



生物多样性 ›› 2021, Vol. 29 ›› Issue (8): 1128-1133. DOI: 10.17520/biods.2021213 cstr: 32101.14.biods.2021213
收稿日期:2021-05-25
接受日期:2021-07-20
出版日期:2021-08-20
发布日期:2021-08-16
通讯作者:
刘勇波
作者简介:* E-mail: liuyb@craes.org.cn基金资助:Received:2021-05-25
Accepted:2021-07-20
Online:2021-08-20
Published:2021-08-16
Contact:
Yongbo Liu
摘要:
基因组多倍化是物种形成和进化的重要驱动力, 几乎所有植物都经历过至少一次基因组加倍。然而, 由于多倍体植株比二倍体表现出更高的死亡率, 多倍化机制被认为是植物进化的“死胡同”。一些植物物种具有自然混合倍性种群, 即同一物种具有不同倍性, 这为揭示多倍体的进化机制提供了最佳途径。本文从基因组加倍形成多倍体植物开始, 综述了混合倍性种群的形成、建立与维持的研究进展, 探讨了多倍体适应自然环境的种群分化而形成多倍体物种的机制。研究自然混合倍性种群的倍性组成、重复基因的功能分化以及多倍体的生态位分化, 有利于明确混合倍性自然种群的生态适应与维持机理, 以及多倍体植物的进化机制。
刘勇波 (2021) 多倍体植物混合倍性种群的建立机制研究进展. 生物多样性, 29, 1128-1133. DOI: 10.17520/biods.2021213.
Yongbo Liu (2021) The mechanism of constructing mixed-ploidy populations in polyploid species. Biodiversity Science, 29, 1128-1133. DOI: 10.17520/biods.2021213.
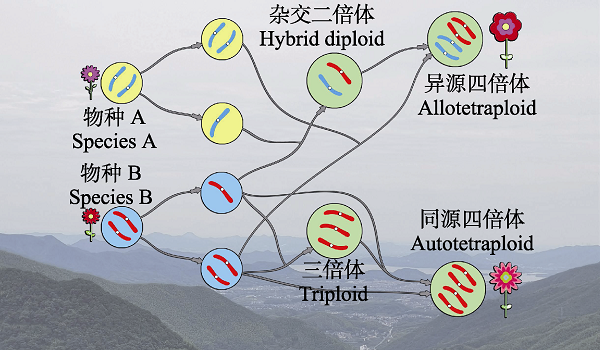
图1 多倍体形成途径的简单示意图。二倍体物种产生正常的单倍体配子和未减数分裂的配子, 未减数分裂配子可以和单体配子产生三倍体, 也可以融合产生同源四倍体; 同时也可以通过体细胞加倍产生同源四倍体; 异源四倍体通过物种之间未减数分裂配子的融合或者通过物种间的杂交再加倍产生。
Fig. 1 Formation paths to polyploid species. Diploid species produce normal haploid gametes and unreduced gametes. Unreduced gametes combine with haploid gametes to give rise to triploids or fuse to be autotetraploid species. Diploids can yield somatically polyploids. Allotetraploids are yielded through the fusion of unreduced gametes or the hybridization of haploid gametes from different species.
| [1] |
Arrigo N, Barker MS (2012) Rarely successful polyploids and their legacy in plant genomes. Current Opinion in Plant Biology, 15, 140-146.
DOI PMID |
| [2] | Babcock EB, Stebbins GL (1938) The American Species of Crepis: Their Interrelationships and Distribution as Affected by Polyploidy and Apomixis. Carnegie Institution of Washington Publication, no. 504, Washington, USA. |
| [3] |
Baduel P, Bray S, Vallejo-Marin M, Kolář F, Yant L (2018) The “Polyploid Hop”: Shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Frontiers in Ecology and Evolution, 6, 117.
DOI URL |
| [4] |
Baniaga AE, Marx HE, Arrigo N, Barker MS (2020) Polyploid plants have faster rates of multivariate niche differentiation than their diploid relatives. Ecology Letters, 23, 68-78.
DOI URL |
| [5] |
Blaine Marchant D, Soltis DE, Soltis PS (2016) Patterns of abiotic niche shifts in allopolyploids relative to their progenitors. New Phytologist, 212, 708-718.
DOI PMID |
| [6] |
Bottani S, Zabet NR, Wendel JF, Veitia RA (2018) Gene Expression dominance in allopolyploids: Hypotheses and models. Trends in Plant Science, 23, 393-402.
DOI PMID |
| [7] |
Castro M, Loureiro J, Figueiredo A, Serrano M, Husband BC, Castro S (2020) Different patterns of ecological divergence between two tetraploids and their diploid counterpart in a parapatric linear coastal distribution polyploid complex. Frontiers in Plant Science, 11, 315.
DOI URL |
| [8] | De Smet R, Adams KL, Vandepoele K, Van Montagu MCE, Maere S, Van de Peer Y (2013) Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proceedings of the National Academy of Sciences, USA, 110, 2898-2903. |
| [9] |
Doyle JJ, Coate JE (2019) Polyploidy, the nucleotype, and novelty: The impact of genome doubling on the biology of the cell. International Journal of Plant Sciences, 180, 1-52.
DOI URL |
| [10] |
Eric Schranz M, Mohammadin S, Edger PP (2012) Ancient whole genome duplications, novelty and diversification: The WGD Radiation Lag-Time Model. Current Opinion in Plant Biology, 15, 147-153.
DOI PMID |
| [11] | Fawcett JA, Maere S, Van de Peer Y (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proceedings of the National Academy of Sciences, USA, 106, 5737-5742. |
| [12] | Folk RA, Siniscalchi CM, Soltis DE (2020) Angiosperms at the edge: Extremity, diversity, and phylogeny. Plant, Cell & Environment, 43, 2871-2893. |
| [13] |
Freeling M (2017) Picking up the Ball at the K/Pg Boundary: The distribution of ancient polyploidies in the plant phylogenetic tree as a spandrel of asexuality with occasional sex. The Plant Cell, 29, 202-206.
DOI PMID |
| [14] |
Gunn BF, Murphy DJ, Walsh NG, Conran JG, Pires JC, MacFarlane TD, Birch JL (2020) Evolution of Lomandroideae: Multiple origins of polyploidy and biome occupancy in Australia. Molecular Phylogenetics and Evolution, 149, 106836.
DOI PMID |
| [15] |
Hias N, Svara A, Keulemans JW (2018) Effect of polyploidisation on the response of apple (Malus ×domestica Borkh.) toVenturia inaequalis infection. European Journal of Plant Pathology, 151, 515-526.
DOI URL |
| [16] |
Hohmann N, Wolf EM, Lysak MA, Koch MA (2015) A time-calibrated road map of Brassicaceae species radiation and evolutionary history. The Plant Cell, 27, 2770-2784.
DOI PMID |
| [17] |
Hülber K, Sonnleitner M, Suda J, Krejčíková J, Schönswetter P, Schneeweiss GM, Winkler M (2015) Ecological differentiation, lack of hybrids involving diploids, and asymmetric gene flow between polyploids in narrow contact zones ofSenecio carniolicus (syn. Jacobaea carniolica, Asteraceae). Ecology and Evolution, 5, 1224-1234.
DOI URL |
| [18] |
Jiao YN, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang HY, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, de Pamphilis CW (2011) Ancestral polyploidy in seed plants and angiosperms. Nature, 473, 97-100.
DOI URL |
| [19] |
Kao RH (2007) Asexuality and the coexistence of cytotypes. New Phytologist, 175, 764-772.
DOI URL |
| [20] |
Kolář F, Čertner M, Suda J, Schönswetter P, Husband BC (2017) Mixed-ploidy species: Progress and opportunities in polyploid research. Trends in Plant Science, 22, 1041-1055.
DOI URL |
| [21] |
Levin DA (1975) Minority cytotype exclusion in local plant populations. Taxon, 24, 35-43.
DOI URL |
| [22] |
Li DW, Liu YF, Zhong CH, Huang HW (2010) Morphological and cytotype variation of wild kiwifruit (Actinidia chinensis complex) along an altitudinal and longitudinal gradient in central-west China. Botanical Journal of the Linnean Society, 164, 72-83.
DOI URL |
| [23] |
Liang SQ, Zhang XC, Wei R (2019) Integrative taxonomy resolved species delimitation in a fern complex: A case study of theAsplenium coenobiale complex. Biodiversity Science, 27, 1205-1220. (in Chinese with English abstract)
DOI |
|
[ 梁思琪, 张宪春, 卫然 (2019) 利用整合分类学方法进行蕨类植物复合体的物种划分: 以线裂铁角蕨复合体为例. 生物多样性, 27, 1205-1220.]
DOI |
|
| [24] |
Mandák B, Vít P, Krak K, Trávníček P, Havrdová A, Hadincová V, Zákravský P, Jarolímová V, Bacles CFE, Douda J (2016) Flow cytometry, microsatellites and niche models reveal the origins and geographical structure ofAlnus glutinosa populations in Europe. Annals of Botany, 117, 107-120.
DOI URL |
| [25] |
Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, Otto SP (2011) Recently formed polyploid plants diversify at lower rates. Science, 333, 1257-1257.
DOI PMID |
| [26] | Morgan C, Zhang HK, Henry CE, Franklin FCH, Bomblies K (2020) Derived alleles of two axis proteins affect meiotic traits in autotetraploidArabidopsis arenosa. Proceedings of the National Academy of Sciences, USA, 117, 8980-8988. |
| [27] |
Mráz P, Španiel S, Keller A, Bowmann G, Farkas A, Šingliarová B, Rohr RP, Broennimann O, Müller-Schärer H (2012) Anthropogenic disturbance as a driver of microspatial and microhabitat segregation of cytotypes ofCentaurea stoebe and cytotype interactions in secondary contact zones. Annals of Botany, 110, 615-627.
DOI URL |
| [28] |
Nuismer SL, Cunningham BM (2005) Selection for phenotypic divergence between diploid and autotetraploidHeuchera grossulariifolia. Evolution, 59, 1928-1935.
PMID |
| [29] |
Ramsey J, Ramsey TS (2014) Ecological studies of polyploidy in the 100 years following its discovery. Philosophical Transactions of the Royal Society B: Biological Sciences, 369, 20130352.
DOI URL |
| [30] |
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics, 29, 467-501.
DOI URL |
| [31] | Rao JY, Liu YF, Huang HW (2012) Analysis of ploidy segregation and genetic variation of progenies of different interploidy crosses inActinidia chinensis. Acta Horticulturae Sinica, 39, 1447-1456. (in Chinese with English abstract) |
| [ 饶静云, 刘义飞, 黄宏文 (2012) 中华猕猴桃不同倍性间杂交后代倍性分离和遗传变异分析. 园艺学报, 39, 1447-1456.] | |
| [32] |
Ren R, Wang HF, Guo CC, Zhang N, Zeng LP, Chen YM, Ma H, Qi J (2018) Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Molecular Plant, 11, 414-428.
DOI PMID |
| [33] |
Rice A, Glick L, Abadi S, Einhorn M, Kopelman NM, Salman-Minkov A, Mayzel J, Chay O, Mayrose I (2015) The Chromosome Counts Database (CCDB)—A community resource of plant chromosome numbers. New Phytologist, 206, 19-26.
DOI URL |
| [34] | Rice A, Šmarda P, Novosolov M, Drori M, Glick L, Sabath N, Meiri S, Belmaker J, Mayrose I (2019) The global biogeography of polyploid plants. Nature Ecology & Evolution, 3, 265-273. |
| [35] |
Rieseberg LH, Willis JH (2007) Plant speciation. Science, 317, 910-914.
PMID |
| [36] |
Sabara HA, Kron P, Husband BC (2013) Cytotype coexistence leads to triploid hybrid production in a diploid-tetraploid contact zone ofChamerion angustifolium (Onagraceae). American Journal of Botany, 100, 962-970.
DOI PMID |
| [37] |
Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng CF, Sankoff Dde Pamphilis CW, Wall PK, Soltis PS (2009) Polyploidy and angiosperm diversification. American Journal of Botany, 96, 336-348.
DOI URL |
| [38] |
Soltis DE, Segovia-Salcedo MC, Jordon-Thaden I, Majure L, Miles NM, Mavrodiev EV, Mei WB, Cortez MB, Soltis PS, Gitzendanner MA (2014) Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al (2011). New Phytologist, 202, 1105-1117.
DOI URL |
| [39] |
Sonnleitner M, Hülber K, Flatscher R, García PE, Winkler M, Suda J, Schönswetter P, Schneeweiss GM (2016) Ecological differentiation of diploid and polyploid cytotypes ofSenecio carniolicus sensu lato (Asteraceae) is stronger in areas of sympatry. Annals of Botany, 117, 269-276.
DOI PMID |
| [40] |
Stevens AV, Nicotra AB, Godfree RC, Guja LK (2020) Polyploidy affects the seed, dormancy and seedling characteristics of a perennial grass, conferring an advantage in stressful climates. Plant Biology, 22, 500-513.
DOI PMID |
| [41] | Stebbins GL (1950) Variation and Evolution in Plants. Columbia University Press, New York. |
| [42] | Stebbins GL (1971) Chromosomal Evolution in Higher Plants. Edward Arnold, London. |
| [43] |
Suda J, Weiss-Schneeweiss H, Tribsch A, Schneeweiss GM, Trávníček P, Schönswetter P (2007) Complex distribution patterns of di-, tetra-, and hexaploid cytotypes in the European high mountain plantSenecio carniolicus (Asteraceae). American Journal of Botany, 94, 1391-1401.
DOI PMID |
| [44] |
Sutherland BL, Galloway LF (2017) Postzygotic isolation varies by ploidy level within a polyploid complex. New Phytologist, 213, 404-412.
DOI PMID |
| [45] |
The Brassica rapa Genome Sequencing Project Consortium (2011) The genome of the mesopolyploid crop speciesBrassica rapa. Nature Genetics, 43, 1035-1039.
DOI URL |
| [46] |
Van de Peer Y, Ashman TL, Soltis PS, Soltis DE (2021) Polyploidy: An evolutionary and ecological force in stressful times. The Plant Cell, 33, 11-26.
DOI URL |
| [47] |
Van de Peer Y, Mizrachi E, Marchal K (2017) The evolutionary significance of polyploidy. Nature Reviews Genetics, 18, 411-424.
DOI URL |
| [48] |
Wang WN, He YH, Cao Z, Deng ZN (2018) Induction of tetraploids inImpatiens (Impatiens walleriana) and characterization of their changes in morphology and resistance to downy mildew. HortScience, 53, 925-931.
DOI URL |
| [49] | Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH (2009) The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences, USA, 106, 13875-13879. |
| [50] |
Wu SD, Han BC, Jiao YN (2020) Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Molecular Plant, 13, 59-71.
DOI URL |
| [51] |
Yant L, Hollister JD, Wright KM, Arnold BJ, Higgins JD, Franklin FCH, Bomblies K (2013) Meiotic adaptation to genome duplication inArabidopsis arenosa. Current Biology, 23, 2151-2156.
DOI URL |
| [52] | Zeng H, Li DW, Huang HW (2009) Distribution pattern of ploidy variation ofActinidia chinensis andA. deliciosa. Journal of Wuhan Botanical Research, 27, 312-317. (in Chinese with English abstract) |
| [ 曾华, 李大卫, 黄宏文 (2009) 中华猕猴桃和美味猕猴桃的倍性变异及地理分布研究. 武汉植物学研究, 27, 312-317.] | |
| [53] | Zhang HK, Bian Y, Gou XW, Dong YZ, Rustgi S, Zhang BJ, Xu CM, Li N, Qi B, Han FP, von Wettstein D, Liu B (2013) Intrinsic karyotype stability and gene copy number variations may have laid the foundation for tetraploid wheat formation. Proceedings of the National Academy of Sciences, USA, 110, 19466-19471. |
| [54] |
Zhang K, Wang XW, Cheng F (2019) Plant polyploidy: Origin, evolution, and its influence on crop domestication. Horticultural Plant Journal, 5, 231-239.
DOI |
| [1] | 易木荣, 卢萍, 彭勇, 汤勇, 许久恒, 尹浩萍, 张路杨, 翁晓东, 底明晓, 雷隽, 卢宸祺, 曹如君, 戴年华, 占德洋, 童媚, 楼智明, 丁永刚, 柴静, 车静. 北潦河金家水支流江西大鲵野外种群现状及栖息地评估[J]. 生物多样性, 2025, 33(4): 24145-. |
| [2] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [3] | 陈嘉珈, 蒲真, 黄中鸿, 于凤琴, 张建军, 许东华, 徐俊泉, 尚鹏, 地里木拉提∙帕尔哈提, 李耀江, Jigme Tshering, 郭玉民. 全球黑颈鹤越冬种群分布与数量[J]. 生物多样性, 2023, 31(6): 22400-. |
| [4] | 李苗, 要晨阳, 陈小勇. 环境RNA技术在水生生物监测中的应用[J]. 生物多样性, 2023, 31(5): 23062-. |
| [5] | 李钊丞, 张燕雪丹. 基于物种濒危状况评价与种群增长的一种新评估方法在水生野生动物保护司法中的应用[J]. 生物多样性, 2023, 31(3): 22319-. |
| [6] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [7] | 李珍珍, 杜梦甜, 朱原辛, 王大伟, 李治霖, 王天明. 基于红外相机的不可个体识别动物种群密度估算方法[J]. 生物多样性, 2023, 31(3): 22422-. |
| [8] | 许再富. 对国家植物园体系建设“统筹原则”的一些见解[J]. 生物多样性, 2023, 31(1): 22470-. |
| [9] | 杨剑焕, 李敬华, 杨浩炫, 欧梓键, 郑玺, Anthony J. Giordano, 陈辈乐. 基于红外相机数据评估华南地区豹猫的种群密度和活动节律[J]. 生物多样性, 2022, 30(9): 21357-. |
| [10] | 韦怡, 姜广顺. 虎豹及有蹄类猎物种群数量监测方法概述[J]. 生物多样性, 2022, 30(9): 21551-. |
| [11] | 夏凡, 杨婧, 李建, 史洋, 盖立新, 黄文华, 张经纬, 杨南, 高福利, 韩莹莹, 鲍伟东. 北京地区四个豹猫亚种群肠道菌群的组成[J]. 生物多样性, 2022, 30(9): 22103-. |
| [12] | 初漠嫣, 梁书洁, 李沛芸, 贾丁, 阿卜杜赛麦提·买尔迪亚力, 李雪阳, 姜楠, 赵翔, 李发祥, 肖凌云, 吕植. 三江源国家级自然保护区内云塔村雪豹种群动态[J]. 生物多样性, 2022, 30(9): 22157-. |
| [13] | 孙哲明, 刘亚恒, 彭秋桐, 徐芷妍, 杨予静, 欧文慧, 李中强. 湖北省极小种群野生植物在原生群落中的竞争地位及保护建议[J]. 生物多样性, 2022, 30(6): 21517-. |
| [14] | 颜文博, 莫燕妮, 曾治高, 薛少亮, 王琦, 梁春生, 黄祝礼, 罗文, 刘大业, 莫世琴, 李晓光, 梁路, 杜鹍鹏. 海南尖峰岭中华穿山甲的分布与保护现状[J]. 生物多样性, 2022, 30(6): 22106-. |
| [15] | 袁智勇, 陈进民, 吴云鹤, 李先琦, 车静. 云南省两栖类物种名录修订[J]. 生物多样性, 2022, 30(4): 21470-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
