



生物多样性 ›› 2020, Vol. 28 ›› Issue (12): 1570-1580. DOI: 10.17520/biods.2019390 cstr: 32101.14.biods.2019390
所属专题: 青藏高原生物多样性与生态安全
• 研究报告 • 上一篇
李明家1,2, 吴凯媛1,3, 孟凡凡1,2, 沈吉1,2, 刘勇勤2,4, 肖能文5,*( ), 王建军1,2,*(
), 王建军1,2,*( )
)
收稿日期:2019-12-07
接受日期:2020-03-08
出版日期:2020-12-20
发布日期:2020-05-25
通讯作者:
肖能文,王建军
作者简介:xiaonw@163.com基金资助:
Mingjia Li1,2, Kaiyuan Wu1,3, Fanfan Meng1,2, Ji Shen1,2, Yongqin Liu2,4, Nengwen Xiao5,*( ), Jianjun Wang1,2,*(
), Jianjun Wang1,2,*( )
)
Received:2019-12-07
Accepted:2020-03-08
Online:2020-12-20
Published:2020-05-25
Contact:
Nengwen Xiao,Jianjun Wang
摘要:
理解沿环境或空间梯度的群落组成变化(即beta多样性)一直是生态学和保护生物学的中心问题, 且beta多样性的形成机制及其对环境的响应已成为当前生物多样性研究的热点问题。本文以西藏横断山区怒江和澜沧江两个流域入江溪流中的细菌为研究对象, 使用Baselga的beta多样性分解方法, 基于Sørensen相异性指数将细菌的beta多样性分解为周转(turnover)和嵌套(nestedness)两个组分, 探究了细菌beta多样性及其分解组分随海拔距离的分布模式, 并且衡量了环境、气候和空间因子的相对重要性。结果表明, 两个流域中细菌的群落结构显著不同。两个流域的细菌总beta多样性和周转组分随海拔距离的增加而增加, 周转组分占总beta多样性的比例较大。气候和环境因子是两个流域中细菌总beta多样性及周转过程的重要预测因子, 并且所有的显著因子均为正相关, 其中环境因子中相关性最高的为海拔距离(R 2= 0.408, P < 0.001), 而气候因子中相关性最高的为年均温差(R 2= 0.417, P < 0.001)。方差分解结果暗示嵌套组分主要受空间扩散的影响; 总beta多样性和周转组分在环境较恶劣的澜沧江主要受环境过滤的影响, 而在环境较温和的怒江主要受空间扩散和环境过滤的共同影响。此外, 较为恶劣的环境条件会增加细菌的总beta多样性和周转率, 并且会形成更强的环境筛选作用去影响细菌群落的物种组成。我们的研究表明对西藏横断山区水体细菌多样性的保护需要从整个流域入手, 而非少量的生物多样性热点地区。
李明家, 吴凯媛, 孟凡凡, 沈吉, 刘勇勤, 肖能文, 王建军 (2020) 西藏横断山区溪流细菌beta多样性组分对气候和水体环境的响应. 生物多样性, 28, 1570-1580. DOI: 10.17520/biods.2019390.
Mingjia Li, Kaiyuan Wu, Fanfan Meng, Ji Shen, Yongqin Liu, Nengwen Xiao, Jianjun Wang (2020) Beta diversity of stream bacteria in Hengduan Mountains: The effects of climatic and environmental variables. Biodiversity Science, 28, 1570-1580. DOI: 10.17520/biods.2019390.
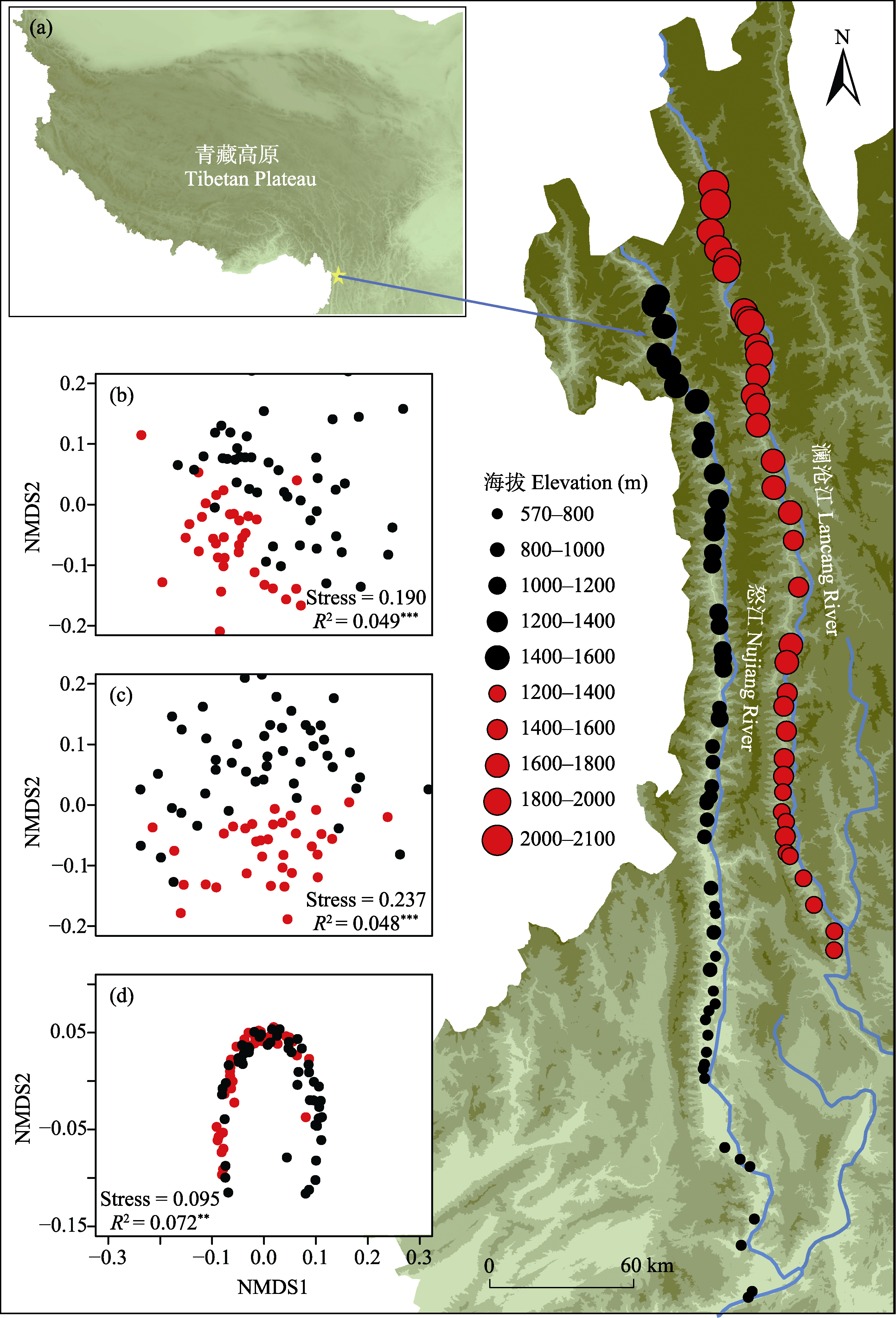
图1 本研究在澜沧江和怒江的89条入江溪流的采样点。(a)研究区位于青藏高原东南部生物多样性热点地区。总共有37条采样溪流流入澜沧江(即澜沧江流域), 52条采样溪流流入怒江(即怒江流域)。NMDS图显示了两个流域中细菌总beta多样性(b)、周转组分(c)和嵌套组分(d)的差异。红色点和黑色点分别代表澜沧江和怒江流域的采样点。* P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001。
Fig. 1 The sample sites of this study along the 89 streams of Lancang River and the Nujiang River. (a) The study area is located in the biodiversity hotspot of south-eastern Tibetan Plateau. In total, 37 streams flow into the Lancang River, while 52 sampled streams flow into the Nujiang River. NMDS plots illustrate the differences in bacterial total beta diversity (b), turnover (c) and nestedness components (d) across the Lancang and Nujiang catchments. The red dots and black dots indicate the sample sites in Langcang and Nujiang catchment, respectively. * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
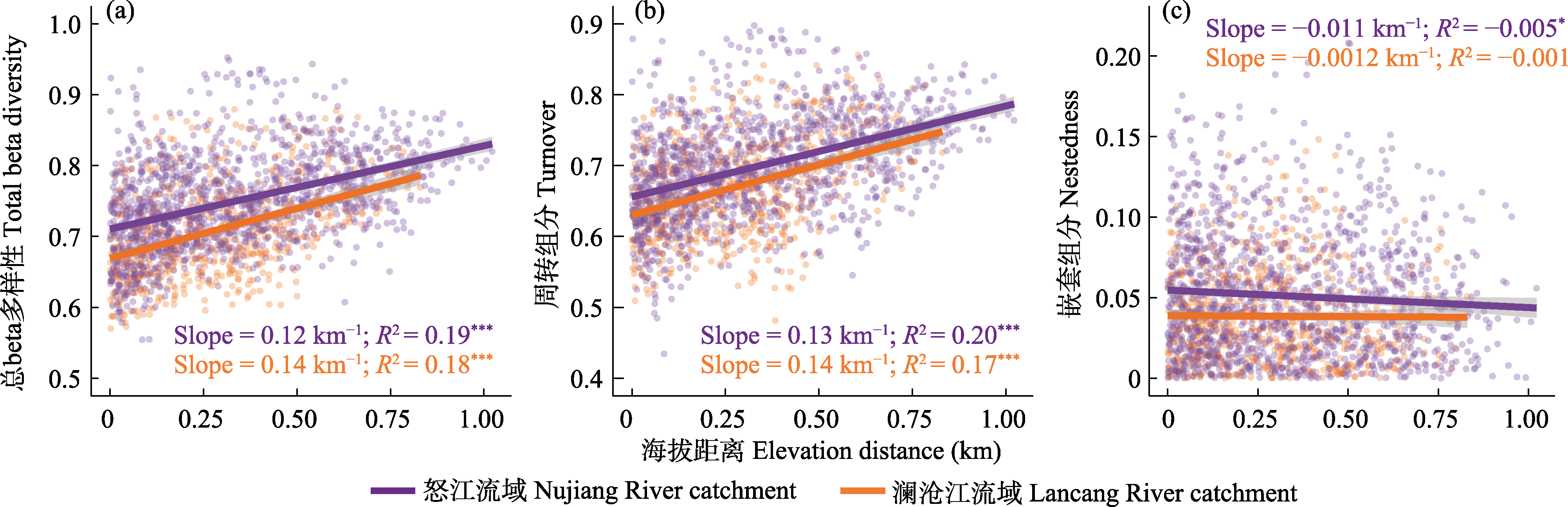
图2 澜沧江和怒江流域细菌总beta多样性(a)、周转组分(b)和嵌套组分(c)与海拔距离的关系。其显著性水平分别为: * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001。
Fig. 2 Plots showing the relationship between (a) bacterial total beta diversity, (b) turnover and (c) nestedness components and elevational distance in Lancang and Nujiang catchments. * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
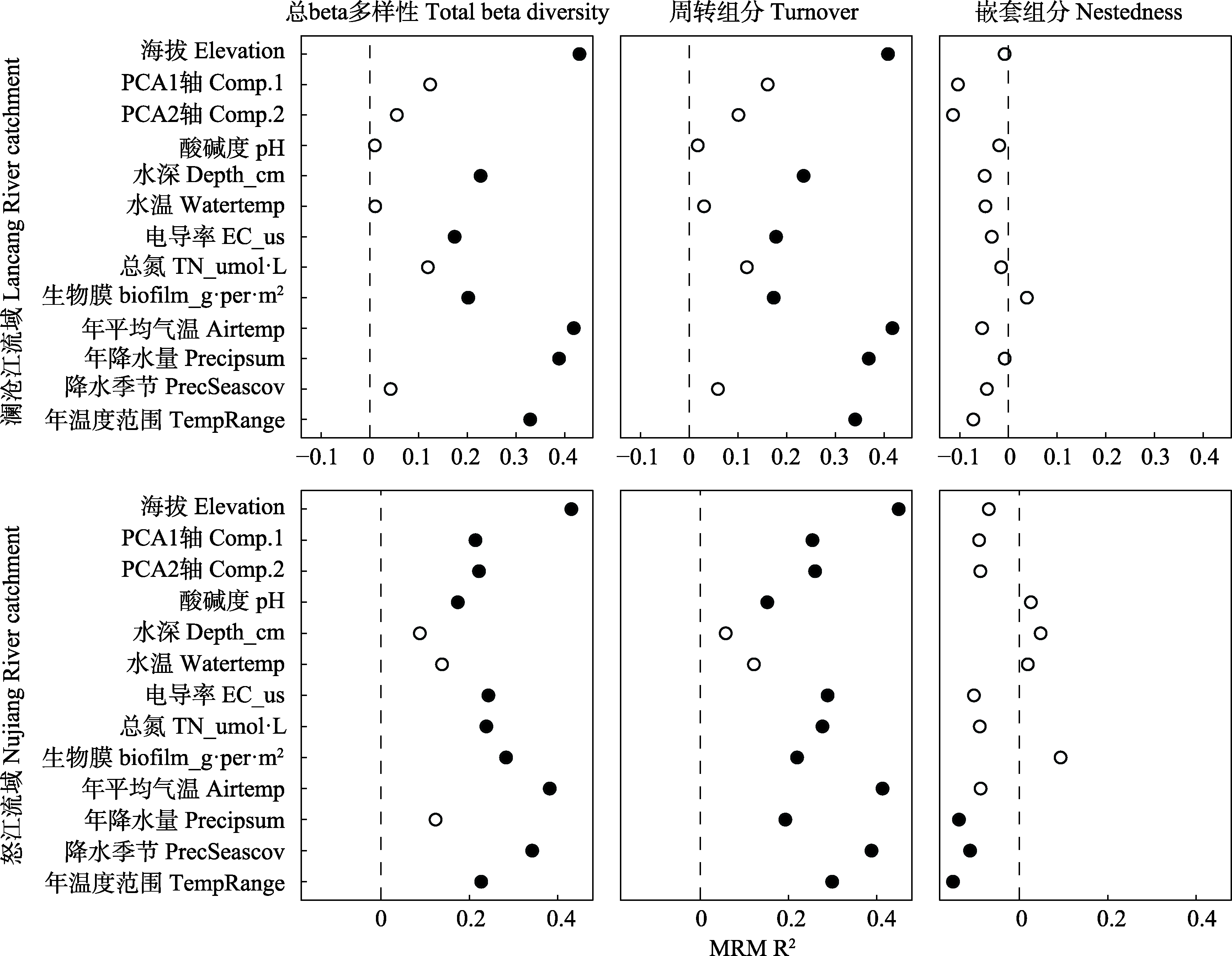
图3 环境和气候因子对澜沧江和怒江两个流域的细菌总beta多样性、周转组分和嵌套组分影响的基于矩阵分析的多元回归(MRM)分析。其中实心和空心的点分别代表显著相关(P < 0.05)和不显著相关性(P > 0.05)。
Fig. 3 The environmental and climate factors related to the variance in bacterial beta diversity and its components in Lancang River and Nujiang River catchments, identified with multiple regression on distance matrices (MRM). Filled shapes and empty shapes indicate significant relationship (P < 0.05) and non-significant relationship (P > 0.05), respectively. Comp.1 and Comp.2 indicate principal component analysis (PCA) was performed to reduce the dimensions of measured metal and metalloid concentrations, and the first two axes were used as proxies for these elements. Depth, Stream depth; EC_ us, Electrical conductivity; TN, Total nitrogen; Airtemp, Mean annual temperature; Precipsum, Annual precipitation; PrecSeascov, Precipitation seasonality conditions; TempRange, Annual temperature range.
| 澜沧江流域 Lancang catchment | 怒江流域 Nujiang catchment | |||||
|---|---|---|---|---|---|---|
| 总beta多样性 Total beta diversity | 周转组分 Turnover | 嵌套组分 Nestedness | 总beta多样性 Total beta diversity | 周转组分 Turnover | 嵌套组分 Nestedness | |
| 环境因子 Local factors | ||||||
| 海拔 Elevation | 0.43*** | 0.41*** | 0.43*** | 0.45*** | ||
| PCA1轴 Comp.1 | 0.21* | 0.25* | ||||
| PCA2轴 Comp.2 | 0.22* | 0.26* | ||||
| 酸碱度 pH | 0.17* | 0.15* | ||||
| 水深 Stream depth | 0.23** | 0.23*** | ||||
| 水温 Water temperature | ||||||
| 电导率 Electrical conductivity | 0.17* | 0.18* | 0.24** | 0.29*** | ||
| 总氮 Total nitrogen | 0.24** | 0.28*** | ||||
| 生物膜 Biofilm | 0.20* | 0.17* | 0.28** | 0.22** | ||
| 气候因子 Climate factors | ||||||
| 年平均气温 Mean annual temperature | 0.42*** | 0.42*** | 0.38*** | 0.41*** | ||
| 年降水量 Annual precipitation | 0.39*** | 0.37*** | 0.19* | -0.14* | ||
| 降水季节 Precipitation seasonality conditions | 0.34*** | 0.39*** | -0.11* | |||
| 年温度范围 Annual temperature range | 0.33* | 0.34** | 0.23** | 0.3** | -0.15** | |
表1 环境和气候因子与最终模型的偏回归系数(R2)解释的两个流域中细菌总beta多样性、周转和嵌套组分
Table 1 The variation of bacterial total beta diversity, turnover or nestedness components in Lancang and Nujiang catchments are explained by the environmental and climate factors and the partial regression coefficients (R2) of the final model is reported.
| 澜沧江流域 Lancang catchment | 怒江流域 Nujiang catchment | |||||
|---|---|---|---|---|---|---|
| 总beta多样性 Total beta diversity | 周转组分 Turnover | 嵌套组分 Nestedness | 总beta多样性 Total beta diversity | 周转组分 Turnover | 嵌套组分 Nestedness | |
| 环境因子 Local factors | ||||||
| 海拔 Elevation | 0.43*** | 0.41*** | 0.43*** | 0.45*** | ||
| PCA1轴 Comp.1 | 0.21* | 0.25* | ||||
| PCA2轴 Comp.2 | 0.22* | 0.26* | ||||
| 酸碱度 pH | 0.17* | 0.15* | ||||
| 水深 Stream depth | 0.23** | 0.23*** | ||||
| 水温 Water temperature | ||||||
| 电导率 Electrical conductivity | 0.17* | 0.18* | 0.24** | 0.29*** | ||
| 总氮 Total nitrogen | 0.24** | 0.28*** | ||||
| 生物膜 Biofilm | 0.20* | 0.17* | 0.28** | 0.22** | ||
| 气候因子 Climate factors | ||||||
| 年平均气温 Mean annual temperature | 0.42*** | 0.42*** | 0.38*** | 0.41*** | ||
| 年降水量 Annual precipitation | 0.39*** | 0.37*** | 0.19* | -0.14* | ||
| 降水季节 Precipitation seasonality conditions | 0.34*** | 0.39*** | -0.11* | |||
| 年温度范围 Annual temperature range | 0.33* | 0.34** | 0.23** | 0.3** | -0.15** | |
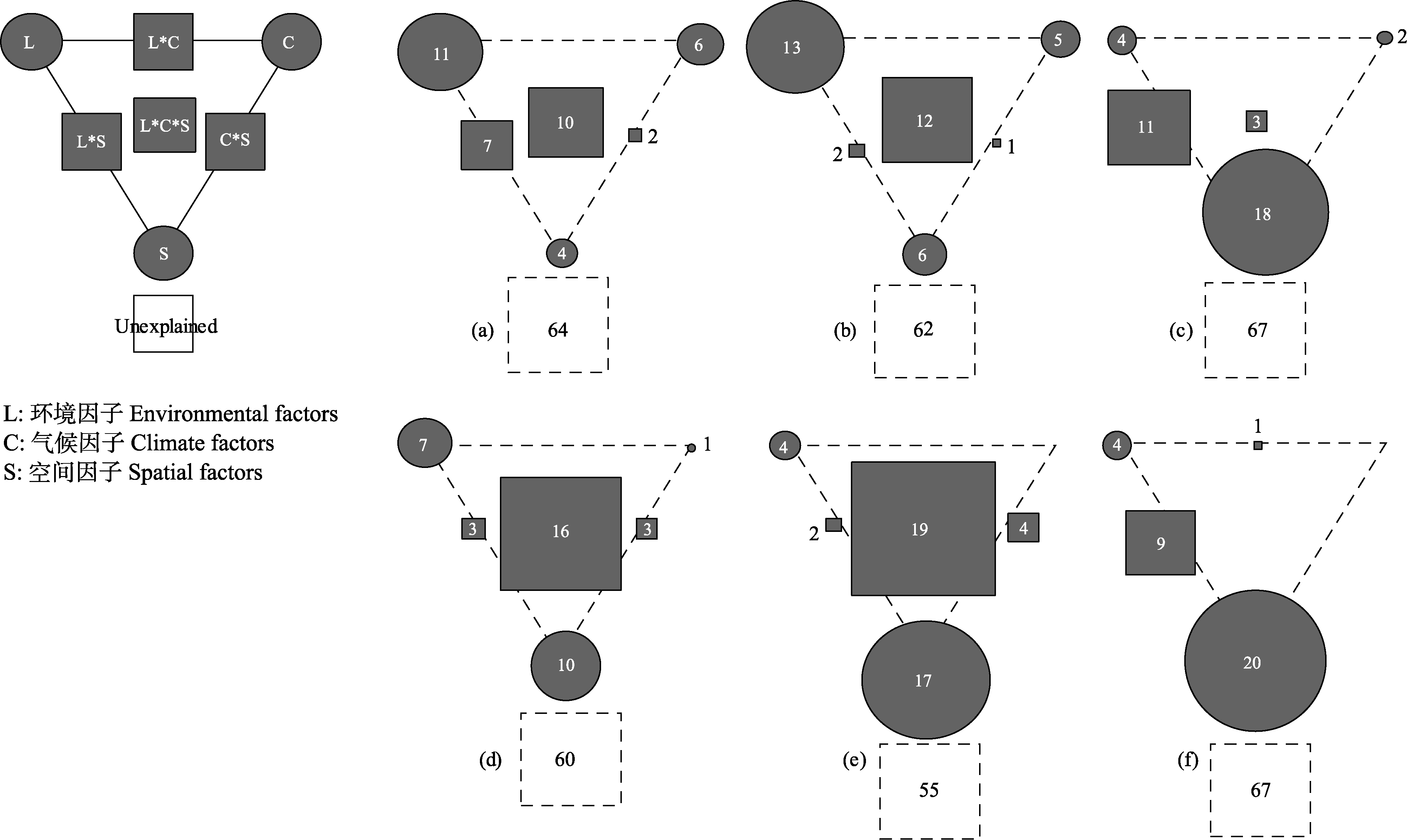
图4 当地环境因子(L)、气候因子(C)和空间因子(S)对两个流域细菌beta多样性、周转组分和嵌套组分的解释比例(小于0的值未显示)。左上方的面板是一般示意图。每类因子的单独解释由三角形的三个角表示。三角形的边和中心分别表示由两个或三个因素共同解释的变异百分比。(a)、(b)、(c)分别为澜沧江流域的细菌总beta多样性、周转组分和嵌套组分; (d)、(e)、(f)分别为怒江流域的细菌总beta多样性、周转组分和嵌套组分。
Fig. 4 The relative importance of environmental (L), climate (C) and spatial (S) factors in explaining the variance in bacterial beta diversity and its components in Nujiang River and Lancang River catchments. The top-left panel is the general outline. The pure variation explained by each factor is represented by the edges of the triangle. The sides and middle of the triangles indicate the percentages of variation explained by interactions of two or all factors, respectively. (a) Total beta diversity, (b) Turnover component and (c) Nestedness component in Lancang catchment. (d) Total beta diversity, (e) Turnover component and (f) Nestedness component in Nujiang catchment.
| [1] | Alahuhta J, Kosten S, Akasaka M, Auderset D, Azzella MM, Bolpagni R, Bove CP, Chambers PA, Chappuis E, Clayton JS ( 2017) Global variation in the beta diversity of lake macrophytes is driven by environmental heterogeneity rather than latitude. Journal of Biogeography, 44, 1758-1769. |
| [2] | Anderson MJ ( 2001) A new method for non‐parametric multivariate analysis of variance. Austral Ecology, 26, 32-46. |
| [3] | Anderson MJ, Cribble NA ( 1998) Partitioning the variation among spatial, temporal and environmental components in a multivariate data set. Austral Ecology, 23, 158-167. |
| [4] | Barberan A, Casamayor EO ( 2010) Global phylogenetic community structure and β-diversity patterns in surface bacterioplankton metacommunities. Aquatic Microbial Ecology, 59, 1-10. |
| [5] |
Bardgett RD, Freeman C, Ostle NJ ( 2008) Microbial contributions to climate change through carbon cycle feedbacks. The ISME Journal, 2, 805-814.
DOI URL PMID |
| [6] | Baselga A ( 2010) Partitioning the turnover and nestedness components of beta diversity. Global Ecology and Biogeography, 19, 134-143. |
| [7] |
Bishop TR, Robertson MP, van Rensburg BJ, Parr CL ( 2015) Contrasting species and functional beta diversity in montane ant assemblages. Journal of Biogeography, 42, 1776-1786.
URL PMID |
| [8] | Borcard D, Legendre P ( 2002) All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling, 153, 51-68. |
| [9] |
Butchart SHM, Walpole M, Collen B, Van SA, Scharlemann JPW, Almond REA, Baillie JEM, Bomhard B, Brown C, Bruno J ( 2010) Global biodiversity: Indicators of recent declines. Science, 328, 1164-1168.
DOI URL PMID |
| [10] |
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI ( 2010) QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7, 335-336.
URL PMID |
| [11] |
Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, Venail P, Narwani A, Mace GM, Tilman D, Wardle DA ( 2012) Biodiversity loss and its impact on humanity. Nature, 486, 59-67.
DOI URL PMID |
| [12] | Carr GM, Morin A, Chambers PA ( 2005) Bacteria and algae in stream periphyton along a nutrient gradient. Freshwater Biology, 50, 1337-1350. |
| [13] | Chase JM ( 2007) Drought mediates the importance of stochastic community assembly. Proceedings of the National Academy of Sciences, USA, 104, 17430-17434. |
| [14] |
Edgar RC ( 2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460-2461.
URL PMID |
| [15] | Forster J, Hirst AG, Atkinson D ( 2012) Warming-induced reductions in body size are greater in aquatic than terrestrial species. Proceedings of the National Academy of Sciences, USA, 109, 19310-19314. |
| [16] | Frauendorf TC, Mackenzie RA, Tingley RW, Frazier AG, Riney MH, Elsabaawi RW ( 2019) Evaluating ecosystem effects of climate change on tropical island streams using high spatial and temporal resolution sampling regimes. Global Change Biology, 25, 1344-1357. |
| [17] | Gutierrezcanovas C, Millan A, Velasco J, Vaughan IP, Ormerod SJ ( 2013) Contrasting effects of natural and anthropogenic stressors on beta diversity in river organisms. Global Ecology and Biogeography, 22, 796-805. |
| [18] | Hanson CA, Fuhrman JA, Hornerdevine MC, Martiny Jennifer BH ( 2012) Beyond biogeographic patterns: Processes shaping the microbial landscape. Nature Reviews Microbiology, 10, 497-506. |
| [19] | Heino J, Tolkkinen M, Pirttila AM, Aisala H, Mykra H ( 2014) Microbial diversity and community-environment relationnships in boreal streams. Journal of Biogeography, 41, 2234-2244. |
| [20] |
Kuypers MMM, Marchant HK, Kartal B ( 2018) The microbial nitrogen-cycling network. Nature Reviews Microbiology, 16, 263-276.
DOI URL PMID |
| [21] |
Lear G, Niyogi DK, Harding JS, Dong Y, Lewis G ( 2009) Biofilm bacterial community structure in streams affected by acid mine drainage. Applied and Environmental Microbiology, 75, 3455-3460.
URL PMID |
| [22] |
Legendre P, Lapointe F, Casgrain P ( 1994) Modeling brain evolution from behavior: A permutational regression approach. Evolution, 48, 1487-1499.
DOI URL PMID |
| [23] |
Magoc T, Salzberg SL ( 2011) FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27, 2957-2963.
DOI URL PMID |
| [24] |
Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreas L, Reysenbach AL, Smith VH, Staley JT ( 2006) Microbial biogeography: Putting microorganisms on the map. Nature Reviews Microbiology, 4, 102-112.
URL PMID |
| [25] | Minchin PR ( 1987) An evaluation of the relative robustness of techniques for ecological ordination. Plant Ecology, 69, 89-107. |
| [26] | Mori A, Shiono T, Haraguchi TF, Ota AT, Koide D, Ohgue T, Kitagawa R, Maeshiro R, Aung TT, Nakamori T ( 2015) Functional redundancy of multiple forest taxa along an elevational gradient: Predicting the consequences of non‐random species loss. Journal of Biogeography, 42, 1383-1396. |
| [27] |
Myers N, Mittermeier RA, Mittermeier CG, Fonseca GABD, Kent J ( 2000) Biodiversity hotspots for conservation priorities. Nature, 403, 853-858.
URL PMID |
| [28] |
Qian H, Ricklefs RE, White PS ( 2004) Beta diversity of angiosperms in temperate floras of eastern Asia and eastern North America. Ecology Letters, 8, 15-22.
DOI URL |
| [29] | R Core Team ( 2018) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. |
| [30] |
Si XF, Zhao YH, Chen CW, Ren P, Zeng D, Wu LB, Ding P ( 2017) Beta-diversity partitioning: Methods, applications and perspectives. Biodiversity Science, 25, 464-480(in Chinese with English abstract).
DOI URL |
| [ 斯幸峰, 赵郁豪, 陈传武, 任鹏, 曾頔, 吴玲兵, 丁平 ( 2017) Beta多样性分解: 方法, 应用与展望. 生物多样性, 25, 464-480.] | |
| [31] |
Socolar JB, Gilroy JJ, Kunin WE, Edwards DP ( 2016) How should beta-diversity inform biodiversity conservation? Trends in Ecology & Evolution, 31, 67-80.
DOI URL PMID |
| [32] | Sun FL, Wang YS, Wu ML, Wang Y, Li QP ( 2011) Spatial heterogeneity of bacterial community structure in the sediments of the Pearl River estuary. Biologia, 66, 574-584. |
| [33] | Swenson NG, Anglada-Cordero P, Barone JA ( 2010) Deterministic tropical tree community turnover: Evidence from patterns of functional beta diversity along an elevational gradient. Proceedings of the Royal Society B: Biological Sciences, 278, 877-884. |
| [34] |
Teittinen A, Kallajoki L, Meier S, Stigzelius T, Soininen J ( 2016) The roles of elevation and local environmental factors as drivers of diatom diversity in subarctic streams. Freshwater Biology, 61, 1509-1521.
DOI URL |
| [35] | Vilmi A, Zhao WQ, Picazo F, Li MJ, Heino J, Soininen J, Wang JJ ( 2019) Ecological processes underlying the community assembly of aquatic microscopic and macroscopic organisms under contrasting climates in the Tibetan Plateau biodiversity hotspot. Science of the Total Environment, 134974. |
| [36] |
Vitousek PM ( 1994) Beyond global warming: Ecology and global change. Ecology, 75, 1861-1876.
DOI URL |
| [37] |
Wang JJ, Meier S, Soininen J, Casamayor EO, Pan FY, Tang XM, Yang XD, Zhang YL, Wu QL, Zhou JZ ( 2017) Regional and global elevational patterns of microbial species richness and evenness. Ecography, 40, 393-402.
DOI URL |
| [38] |
Wang JJ, Soininen J, Zhang Y, Wang BX, Yang XD, Shen J ( 2011) Contrasting patterns in elevational diversity between microorganisms and macroorganisms. Journal of Biogeography, 38, 595-603.
DOI URL |
| [39] |
Wang JJ, Soininen J, Zhang Y, Wang BX, Yang XD, Shen J ( 2012) Patterns of elevational beta diversity in micro- and macroorganisms. Global Ecology and Biogeography, 21, 743-750.
DOI URL |
| [40] |
Wang Q, Garrity GM, Tiedje JM, Cole JR ( 2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73, 5261-5267.
DOI URL PMID |
| [41] | Waters CN, Zalasiewicz J, Summerhayes C, Barnosky AD, Poirier C, Gałuszka A, Cearreta A, Edgeworth M, Ellis EC, Ellis M ( 2016) The Anthropocene is functionally and stratigraphically distinct from the Holocene. Science, 351, 137-147. |
| [42] |
Whittaker RH ( 1960) Vegetation of the Siskiyou Mountains, Oregon and California. Ecological Monographs, 30, 279-338.
DOI URL |
| [43] | Wieczynski DJ, Boyle B, Buzzard V, Duran SM, Henderson AN, Hulshof CM, Kerkhoff AJ, Mccarthy MC, Michaletz ST, Swenson NG ( 2019) Climate shapes and shifts functional biodiversity in forests worldwide. Proceedings of the National Academy of Sciences, USA, 116, 587-592. |
| [44] |
Woodward G, Perkins DM, Brown LE ( 2010) Climate change and freshwater ecosystems: Impacts across multiple levels of organization. Philosophical Transactions of the Royal Society B, 365, 2093-2106.
DOI URL |
| [45] | Wu YJ, Lei FM ( 2013) Species richness patterns and mechanisms along the elevational gradients. Chinese Journal of Zoology, 48, 797-807(in Chinese with English abstract). |
| [ 吴永杰, 雷富民 ( 2013) 物种丰富度垂直分布格局及影响机制. 动物学杂志, 48, 797-807.] | |
| [46] |
Zhou JZ, Bruns MA, Tiedje JM ( 1996) DNA recovery from soils of diverse composition. Applied and Environmental Microbiology, 62, 316-322.
URL PMID |
| [1] | 顾婧婧, 刘宜卓, 苏杨. 基层地方政府在完成《昆蒙框架》中的作用和难点: 基于《联合国气候变化框架公约》任务的比较[J]. 生物多样性, 2025, 33(3): 24585-. |
| [2] | 李蓉姣, 董江海, 郑文芳, 刘入源, 赵立娟, 高瑞贺. 关帝山不同海拔杨桦混交林土壤动物多样性特征及其影响因素[J]. 生物多样性, 2024, 32(9): 24070-. |
| [3] | 李斌, 宋鹏飞, 顾海峰, 徐波, 刘道鑫, 江峰, 梁程博, 张萌, 高红梅, 蔡振媛, 张同作. 昆仑山青海片区鸟类群落多样性格局及其驱动因素[J]. 生物多样性, 2024, 32(4): 23406-. |
| [4] | 吴琪, 张晓青, 杨雨婷, 周艺博, 马毅, 许大明, 斯幸峰, 王健. 浙江钱江源-百山祖国家公园庆元片区叶附生苔多样性及其时空变化[J]. 生物多样性, 2024, 32(4): 24010-. |
| [5] | 王鹏, 隋佳容, 丁欣瑶, 王伟中, 曹雪倩, 赵海鹏, 王彦平. 郑州城市公园鸟类群落嵌套分布格局及其影响因素[J]. 生物多样性, 2024, 32(3): 23359-. |
| [6] | 曹可欣, 王敬雯, 郑国, 武鹏峰, 李英滨, 崔淑艳. 降水格局改变及氮沉降对北方典型草原土壤线虫多样性的影响[J]. 生物多样性, 2024, 32(3): 23491-. |
| [7] | 施国杉, 刘峰, 曹光宏, 陈典, 夏尚文, 邓云, 王彬, 杨效东, 林露湘. 西双版纳热带季节雨林木本植物的beta多样性: 空间、环境与林分结构的作用[J]. 生物多样性, 2024, 32(12): 24285-. |
| [8] | 杜聪聪, 冯学宇, 陈志林. 桥头堡效应中气候生态位差异的缩小促进了红火蚁的入侵[J]. 生物多样性, 2024, 32(11): 24276-. |
| [9] | 原雪姣, 张渊媛, 张衍亮, 胡璐祎, 桑卫国, 杨峥, 陈颀. 基于飞机草历史分布数据拟合的物种分布模型及其预测能力[J]. 生物多样性, 2024, 32(11): 24288-. |
| [10] | 罗小燕, 李强, 黄晓磊. 戴云山国家级自然保护区访花昆虫DNA条形码数据集[J]. 生物多样性, 2023, 31(8): 23236-. |
| [11] | 刘志发, 王新财, 龚粤宁, 陈道剑, 张强. 基于红外相机监测的广东南岭国家级自然保护区鸟兽多样性及其垂直分布特征[J]. 生物多样性, 2023, 31(8): 22689-. |
| [12] | 冯莉. 国际法视野下生物多样性和气候变化的协同治理[J]. 生物多样性, 2023, 31(7): 23110-. |
| [13] | 魏庐潞, 徐婷婷, 李媛媛, 艾喆, 马飞. 同质园环境和遗传分化影响锦鸡儿属植物根际土壤固氮菌多样性和群落结构[J]. 生物多样性, 2023, 31(4): 22477-. |
| [14] | 姚雪, 陈星, 戴尊, 宋坤, 邢诗晨, 曹宏彧, 邹璐, 王健. 采集策略对叶附生苔类植物发现概率及物种多样性的重要性[J]. 生物多样性, 2023, 31(4): 22685-. |
| [15] | 邵雯雯, 范国祯, 何知舟, 宋志平. 多地同质园实验揭示普通野生稻的表型可塑性与本地适应性[J]. 生物多样性, 2023, 31(3): 22311-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()