海南鲌群体可能在0.010-0.025 Ma经历了群体扩张, 表明更新世的气候波动也影响了海南鲌的群体大小和分布。综上所述, 海南鲌群体由3个谱系组成, 更新世气候变化是导致3个谱系分化和影响海南鲌群体动态历史的重要因素。此外, 海南鲌群体之间的遗传分化也可能受到了空间距离的影响。
向登高, 李跃飞, 李新辉, 陈蔚涛, 马秀慧 (2021) 多基因联合揭示海南鲌的遗传结构与遗传多样性. 生物多样性, 29, 1505-1512. DOI: 10.17520/biods.2021166.
Denggao Xiang, Yuefei Li, Xinhui Li, Weitao Chen, Xiuhui Ma (2021) Population structure and genetic diversity of Culter recurviceps revealed by multi-loci. Biodiversity Science, 29, 1505-1512. DOI: 10.17520/biods.2021166.
| 群体 Population | 样本量 Sample size | 经纬度 Locality |
|---|---|---|
| 海南岛水系 Hainan Island drainage | ||
| 白沙 Baisha (BS) | 33 | 19˚13′ N/109˚26′ E |
| 南丰 Nanfeng (NF) | 15 | 19˚25′ N/109˚33′ E |
| 定安 Dingan (DA) | 4 | 19˚40′ N/110˚21′ E |
| 珠江水系 Pearl River drainage | ||
| 从江 Congjiang (CJ) | 9 | 25˚44′ N/108˚55′ E |
| 崇左 Chongzuo (CZ) | 14 | 22˚16′ N/107˚22′ E |
| 桂平 Guiping (GP) | 14 | 23˚23′ N/110˚04′ E |
| 武宣 Wuxuan (WX) | 2 | 23˚35′ N/109˚40′ E |
| 古竹 Guzhu (GZ) | 16 | 23˚30′ N/114˚42′ E |
| 惠州 Huizhou (HZ) | 11 | 23˚06′ N/114˚24′ E |
| 江门 Jiangmen (JM) | 4 | 22˚34′ N/113˚07′ E |
| 横沥 Hengli (HL) | 1 | 22˚43′ N/113˚29′ E |
| 开平 Kaiping (KP) | 5 | 22˚22′ N/112˚41′ E |
| 柳州 Liuzhou (LZ) | 5 | 24˚18′ N/109˚23′ E |
| 柳城 Liucheng (LC) | 8 | 24˚38′ N/109˚14′ E |
| 南宁 Nanning (NN) | 7 | 22˚46′ N/108˚21′ E |
| 清远 Qingyuan (QY) | 5 | 23˚37′ N/113˚03′ E |
| 梧州 Wuzhou (WZ) | 13 | 23˚25′ N/111˚16′ E |
| 濛江 Mengjiang (MJ) | 1 | 23˚27′ N/110˚44′ E |
| 永福 Yongfu (YF) | 12 | 24˚59′ N/109˚59′ E |
| 郁南 Yunan (YN) | 11 | 23˚14′ N/111˚31′ E |
| 思林 Silin (SL) | 12 | 23˚30′ N/107˚20′ E |
| 韶关 Shaoguan (SG) | 2 | 24˚47′ N/113˚35′ E |
| 平乐 Pingle (PL) | 3 | 24˚37′ N/110˚38′ E |
| 总计 Total | 207 | |
表1 海南鲌样本采集信息
Table 1 Sampling information of Culter recurviceps specimens
| 群体 Population | 样本量 Sample size | 经纬度 Locality |
|---|---|---|
| 海南岛水系 Hainan Island drainage | ||
| 白沙 Baisha (BS) | 33 | 19˚13′ N/109˚26′ E |
| 南丰 Nanfeng (NF) | 15 | 19˚25′ N/109˚33′ E |
| 定安 Dingan (DA) | 4 | 19˚40′ N/110˚21′ E |
| 珠江水系 Pearl River drainage | ||
| 从江 Congjiang (CJ) | 9 | 25˚44′ N/108˚55′ E |
| 崇左 Chongzuo (CZ) | 14 | 22˚16′ N/107˚22′ E |
| 桂平 Guiping (GP) | 14 | 23˚23′ N/110˚04′ E |
| 武宣 Wuxuan (WX) | 2 | 23˚35′ N/109˚40′ E |
| 古竹 Guzhu (GZ) | 16 | 23˚30′ N/114˚42′ E |
| 惠州 Huizhou (HZ) | 11 | 23˚06′ N/114˚24′ E |
| 江门 Jiangmen (JM) | 4 | 22˚34′ N/113˚07′ E |
| 横沥 Hengli (HL) | 1 | 22˚43′ N/113˚29′ E |
| 开平 Kaiping (KP) | 5 | 22˚22′ N/112˚41′ E |
| 柳州 Liuzhou (LZ) | 5 | 24˚18′ N/109˚23′ E |
| 柳城 Liucheng (LC) | 8 | 24˚38′ N/109˚14′ E |
| 南宁 Nanning (NN) | 7 | 22˚46′ N/108˚21′ E |
| 清远 Qingyuan (QY) | 5 | 23˚37′ N/113˚03′ E |
| 梧州 Wuzhou (WZ) | 13 | 23˚25′ N/111˚16′ E |
| 濛江 Mengjiang (MJ) | 1 | 23˚27′ N/110˚44′ E |
| 永福 Yongfu (YF) | 12 | 24˚59′ N/109˚59′ E |
| 郁南 Yunan (YN) | 11 | 23˚14′ N/111˚31′ E |
| 思林 Silin (SL) | 12 | 23˚30′ N/107˚20′ E |
| 韶关 Shaoguan (SG) | 2 | 24˚47′ N/113˚35′ E |
| 平乐 Pingle (PL) | 3 | 24˚37′ N/110˚38′ E |
| 总计 Total | 207 | |
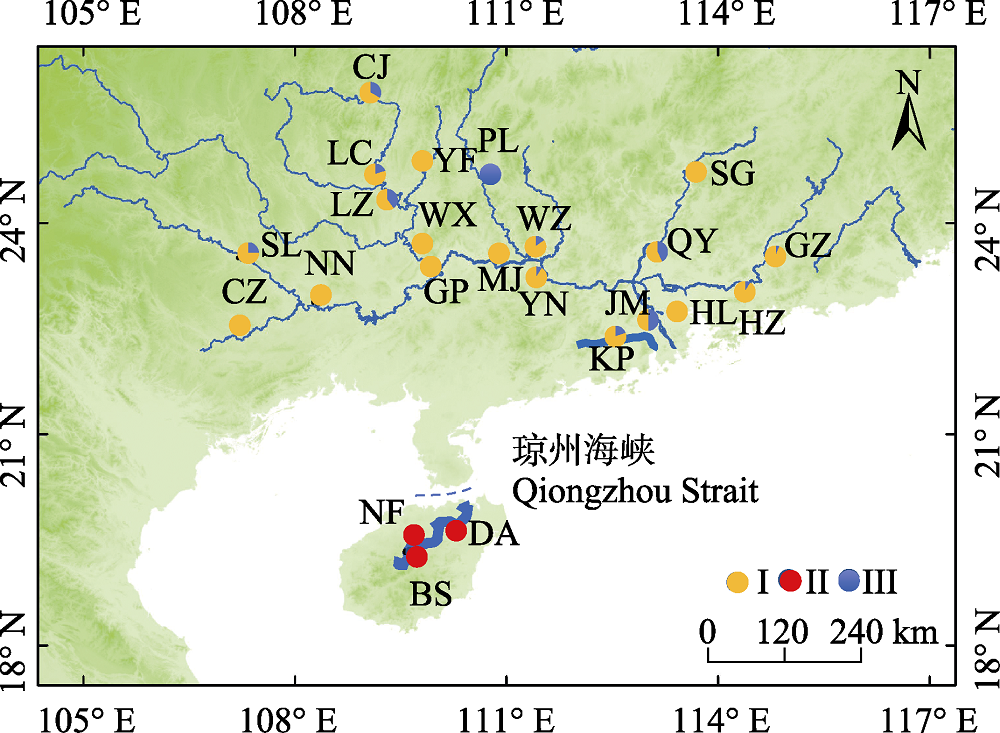
图1 海南鲌样本采集地点示意图。不同颜色代表不同谱系。群体代号含义见表1。
Fig. 1 Map of sample sites of Culter recurviceps specimens. Different colors represent different lineages. Population codes see Table 1.
| 群体 Population | 单倍型数 Number of haplotype | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity | 中性检测 Neutrality tests | |
|---|---|---|---|---|---|
| Tajima’s D | Fu’s Fs | ||||
| BS | 7 | 0.383 (± 0.107) | 0.0002 (±0.0001) | -1.75856** | -6.19981*** |
| CJ | 4 | 0.806 (± 0.089) | 0.0021 (±0.0003) | 1.29983 | 2.70828 |
| CZ | 7 | 0.879 (± 0.058) | 0.0009 (±0.0001) | 0.01278 | -1.77070 |
| GP | 10 | 0.923 (± 0.060) | 0.0011 (±0.0001) | -0.27622 | -5.21979*** |
| GZ | 10 | 0.917 (± 0.049) | 0.0013 (±0.0003) | -1.10830 | -3.75174* |
| HZ | 7 | 0.891 (± 0.074) | 0.0014 (±0.0002) | -1.41142 | -1.25974 |
| KP | 4 | 0.900 (± 0.161) | 0.0019 (±0.0008) | -0.92693 | 0.35685 |
| LZ | 3 | 0.800 (± 0.164) | 0.0026 (±0.0006) | 1.25408 | 2.74649 |
| LC | 5 | 0.893 (± 0.086) | 0.0018 (±0.0005) | -0.66317 | 0.49798 |
| NF | 2 | 0.133 (± 0.112) | 0.0001 (±0.0001) | -1.15945 | -0.64899 |
| NN | 3 | 0.762 (± 0.115) | 0.0006 (±0.0002) | 1.10686 | 0.78880 |
| QY | 5 | 1.000 (± 0.126) | 0.0031 (±0.0006) | 0.40617 | -0.71434 |
| WZ | 12 | 0.987 (± 0.035) | 0.0022 (±0.0004) | -1.15138 | -6.16071*** |
| YF | 2 | 0.485 (± 0.106) | 0.0004 (±0.0001) | 1.35640 | 2.32787 |
| YN | 5 | 0.782 (± 0.107) | 0.0011 (±0.0004) | -1.13653 | 0.36244 |
| SL | 9 | 0.909 (± 0.079) | 0.0020 (±0.0004) | -0.32946 | -2.27867 |
| DA | 1 | NA | NA | NA | NA |
| SG | 2 | NA | NA | NA | NA |
| PL | 1 | NA | NA | NA | NA |
| WX | 2 | NA | NA | NA | NA |
| JM | 4 | NA | NA | NA | NA |
| MJ | 1 | NA | NA | NA | NA |
| HL | 1 | NA | NA | NA | NA |
| 总计 Total | 52 | 0.912 (± 0.012) | 0.0023 (±0.0001) | -0.91788 | -24.88587*** |
表2 基于CCN序列单倍型数、单倍型多样性、核苷酸多样性和中性检测。群体代号见表1。
Table 2 Number of haplotype, haplotype diversity, nucleotide diversity and neutrality tests based on CCN sequences. Population codes see Table 1.
| 群体 Population | 单倍型数 Number of haplotype | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity | 中性检测 Neutrality tests | |
|---|---|---|---|---|---|
| Tajima’s D | Fu’s Fs | ||||
| BS | 7 | 0.383 (± 0.107) | 0.0002 (±0.0001) | -1.75856** | -6.19981*** |
| CJ | 4 | 0.806 (± 0.089) | 0.0021 (±0.0003) | 1.29983 | 2.70828 |
| CZ | 7 | 0.879 (± 0.058) | 0.0009 (±0.0001) | 0.01278 | -1.77070 |
| GP | 10 | 0.923 (± 0.060) | 0.0011 (±0.0001) | -0.27622 | -5.21979*** |
| GZ | 10 | 0.917 (± 0.049) | 0.0013 (±0.0003) | -1.10830 | -3.75174* |
| HZ | 7 | 0.891 (± 0.074) | 0.0014 (±0.0002) | -1.41142 | -1.25974 |
| KP | 4 | 0.900 (± 0.161) | 0.0019 (±0.0008) | -0.92693 | 0.35685 |
| LZ | 3 | 0.800 (± 0.164) | 0.0026 (±0.0006) | 1.25408 | 2.74649 |
| LC | 5 | 0.893 (± 0.086) | 0.0018 (±0.0005) | -0.66317 | 0.49798 |
| NF | 2 | 0.133 (± 0.112) | 0.0001 (±0.0001) | -1.15945 | -0.64899 |
| NN | 3 | 0.762 (± 0.115) | 0.0006 (±0.0002) | 1.10686 | 0.78880 |
| QY | 5 | 1.000 (± 0.126) | 0.0031 (±0.0006) | 0.40617 | -0.71434 |
| WZ | 12 | 0.987 (± 0.035) | 0.0022 (±0.0004) | -1.15138 | -6.16071*** |
| YF | 2 | 0.485 (± 0.106) | 0.0004 (±0.0001) | 1.35640 | 2.32787 |
| YN | 5 | 0.782 (± 0.107) | 0.0011 (±0.0004) | -1.13653 | 0.36244 |
| SL | 9 | 0.909 (± 0.079) | 0.0020 (±0.0004) | -0.32946 | -2.27867 |
| DA | 1 | NA | NA | NA | NA |
| SG | 2 | NA | NA | NA | NA |
| PL | 1 | NA | NA | NA | NA |
| WX | 2 | NA | NA | NA | NA |
| JM | 4 | NA | NA | NA | NA |
| MJ | 1 | NA | NA | NA | NA |
| HL | 1 | NA | NA | NA | NA |
| 总计 Total | 52 | 0.912 (± 0.012) | 0.0023 (±0.0001) | -0.91788 | -24.88587*** |
| BS | CJ | CZ | WG | GZ | JH | NF | HZ | KP | ZC | MZ | NN | QY | SL | YF | YN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BS | 0.000 | *** | *** | *** | *** | *** | NS | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| CJ | 0.816 | 0.000 | * | * | ** | NS | *** | NS | NS | NS | NS | NS | NS | NS | * | NS |
| CZ | 0.859 | 0.160 | 0.000 | NS | *** | NS | *** | *** | NS | * | NS | NS | *** | NS | NS | NS |
| WG | 0.838 | 0.157 | 0.000 | 0.000 | ** | * | *** | ** | NS | NS | NS | NS | * | * | NS | NS |
| GZ | 0.842 | 0.260 | 0.211 | 0.111 | 0.000 | * | *** | NS | * | * | NS | *** | *** | ** | *** | *** |
| JH | 0.849 | 0.000 | 0.237 | 0.234 | 0.307 | 0.000 | *** | * | NS | NS | NS | NS | NS | NS | * | * |
| NF | 0.000 | 0.753 | 0.834 | 0.802 | 0.803 | 0.795 | 0.000 | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| HZ | 0.868 | 0.264 | 0.288 | 0.175 | 0.025 | 0.292 | 0.830 | 0.000 | NS | * | NS | *** | ** | * | *** | *** |
| KP | 0.876 | 0.000 | 0.021 | 0.021 | 0.142 | 0.000 | 0.847 | 0.150 | 0.000 | NS | NS | NS | NS | NS | NS | NS |
| ZC | 0.747 | 0.000 | 0.091 | 0.043 | 0.103 | 0.000 | 0.664 | 0.119 | 0.000 | 0.000 | NS | NS | NS | NS | * | NS |
| MZ | 0.765 | 0.050 | 0.010 | 0.000 | 0.036 | 0.058 | 0.691 | 0.057 | 0.000 | 0.000 | 0.000 | NS | * | NS | NS | NS |
| NN | 0.899 | 0.100 | 0.083 | 0.061 | 0.290 | 0.192 | 0.906 | 0.372 | 0.136 | 0.020 | 0.078 | 0.000 | * | NS | NS | NS |
| QY | 0.841 | 0.007 | 0.351 | 0.344 | 0.373 | 0.000 | 0.779 | 0.361 | 0.046 | 0.062 | 0.146 | 0.299 | 0.000 | NS | ** | * |
| SL | 0.800 | 0.000 | 0.068 | 0.085 | 0.168 | 0.000 | 0.735 | 0.185 | 0.000 | 0.003 | 0.000 | 0.105 | 0.102 | 0.000 | NS | NS |
| YF | 0.913 | 0.211 | 0.000 | 0.024 | 0.209 | 0.324 | 0.921 | 0.314 | 0.053 | 0.113 | 0.028 | 0.227 | 0.429 | 0.066 | 0.000 | NS |
| YN | 0.854 | 0.093 | 0.000 | 0.000 | 0.161 | 0.146 | 0.821 | 0.228 | 0.000 | 0.043 | 0.000 | 0.107 | 0.218 | 0.013 | 0.000 | 0.000 |
表3 两两群体之间FST值(对角线下)和显著性检验(对角线上)。群体代号见表1。
Table 3 Pairwise population FST values (below diagonal) and significance probability estimates (above diagonal). Population codes see Table 1.
| BS | CJ | CZ | WG | GZ | JH | NF | HZ | KP | ZC | MZ | NN | QY | SL | YF | YN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BS | 0.000 | *** | *** | *** | *** | *** | NS | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| CJ | 0.816 | 0.000 | * | * | ** | NS | *** | NS | NS | NS | NS | NS | NS | NS | * | NS |
| CZ | 0.859 | 0.160 | 0.000 | NS | *** | NS | *** | *** | NS | * | NS | NS | *** | NS | NS | NS |
| WG | 0.838 | 0.157 | 0.000 | 0.000 | ** | * | *** | ** | NS | NS | NS | NS | * | * | NS | NS |
| GZ | 0.842 | 0.260 | 0.211 | 0.111 | 0.000 | * | *** | NS | * | * | NS | *** | *** | ** | *** | *** |
| JH | 0.849 | 0.000 | 0.237 | 0.234 | 0.307 | 0.000 | *** | * | NS | NS | NS | NS | NS | NS | * | * |
| NF | 0.000 | 0.753 | 0.834 | 0.802 | 0.803 | 0.795 | 0.000 | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| HZ | 0.868 | 0.264 | 0.288 | 0.175 | 0.025 | 0.292 | 0.830 | 0.000 | NS | * | NS | *** | ** | * | *** | *** |
| KP | 0.876 | 0.000 | 0.021 | 0.021 | 0.142 | 0.000 | 0.847 | 0.150 | 0.000 | NS | NS | NS | NS | NS | NS | NS |
| ZC | 0.747 | 0.000 | 0.091 | 0.043 | 0.103 | 0.000 | 0.664 | 0.119 | 0.000 | 0.000 | NS | NS | NS | NS | * | NS |
| MZ | 0.765 | 0.050 | 0.010 | 0.000 | 0.036 | 0.058 | 0.691 | 0.057 | 0.000 | 0.000 | 0.000 | NS | * | NS | NS | NS |
| NN | 0.899 | 0.100 | 0.083 | 0.061 | 0.290 | 0.192 | 0.906 | 0.372 | 0.136 | 0.020 | 0.078 | 0.000 | * | NS | NS | NS |
| QY | 0.841 | 0.007 | 0.351 | 0.344 | 0.373 | 0.000 | 0.779 | 0.361 | 0.046 | 0.062 | 0.146 | 0.299 | 0.000 | NS | ** | * |
| SL | 0.800 | 0.000 | 0.068 | 0.085 | 0.168 | 0.000 | 0.735 | 0.185 | 0.000 | 0.003 | 0.000 | 0.105 | 0.102 | 0.000 | NS | NS |
| YF | 0.913 | 0.211 | 0.000 | 0.024 | 0.209 | 0.324 | 0.921 | 0.314 | 0.053 | 0.113 | 0.028 | 0.227 | 0.429 | 0.066 | 0.000 | NS |
| YN | 0.854 | 0.093 | 0.000 | 0.000 | 0.161 | 0.146 | 0.821 | 0.228 | 0.000 | 0.043 | 0.000 | 0.107 | 0.218 | 0.013 | 0.000 | 0.000 |
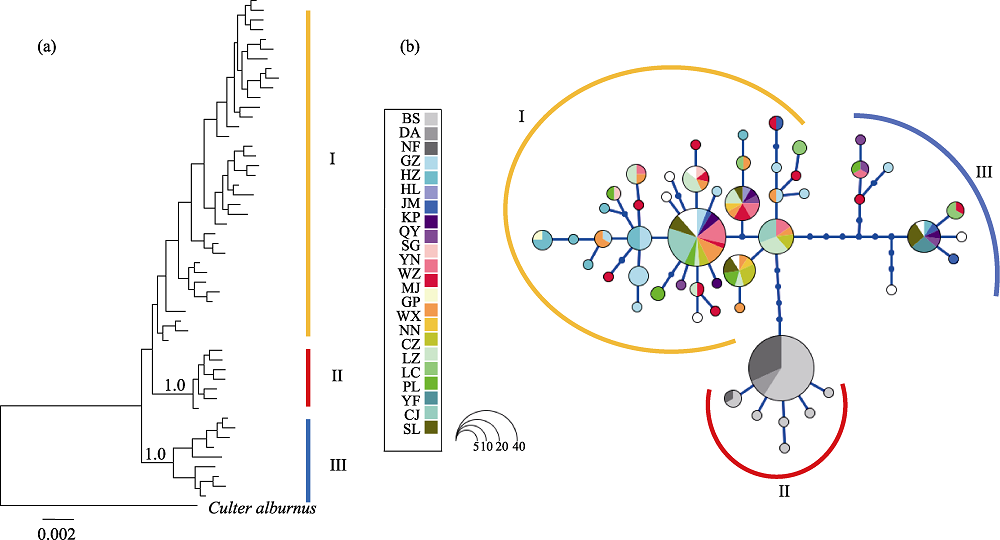
图2 分子系统树和单倍型网络图。(a)单倍型的分子系统树(数字代表后验概率); (b)海南鲌单倍型网络图; 圆圈的大小对应单倍型频率。群体代号见表1。
Fig. 2 Bayesian tree and haplotype network. (a) Bayesian tree for Culter recurviceps (numbers indicated posterior probability); (b) Haplotype network of C. recurviceps. Cycle size is roughly proportional to the haplotype frequency. Population codes see Table 1.
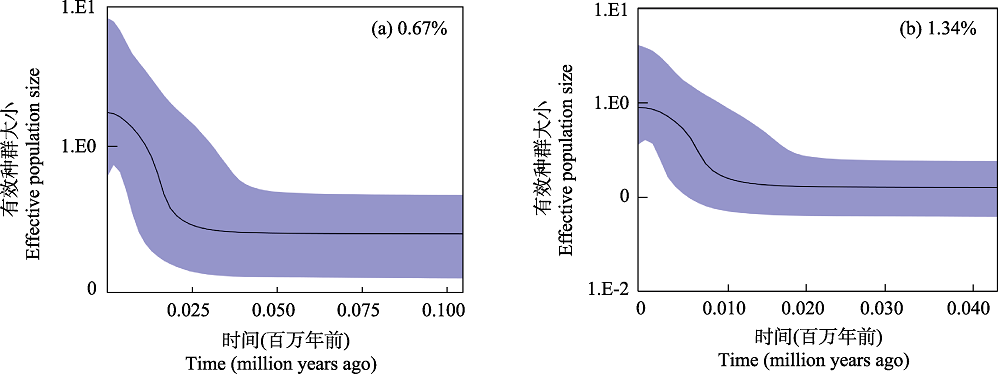
图3 海南鲌贝叶斯天际线点图。(a)基于0.67%的进化速率; (b)基于1.34%的进化速率。
Fig. 3 Bayesian skyline plots of Culter recurviceps. (a) Based on 0.67% evolutionary rate; (b) Based on 1.34% evolutionary rate.
| [1] |
Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC (1987) Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Annual Review of Ecology and Systematics, 18, 489-522.
DOI URL |
| [2] | Chen WT, Li C, Chen FC, Li YF, Yang JP, Li J, Li XH (2020) Phylogeographic analyses of a migratory freshwater fish (Megalobrama terminalis) reveal a shallow genetic structure and pronounced effects of sea-level changes. Gene, 737, 144478. |
| [3] |
Chen XL, Chiang TY, Lin HD, Zheng HS, Shao KT, Zhang Q, Hsu KC (2007) Mitochondrial DNA phylogeography of Glyptothorax fokiensis and Glyptothorax hainanensis in Asia. Journal of Fish Biology, 70, 75-93.
DOI URL |
| [4] |
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214.
PMID |
| [5] |
Durand JD, Tsigenopoulos CS, Ünlü E, Berrebi P (2002) Erratum to “Phylogeny and Biogeography of the Family Cyprinidae in the Middle East Inferred from Cytochrome b DNA-Evolutionary Significance of This Region”. Molecular Phylogenetics and Evolution, 22, 91-100.
PMID |
| [6] |
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI PMID |
| [7] |
Fu YX (1996) Estimating the age of the common ancestor of a DNA sample using the number of segregating sites. Genetics, 144, 829-838.
PMID |
| [8] |
Gascoyne M, Benjamin GJ, Schwarcz HP, Ford DC (1979) Sea-level lowering during the Illinoian glaciation: Evidence from a Bahama “Blue Hole”. Science, 205, 806-808.
PMID |
| [9] |
Hey J (2010) Isolation with migration models for more than two populations. Molecular Biology and Evolution, 27, 905-920.
DOI URL |
| [10] |
Ketmaier V, Bianco PG, Cobolli M, Krivokapic M, Caniglia R,De Matthaeis E (2004) Molecular phylogeny of two lineages of Leuciscinae cyprinids (Telestes and Scardinius) from the peri-Mediterranean area based on cytochrome b data. Molecular Phylogenetics and Evolution, 32, 1061-1071.
PMID |
| [11] |
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16, 111-120.
PMID |
| [12] |
Li J, Li XH, Tan XC, Li YF, He MF, Luo JR, Lin JZ, Su SF (2009) Species diversity of fish community of Provincial Xijiang River Rare Fishes Natural Reserve in Zhaoqing City, Guangdong Province. Journal of Lake Sciences, 21, 556-562. (in Chinese with English abstract)
DOI URL |
| [ 李捷, 李新辉, 谭细畅, 李跃飞, 何美峰, 罗建仁, 林建志, 苏少芳 (2009) 广东肇庆西江珍稀鱼类省级自然保护区鱼类多样性. 湖泊科学, 21, 556-562.] | |
| [13] |
Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452.
DOI PMID |
| [14] |
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: Combining landscape ecology and population genetics. Trends in Ecology & Evolution, 18, 189-197.
DOI URL |
| [15] |
Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey J (2004) Bayesian phylogenetic analysis of combined data. Systematic Biology, 53, 47-67.
PMID |
| [16] | Qiu CF, Lin YG, Qing N, Zhao J, Chen XL (2008) Genetic variation and phylogeography of Micronoemacheilus pulcher populations among basin systems between western South China and Hainan Island. Acta Zoologica Sinica, 54, 805-813. (in Chinese with English abstract) |
| [ 丘城锋, 林岳光, 庆宁, 赵俊, 陈湘粦 (2008) 华南西部及海南岛美丽小条鳅种群遗传变异与亲缘地理. 动物学报, 54, 805-813.] | |
| [17] |
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572-1574.
PMID |
| [18] | Salzburger W, Ewing GB, Von Haeseler A (2011) The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Molecular Ecology, 20, 1952-1963. |
| [19] | Shuai FM, Li XF, Liu QF, Li YF, Yang JP, Li J, Chen FC (2017) Spatial patterns of fish diversity and distribution in the Pearl River. Acta Ecologica Sinica, 37, 3182-3192. (in Chinese with English abstract) |
| [ 帅方敏, 李新辉, 刘乾甫, 李跃飞, 杨计平, 李捷, 陈方灿 (2017) 珠江水系鱼类群落多样性空间分布格局. 生态学报, 37, 3182-3192.] | |
| [20] |
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585-595.
PMID |
| [21] |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725-2729.
DOI URL |
| [22] |
Wright S (1943) Isolation by distance. Genetics, 28, 114-138.
PMID |
| [23] | Xie G, Pang SX, Xu SY, Ye X, Qi BL, Pan DB (1998) Artificial reproduction and embryo development of Culter recurviceps. Fisheries Science and Technology, (5), 23-25. (in Chinese) |
| [ 谢刚, 庞世勋, 许淑英, 叶星, 祁宝仑, 潘德博 (1998) 海南红鲌人工繁殖和胚胎发育. 水产科技, (5), 23-25.] | |
| [24] | Xu SY, Xie G, Qi BL, Ye X, Pang SX (2001) Study on the sensitivity of Culter recurviceps to aquatic drugs. Reservoir Fisheries, 21(1), 36-36. (in Chinese) |
| [ 许淑英, 谢刚, 祁宝仑, 叶星, 庞世勋 (2001) 海南红鲌对水产药物的敏感性研究. 水利渔业, 21(1), 36-37.] | |
| [25] | Xuan ZY, Jiang T, Liu HB, Chen XB, Yang J (2020) Genetic divergence of Coilia nasus taihuensis and Coilia brachygnathus populations based on the mitochondrial Cyt-b gene. Progress in Fishery Sciences, 41(4), 33-40. (in Chinese with English abstract) |
| [ 轩中亚, 姜涛, 刘洪波, 陈修报, 杨健 (2020) 基于线粒体Cyt-b序列的太湖湖鲚与短颌鲚种群遗传分析. 渔业科学进展, 41(4), 33-40.] | |
| [26] |
Yang JQ, Hsu KC, Liu ZZ, Su LW, Kuo PH, Tang WQ, Zhou ZC, Liu D, Bao BL, Lin HD (2016) The population history of Garra orientalis (Teleostei: Cyprinidae) using mitochondrial DNA and microsatellite data with approximate Bayesian computation. BMC Evolutionary Biology, 16, 73.
DOI URL |
| [27] | Zheng CY (1989) The Ichthyography of the Pearl River. Science Press, Beijing. (in Chinese) |
| [ 郑慈英 (1989) 珠江鱼类志. 科学出版社, 北京.] | |
| [28] | Zhou ZX, Liu B, Gong J, Bai YL, Yang JY, Xu P (2020) Phylogeny and population genetics of species in Takifugu genus based on mitochondrial genome. Journal of Fisheries of China, 44, 1792-1803. (in Chinese with English abstract) |
| [ 周志雄, 刘波, 宫杰, 白玉麟, 阳俊逸, 徐鹏 (2020) 基于线粒体基因组的东方鲀属系统发育学及群体遗传学. 水产学报, 44, 1792-1803.] |
| [1] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [2] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [3] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [4] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [5] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [6] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [7] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [8] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [9] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [10] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [11] | 崔静, 徐明芳, 章群, 李瑶, 曾晓舒, 李莎. 基于3种线粒体标记探讨中日沿海角木叶鲽遗传多样性差异[J]. 生物多样性, 2022, 30(5): 21485-. |
| [12] | 孙军, 宋煜尧, 施义锋, 翟键, 燕文卓. 近十年中国海洋生物多样性研究进展[J]. 生物多样性, 2022, 30(10): 22526-. |
| [13] | 栗冬梅, 杨卫红, 李庆多, 韩茜, 宋秀平, 潘虹, 冯云. 巴尔通体在滇西南蝙蝠中高度流行并具有丰富的遗传变异特征[J]. 生物多样性, 2021, 29(9): 1245-1255. |
| [14] | 姚志, 郭军, 金晨钟, 刘勇波. 中国纳入一级保护的极小种群野生植物濒危机制[J]. 生物多样性, 2021, 29(3): 394-408. |
| [15] | 叶俊伟, 田斌. 中国西南地区重要木本油料植物扁核木的遗传结构及成因[J]. 生物多样性, 2021, 29(12): 1629-1637. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn