



生物多样性 ›› 2019, Vol. 27 ›› Issue (6): 667-676. DOI: 10.17520/biods.2019013 cstr: 32101.14.biods.2019013
翁茁先1,2,3,黄佳琼2,张仕豪2,余锴纯2,钟福生1,2,3,黄勋和2,3,*( ),张彬1,*(
),张彬1,*( )
)
收稿日期:2019-01-17
接受日期:2019-05-20
出版日期:2019-06-20
发布日期:2019-06-14
通讯作者:
黄勋和,张彬
基金资助:
Weng Zhuoxian1,2,3,Huang Jiaqiong2,Zhang Shihao2,Yu Kaichun2,Zhong Fusheng1,2,3,Huang Xunhe2,3,*( ),Zhang Bin1,*(
),Zhang Bin1,*( )
)
Received:2019-01-17
Accepted:2019-05-20
Online:2019-06-20
Published:2019-06-14
Contact:
Huang Xunhe,Zhang Bin
摘要:
全面了解中国乌骨鸡的遗传背景有利于保护和开发利用其种质资源。本研究测定了中国12个乌骨鸡品种线粒体细胞色素c氧化酶亚基I (cytochrome c oxidase subunit I, COI)基因, 比较分析其遗传多样性和群体遗传结构。255份乌骨鸡样品共检测到22个变异位点, 占分析位点的3.17%; 核苷酸多样性为0.00142-0.00339, 单倍型多样性为0.380-0.757, 其中略阳乌鸡核苷酸多样性最高, 德化黑鸡最低。检测到7个氨基酸变异位点, 来自6个品种共11个个体。定义了24种单倍型, 其中单倍型H1和H3为12个乌骨鸡品种共享, 出现频率分别为115次和64次; 盐津乌骨鸡单倍型数最多, 广西乌鸡最少。中性检验与错配分析显示实验种群未经历显著的群体扩张事件。分子变异分析显示81.06%的变异来自群体内; 品种间遗传距离为0.002-0.004, 品种间遗传分化系数Fst值为-0.035至0.594, 雪峰乌骨鸡与其他种群间的遗传分化程度最高。邻接树显示, 乌骨鸡未能独立形成分支, 不能从家鸡和红原鸡中有效区分开来。中国乌骨鸡中介网络图将24个单倍型分为3条进化主支, 呈现出一定的品种特异性, 由无量山乌骨鸡、云南盐津乌骨鸡和雪峰乌骨鸡组成单倍型H8、H9、H11、H12游离于这3条进化主支之外。增加其他家鸡和红原鸡COI基因的中介网络图主体结构与中国乌骨鸡的相同。结果表明中国乌骨鸡品种遗传多样性较低, 但品种间遗传分化显著, 可能是从当地家鸡中选育而来, 需要加强种质资源的保护。
翁茁先, 黄佳琼, 张仕豪, 余锴纯, 钟福生, 黄勋和, 张彬 (2019) 利用线粒体COI基因揭示中国乌骨鸡遗传多样性和群体遗传结构. 生物多样性, 27, 667-676. DOI: 10.17520/biods.2019013.
Weng Zhuoxian, Huang Jiaqiong, Zhang Shihao, Yu Kaichun, Zhong Fusheng, Huang Xunhe, Zhang Bin (2019) Genetic diversity and population structure of black-bone chickens in China revealed by mitochondrial COI gene sequences. Biodiversity Science, 27, 667-676. DOI: 10.17520/biods.2019013.
| 代号 Code | 品种名 Breed | 产地 Origin | 样本量 Sample size |
|---|---|---|---|
| LY | 略阳乌鸡 Lueyang chicken | 陕西省略阳县 Lueyang, Shaanxi | 10 |
| WLS | 无量山乌骨鸡 Wuliangshan black-bone chicken | 云南省南涧县 Nanjian, Yunnan | 25 |
| YJ | 盐津乌骨鸡 Yanjin black-bone chicken | 云南省盐津县 Yanjin, Yunnan | 24 |
| ZX | 竹乡鸡 Zhuxiang chicken | 贵州省赤水市 Chishui, Guizhou | 32 |
| XF | 雪峰乌骨鸡 Xuefeng black-bone chicken | 湖南省洪江市 Hongjiang, Hunan | 19 |
| JS | 江山乌骨鸡 Jiangshan black-bone chicken | 浙江省江山市 Jiangshan, Zhejiang | 21 |
| SK | 丝羽乌骨鸡 Silkies | 江西省泰和县 Taihe, Jiangxi | 25 |
| YG | 余干乌骨鸡 Yugan black-bone chicken | 江西省余干县 Yugan, Jiangxi | 18 |
| HY | 黄羽黑鸡 Huangyu black chicken | 江西省宜春市 Yichun, Jiangxi | 18 |
| DH | 德化黑鸡 Dehua black chicken | 福建省德化县 Dehua, Fujian | 22 |
| JH | 金湖乌凤鸡 Jinhu black-bone chicken | 福建省泰宁县 Taining, Fujian | 24 |
| GX | 广西乌鸡 Guangxi black-bone chicken | 广西东兰县 Donglan, Guangxi | 17 |
表1 本研究样品信息
Table 1 Sample information of this study
| 代号 Code | 品种名 Breed | 产地 Origin | 样本量 Sample size |
|---|---|---|---|
| LY | 略阳乌鸡 Lueyang chicken | 陕西省略阳县 Lueyang, Shaanxi | 10 |
| WLS | 无量山乌骨鸡 Wuliangshan black-bone chicken | 云南省南涧县 Nanjian, Yunnan | 25 |
| YJ | 盐津乌骨鸡 Yanjin black-bone chicken | 云南省盐津县 Yanjin, Yunnan | 24 |
| ZX | 竹乡鸡 Zhuxiang chicken | 贵州省赤水市 Chishui, Guizhou | 32 |
| XF | 雪峰乌骨鸡 Xuefeng black-bone chicken | 湖南省洪江市 Hongjiang, Hunan | 19 |
| JS | 江山乌骨鸡 Jiangshan black-bone chicken | 浙江省江山市 Jiangshan, Zhejiang | 21 |
| SK | 丝羽乌骨鸡 Silkies | 江西省泰和县 Taihe, Jiangxi | 25 |
| YG | 余干乌骨鸡 Yugan black-bone chicken | 江西省余干县 Yugan, Jiangxi | 18 |
| HY | 黄羽黑鸡 Huangyu black chicken | 江西省宜春市 Yichun, Jiangxi | 18 |
| DH | 德化黑鸡 Dehua black chicken | 福建省德化县 Dehua, Fujian | 22 |
| JH | 金湖乌凤鸡 Jinhu black-bone chicken | 福建省泰宁县 Taining, Fujian | 24 |
| GX | 广西乌鸡 Guangxi black-bone chicken | 广西东兰县 Donglan, Guangxi | 17 |
| 品种名 Breed | 变异位点数 No. of variable sites | 平均核苷酸差异 Average K (k) | 核苷酸多样性 Nucleotide diversity (π) | 单倍型多样性 Haplotype diversity (h) |
|---|---|---|---|---|
| 略阳乌鸡 LY | 6 | 2.356 | 0.00339 ± 0.00064 | 0.733 ± 0.120 |
| 无量山乌骨鸡 WLS | 6 | 1.653 | 0.00238 ± 0.00026 | 0.743 ± 0.051 |
| 盐津乌骨鸡 YJ | 10 | 2.036 | 0.00293 ± 0.00042 | 0.757 ± 0.075 |
| 竹乡鸡 ZX | 7 | 2.018 | 0.00290 ± 0.00028 | 0.730 ± 0.056 |
| 雪峰乌骨鸡 XF | 6 | 1.310 | 0.00188 ± 0.00069 | 0.380 ± 0.134 |
| 江山乌骨鸡 JS | 7 | 1.952 | 0.00281 ± 0.00048 | 0.729 ± 0.078 |
| 丝羽乌骨鸡 SK | 7 | 0.993 | 0.00143 ± 0.00044 | 0.537 ± 0.115 |
| 余干乌骨鸡 YG | 5 | 1.804 | 0.00260 ± 0.00025 | 0.680 ± 0.074 |
| 黄羽黑鸡 HY | 5 | 1.523 | 0.00219 ± 0.00051 | 0.634 ± 0.093 |
| 德化黑鸡 DH | 4 | 0.987 | 0.00142 ± 0.00047 | 0.455 ± 0.114 |
| 金湖乌凤鸡 JH | 5 | 1.486 | 0.00214 ± 0.00044 | 0.572 ± 0.095 |
| 广西乌鸡 GX | 3 | 1.456 | 0.00209 ± 0.00034 | 0.485 ± 0.079 |
表2 12个乌骨鸡品种COI基因序列变异位点数、平均核苷酸差异、核苷酸多样性和单倍型多样性。品种代号同表1。
Table 2 The number of variable sites, average K (k), nucleotide diversity (π) and haplotype diversity (h) of COI gene in the 12 black-bone chicken breeds. Breed codes are the same as in Table 1.
| 品种名 Breed | 变异位点数 No. of variable sites | 平均核苷酸差异 Average K (k) | 核苷酸多样性 Nucleotide diversity (π) | 单倍型多样性 Haplotype diversity (h) |
|---|---|---|---|---|
| 略阳乌鸡 LY | 6 | 2.356 | 0.00339 ± 0.00064 | 0.733 ± 0.120 |
| 无量山乌骨鸡 WLS | 6 | 1.653 | 0.00238 ± 0.00026 | 0.743 ± 0.051 |
| 盐津乌骨鸡 YJ | 10 | 2.036 | 0.00293 ± 0.00042 | 0.757 ± 0.075 |
| 竹乡鸡 ZX | 7 | 2.018 | 0.00290 ± 0.00028 | 0.730 ± 0.056 |
| 雪峰乌骨鸡 XF | 6 | 1.310 | 0.00188 ± 0.00069 | 0.380 ± 0.134 |
| 江山乌骨鸡 JS | 7 | 1.952 | 0.00281 ± 0.00048 | 0.729 ± 0.078 |
| 丝羽乌骨鸡 SK | 7 | 0.993 | 0.00143 ± 0.00044 | 0.537 ± 0.115 |
| 余干乌骨鸡 YG | 5 | 1.804 | 0.00260 ± 0.00025 | 0.680 ± 0.074 |
| 黄羽黑鸡 HY | 5 | 1.523 | 0.00219 ± 0.00051 | 0.634 ± 0.093 |
| 德化黑鸡 DH | 4 | 0.987 | 0.00142 ± 0.00047 | 0.455 ± 0.114 |
| 金湖乌凤鸡 JH | 5 | 1.486 | 0.00214 ± 0.00044 | 0.572 ± 0.095 |
| 广西乌鸡 GX | 3 | 1.456 | 0.00209 ± 0.00034 | 0.485 ± 0.079 |
| 单倍型 Haplotype | 变异位点 Variation sites | 单倍型在品种的分布(频率) Haplotype distribution in breeds (frequency) | 合计 Total |
|---|---|---|---|
AP003321 | 6666666666677777777777 7777788889900012233333 1146704890012331537788 0255272216776245331467 ACATGTTTAATGAGACAGTCCA | ||
| H1 | ...................... | LY5, WLS10, YJ4, ZX14, XF1, JS10, SK17, YG7, HY10, DH16, JH15, GX6 | 115 |
| H2 | T..................... | ZX1 | 1 |
| H3 | ...C.C...G............ | LY2, WLS2, YJ1, ZX9, XF15, JS4, SK2, YG8, HY2, DH3, JH5, GX11 | 64 |
| H4 | T..C.C...G............ | ZX2 | 2 |
| H5 | C..............T...... | ZX1 | 1 |
| H6 | ..GC.C...G............ | ZX1 | 1 |
| H7 | ...............T...... | WLS1, YJ1, ZX4, JS4, SK2, YG1, HY1, DH3, JH3 | 20 |
| H8 | .........G...A........ | WLS7, YJ11, XF2 | 20 |
| H9 | .........G..........T. | WLS5, YJ1 | 6 |
| H10 | ......C............... | YJ4 | 4 |
| H11 | ..G....C.G...A........ | YJ1 | 1 |
| H12 | .GG......G..........T. | YJ1 | 1 |
| H13 | ..................C... | SK1 | 1 |
| H14 | ................G..... | SK2 | 2 |
| H15 | ................GC.... | SK1 | 1 |
| H16 | ....................TG | LY1 | 1 |
| H17 | ...C.C...G..G......... | LY2 | 2 |
| H18 | ...............T...T.. | XF1 | 1 |
| H19 | ...C.C...G....G....... | JS1, JH1 | 2 |
| H20 | ...CAC...G............ | JS1 | 1 |
| H21 | ...........A...T...... | JS1 | 1 |
| H22 | .....C...G............ | YG1 | 1 |
| H23 | ..........A........... | YG1 | 1 |
| H24 | ........G......T...... | HY5 | 5 |
表3 12个乌骨鸡品种线粒体COI基因的单倍型及其在不同品种的分布
Table 3 COI haplotypes and their distributions in the 12 black-bone chicken breeds
| 单倍型 Haplotype | 变异位点 Variation sites | 单倍型在品种的分布(频率) Haplotype distribution in breeds (frequency) | 合计 Total |
|---|---|---|---|
AP003321 | 6666666666677777777777 7777788889900012233333 1146704890012331537788 0255272216776245331467 ACATGTTTAATGAGACAGTCCA | ||
| H1 | ...................... | LY5, WLS10, YJ4, ZX14, XF1, JS10, SK17, YG7, HY10, DH16, JH15, GX6 | 115 |
| H2 | T..................... | ZX1 | 1 |
| H3 | ...C.C...G............ | LY2, WLS2, YJ1, ZX9, XF15, JS4, SK2, YG8, HY2, DH3, JH5, GX11 | 64 |
| H4 | T..C.C...G............ | ZX2 | 2 |
| H5 | C..............T...... | ZX1 | 1 |
| H6 | ..GC.C...G............ | ZX1 | 1 |
| H7 | ...............T...... | WLS1, YJ1, ZX4, JS4, SK2, YG1, HY1, DH3, JH3 | 20 |
| H8 | .........G...A........ | WLS7, YJ11, XF2 | 20 |
| H9 | .........G..........T. | WLS5, YJ1 | 6 |
| H10 | ......C............... | YJ4 | 4 |
| H11 | ..G....C.G...A........ | YJ1 | 1 |
| H12 | .GG......G..........T. | YJ1 | 1 |
| H13 | ..................C... | SK1 | 1 |
| H14 | ................G..... | SK2 | 2 |
| H15 | ................GC.... | SK1 | 1 |
| H16 | ....................TG | LY1 | 1 |
| H17 | ...C.C...G..G......... | LY2 | 2 |
| H18 | ...............T...T.. | XF1 | 1 |
| H19 | ...C.C...G....G....... | JS1, JH1 | 2 |
| H20 | ...CAC...G............ | JS1 | 1 |
| H21 | ...........A...T...... | JS1 | 1 |
| H22 | .....C...G............ | YG1 | 1 |
| H23 | ..........A........... | YG1 | 1 |
| H24 | ........G......T...... | HY5 | 5 |
| 品种名 Breed | 变异位点 Variation sites | 氨基酸变异 Amino acid variation | 变异数 Variation number |
|---|---|---|---|
AP003321 | 0000122 1235124 9010875 YLTEVDI | ||
| 竹乡鸡 ZX | F...... | 酪氨酸→苯丙氨酸 Tyrosine → Phenylalanine | 3 |
| 竹乡鸡 ZX | S...... | 酪氨酸→丝氨酸 Tyrosine → Serine | 1 |
| 竹乡鸡 ZX | ..A.... | 苏氨酸→丙氨酸 Threonine → Alanine | 1 |
| 盐津乌骨鸡 YJ | ..A.... | 苏氨酸→丙氨酸 Threonine → Alanine | 1 |
| 盐津乌骨鸡 YJ | .VA.... | 亮氨酸→缬氨酸, 苏氨酸→丙氨酸 Leucine → Valine, Threonine → Alanine | 1 |
| 丝羽乌骨鸡 SK | .....H. | 天冬氨酸→组氨酸 Asparagine → Histidine | 1 |
| 略阳乌鸡 LY | ......V | 异亮氨酸→缬氨酸 Isoleucine → Valine | 1 |
| 江山乌骨鸡 JS | ...K... | 谷氨酸→赖氨酸 Glutamic acid → Lysine | 1 |
| 余干乌骨鸡 YG | ....E.. | 缬氨酸→谷氨酸 Valine → Glutamic acid | 1 |
表4 COI基因编码的氨基酸变异及其在不同品种的分布。品种代号同表1。
Table 4 Amino acid variation of COI gene and their distribution in these black-bone chicken breeds. Breed codes are the same as in Table 1.
| 品种名 Breed | 变异位点 Variation sites | 氨基酸变异 Amino acid variation | 变异数 Variation number |
|---|---|---|---|
AP003321 | 0000122 1235124 9010875 YLTEVDI | ||
| 竹乡鸡 ZX | F...... | 酪氨酸→苯丙氨酸 Tyrosine → Phenylalanine | 3 |
| 竹乡鸡 ZX | S...... | 酪氨酸→丝氨酸 Tyrosine → Serine | 1 |
| 竹乡鸡 ZX | ..A.... | 苏氨酸→丙氨酸 Threonine → Alanine | 1 |
| 盐津乌骨鸡 YJ | ..A.... | 苏氨酸→丙氨酸 Threonine → Alanine | 1 |
| 盐津乌骨鸡 YJ | .VA.... | 亮氨酸→缬氨酸, 苏氨酸→丙氨酸 Leucine → Valine, Threonine → Alanine | 1 |
| 丝羽乌骨鸡 SK | .....H. | 天冬氨酸→组氨酸 Asparagine → Histidine | 1 |
| 略阳乌鸡 LY | ......V | 异亮氨酸→缬氨酸 Isoleucine → Valine | 1 |
| 江山乌骨鸡 JS | ...K... | 谷氨酸→赖氨酸 Glutamic acid → Lysine | 1 |
| 余干乌骨鸡 YG | ....E.. | 缬氨酸→谷氨酸 Valine → Glutamic acid | 1 |
| 品种名 Breed | Tajima’s D | Fu’s Fs | ||
|---|---|---|---|---|
| D | P | Fs | P | |
| 略阳乌鸡 LY | 0.45768 | 0.70000 | 0.96375 | 0.68900 |
| 无量山乌骨鸡 WLS | 0.12109 | 0.56400 | 0.49464 | 0.62700 |
| 盐津乌骨鸡 YJ | -0.79944 | 0.25300 | -1.69126 | 0.17200 |
| 竹乡鸡 ZX | 0.99606 | 0.85100 | -0.31381 | 0.45400 |
| 雪峰乌骨鸡 XF | -0.76698 | 0.25300 | 0.57269 | 0.65900 |
| 江山乌骨鸡 JS | 0.01119 | 0.54100 | -0.25147 | 0.44900 |
| 丝羽乌骨鸡 SK | -1.43529 | 0.07300 | -1.92588 | 0.07000 |
| 余干乌骨鸡 YG | 0.76262 | 0.79100 | 0.24173 | 0.59100 |
| 黄羽黑鸡 HY | 0.15067 | 0.60000 | 0.87825 | 0.69200 |
| 德化黑鸡 DH | -0.28222 | 0.43600 | 1.28651 | 0.75700 |
| 金湖乌凤鸡 JH | 0.31722 | 0.67500 | 1.19223 | 0.77300 |
| 广西乌鸡 GX | 1.81444 | 0.97700 | 3.86919 | 0.94800 |
| 平均 Average | 0.11225 | 0.55950 | 0.44305 | 0.57342 |
| 方差 Squared deviation | 0.87539 | 0.26761 | 1.50012 | 0.25155 |
表5 12个乌骨鸡品种的COI序列中性检验。品种代号同表1。
Table 5 Neutrality test in 12 breeds based on COI sequences. Breed codes are the same as in Table 1.
| 品种名 Breed | Tajima’s D | Fu’s Fs | ||
|---|---|---|---|---|
| D | P | Fs | P | |
| 略阳乌鸡 LY | 0.45768 | 0.70000 | 0.96375 | 0.68900 |
| 无量山乌骨鸡 WLS | 0.12109 | 0.56400 | 0.49464 | 0.62700 |
| 盐津乌骨鸡 YJ | -0.79944 | 0.25300 | -1.69126 | 0.17200 |
| 竹乡鸡 ZX | 0.99606 | 0.85100 | -0.31381 | 0.45400 |
| 雪峰乌骨鸡 XF | -0.76698 | 0.25300 | 0.57269 | 0.65900 |
| 江山乌骨鸡 JS | 0.01119 | 0.54100 | -0.25147 | 0.44900 |
| 丝羽乌骨鸡 SK | -1.43529 | 0.07300 | -1.92588 | 0.07000 |
| 余干乌骨鸡 YG | 0.76262 | 0.79100 | 0.24173 | 0.59100 |
| 黄羽黑鸡 HY | 0.15067 | 0.60000 | 0.87825 | 0.69200 |
| 德化黑鸡 DH | -0.28222 | 0.43600 | 1.28651 | 0.75700 |
| 金湖乌凤鸡 JH | 0.31722 | 0.67500 | 1.19223 | 0.77300 |
| 广西乌鸡 GX | 1.81444 | 0.97700 | 3.86919 | 0.94800 |
| 平均 Average | 0.11225 | 0.55950 | 0.44305 | 0.57342 |
| 方差 Squared deviation | 0.87539 | 0.26761 | 1.50012 | 0.25155 |
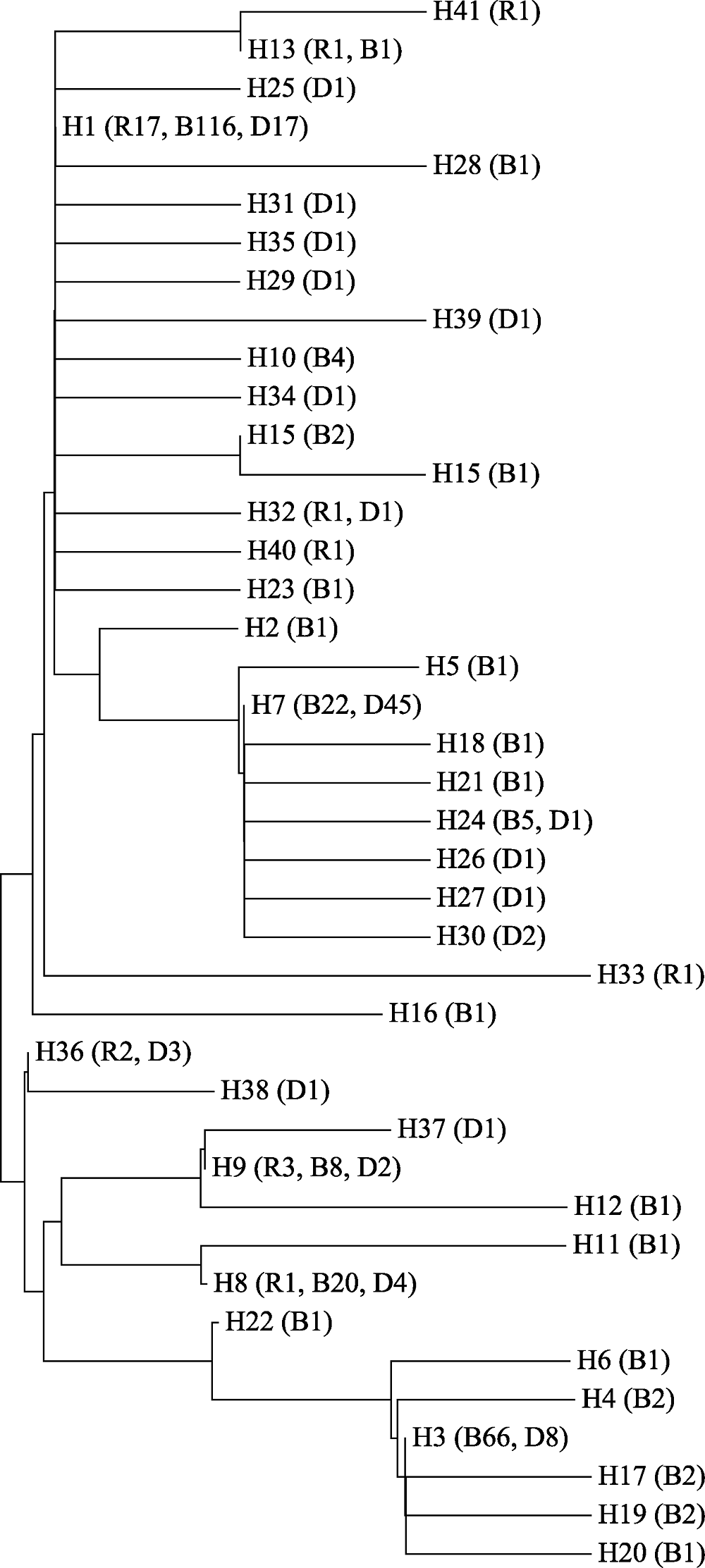
图2 384只鸡的41个COI单倍型邻接树。R为红原鸡, B为乌骨鸡, D为家鸡, 数字表示样品数量。品种代号同表1。
Fig. 2 Neighbor-joining tree of 41 COI haplotypes deteced in the samples of 384 chickens. R, Red junglefowl; B, Black-bone chicken; D, Domestic chicken. Numbers represent the sample size. Breed codes are the same as in Table 1.
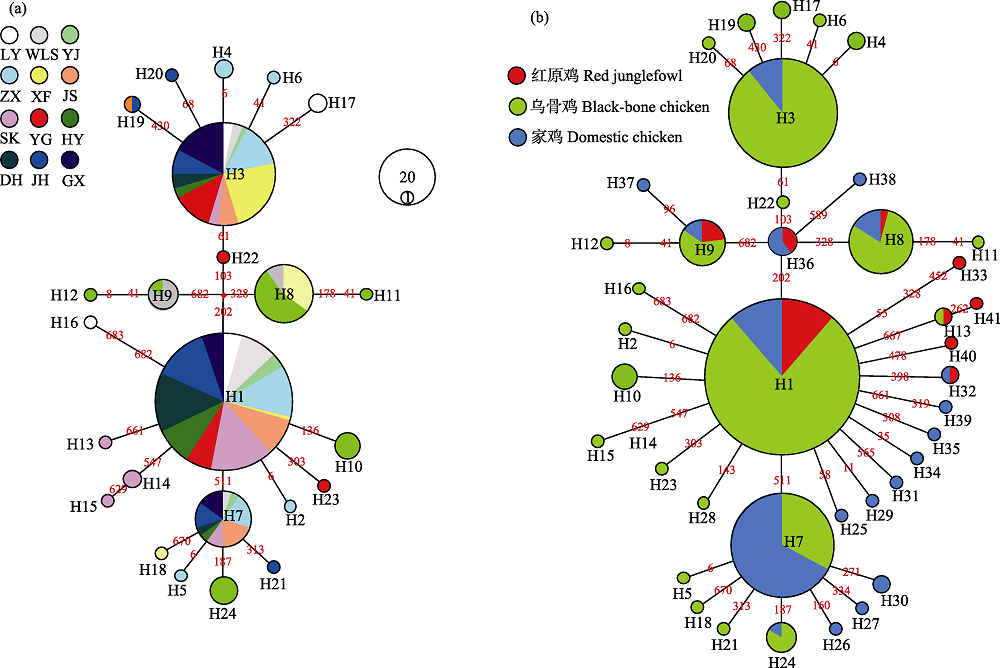
图3 COI基因中介网络图。连接点数字表示核苷酸转换的位置; 圆的大小对应单倍型频率, 不同鸡品种用不同颜色标注, 品种代号同表1。(a)中国12种255只乌骨鸡个体的24个COI单倍型。(b) 384只鸡个体的41个COI单倍型中介网络图。
Fig. 3 Median-joining network based on COI gene. The links are labeled by the nucleotide positions to designate transitions. Cycle size is roughly proportional to the haplotype frequency, the breeds are indicated by different colors, breed codes are the same as in Table 1. (a) Median-joining network of 24 COI haplotypes deteced in the samples of 255 individuals of 12 Chinese black-bone breeds. (b) Median-joining network of 41 COI haplotypes deteced in the samples of 384 individuals.
| 变异起源 Source of variation | 自由度 df | 平方和 Sum of squares | 方差组分 Variance components | 方差比例 Percentage of variance (%) |
|---|---|---|---|---|
| 群体间 Among populations | 11 | 52.622 | 0.18826 | 18.94 |
| 群体内 Within populations | 243 | 195.743 | 0.805553 | 81.06 |
| 总变异 Total variation | 254 | 248.365 | 0.99378 | 100 |
表6 12种乌骨鸡种群遗传变异的分子变异分析
Table 6 AMOVA analysis of genetic variation of 12 black-bone chicken breeds
| 变异起源 Source of variation | 自由度 df | 平方和 Sum of squares | 方差组分 Variance components | 方差比例 Percentage of variance (%) |
|---|---|---|---|---|
| 群体间 Among populations | 11 | 52.622 | 0.18826 | 18.94 |
| 群体内 Within populations | 243 | 195.743 | 0.805553 | 81.06 |
| 总变异 Total variation | 254 | 248.365 | 0.99378 | 100 |
| Breed | LY | WLS | YJ | ZX | XF | JS | SK | YG | HY | DH | JH | GX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LY | 0.003 | 0.004 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | |
| WLS | 0.116* | 0.003 | 0.003 | 0.004 | 0.004 | 0.002 | 0.002 | 0.003 | 0.002 | 0.003 | 0.003 | |
| YJ | 0.189* | 0.017 | 0.004 | 0.004 | 0.004 | 0.003 | 0.004 | 0.004 | 0.003 | 0.003 | 0.004 | |
| ZX | -0.018 | 0.134* | 0.207* | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.002 | 0.003 | 0.003 | |
| XF | 0.231* | 0.427* | 0.433* | 0.248* | 0.004 | 0.004 | 0.003 | 0.004 | 0.004 | 0.003 | 0.002 | |
| JS | 0.004 | 0.121* | 0.196* | -0.014 | 0.341* | 0.002 | 0.003 | 0.003 | 0.003 | 0.002 | 0.003 | |
| SK | 0.178* | 0.192* | 0.269* | 0.134* | 0.594* | 0.074* | 0.003 | 0.002 | 0.001 | 0.002 | 0.003 | |
| YG | -0.035 | 0.170* | 0.234* | -0.011 | 0.158* | 0.032 | 0.250* | 0.003 | 0.002 | 0.003 | 0.002 | |
| HY | 0.165* | 0.204* | 0.261* | 0.120* | 0.535* | 0.051 | 0.079* | 0.223* | 0.002 | 0.002 | 0.004 | |
| DH | 0.117 | 0.160* | 0.243* | 0.072 | 0.551* | 0.011 | -0.012 | 0.182* | 0.044 | 0.002 | 0.003 | |
| JH | 0.013 | 0.121* | 0.206* | -0.001 | 0.403* | -0.033 | 0.445 | 0.546 | 0.067 | -0.011 | 0.003 | |
| GX | 0.054 | 0.305* | 0.343* | 0.089* | 0.029 | 0.173* | 0.441* | -0.002 | 0.393* | 0.383* | 0.220* |
表7 12个乌骨鸡品种间Kiumura遗传双参数距离(上三角)和遗传分化系数(Fst) (下三角)。品种代号同表1。
Table 7 K2P distance (above diagonal) and fixation index (Fst) (below diagonal) among 12 black-bone chicken breeds. Breed codes are the same as in Table 1.
| Breed | LY | WLS | YJ | ZX | XF | JS | SK | YG | HY | DH | JH | GX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LY | 0.003 | 0.004 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | |
| WLS | 0.116* | 0.003 | 0.003 | 0.004 | 0.004 | 0.002 | 0.002 | 0.003 | 0.002 | 0.003 | 0.003 | |
| YJ | 0.189* | 0.017 | 0.004 | 0.004 | 0.004 | 0.003 | 0.004 | 0.004 | 0.003 | 0.003 | 0.004 | |
| ZX | -0.018 | 0.134* | 0.207* | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.002 | 0.003 | 0.003 | |
| XF | 0.231* | 0.427* | 0.433* | 0.248* | 0.004 | 0.004 | 0.003 | 0.004 | 0.004 | 0.003 | 0.002 | |
| JS | 0.004 | 0.121* | 0.196* | -0.014 | 0.341* | 0.002 | 0.003 | 0.003 | 0.003 | 0.002 | 0.003 | |
| SK | 0.178* | 0.192* | 0.269* | 0.134* | 0.594* | 0.074* | 0.003 | 0.002 | 0.001 | 0.002 | 0.003 | |
| YG | -0.035 | 0.170* | 0.234* | -0.011 | 0.158* | 0.032 | 0.250* | 0.003 | 0.002 | 0.003 | 0.002 | |
| HY | 0.165* | 0.204* | 0.261* | 0.120* | 0.535* | 0.051 | 0.079* | 0.223* | 0.002 | 0.002 | 0.004 | |
| DH | 0.117 | 0.160* | 0.243* | 0.072 | 0.551* | 0.011 | -0.012 | 0.182* | 0.044 | 0.002 | 0.003 | |
| JH | 0.013 | 0.121* | 0.206* | -0.001 | 0.403* | -0.033 | 0.445 | 0.546 | 0.067 | -0.011 | 0.003 | |
| GX | 0.054 | 0.305* | 0.343* | 0.089* | 0.029 | 0.173* | 0.441* | -0.002 | 0.393* | 0.383* | 0.220* |
| [1] |
Bandelt HJ, Forster P, Röhl A ( 1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48.
DOI URL |
| [2] | China National Commission of Animal Genetic Resources( 2011) Animal Genetic Resources in China: Poultry. China Agriculture Press, Beijing. (in Chinese) |
| [ 国家畜禽遗传资源委员会( 2011) 中国畜禽遗传资源志: 家禽志. 中国农业出版社, 北京.] | |
| [3] | Dharmayanthi AB, Terai Y, Sulandari S, Zein MSA, Akiyama T, Satta Y ( 2017) The origin and evolution of fibromelanosis in domesticated chickens: Genomic comparison of Indonesian Cemani and Chinese Silkie breeds. PLoS ONE, 4, e0173147. |
| [4] | Ekarius C ( 2007) Storey’s Illustrated Guide to Poultry Breeds. Storey Publishing, Massachusetts. |
| [5] |
Excoffier L, Lischer HE ( 2010) Arlequin suite ver. 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI URL |
| [6] | Feng CG, Gao Y, Dorshorst B, Song C, Gu XR, Li QY, Li JX, Liu TX, Rubin CJ, Zhao YQ, Wang YQ, Fei J, Li HF, Chen KW, Qu H, Shu DM, Ashwell C, Da Y, Andersson L, Hu X, Li N ( 2014) A cis-regulatory mutation of PDSS2 causes silky-feather in chickens. PLoS Genetics, 8, e1004576. |
| [7] |
Grant W, Bowen B ( 1998) Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89, 415-426.
DOI URL |
| [8] |
Groeneveld LF, Lenstra JA, Eding H, Toro MA, Scherf B, Pilling D, Negrini R, Finlay EK, Han JL, Groeneveld E, Weigend S, GLOBALDIV Consortium ( 2010) Genetic diversity in farm animals—A review. Animal Genetics, 41, 6-31.
DOI URL |
| [9] | Hall TA ( 1999) Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98. |
| [10] | Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM ( 2004) Identification of birds through DNA barcodes. PLoS Biology, 2, 1657-1663. |
| [11] | Huang XH, Chen JB, He DL, Zhang XQ, Zhong FS ( 2016) DNA barcoding of indigenous chickens in China: A reevaluation. Scientia Agricultura Sinica, 49, 2622-2633. (in Chinese with English abstract) |
| [ 黄勋和, 陈洁波, 何丹林, 张细权, 钟福生 ( 2016) DNA条形码技术鉴定中国地方鸡品种的重新评估. 中国农业科学, 49, 2622-2633.] | |
| [12] |
Huang XH, Wu YJ, Miao YW, Peng MS, Chen X, He DL, Suwannapoom C, Du BW, Li XY, Weng ZX, Jin SH, Song JJ, Wang MS, Chen JB, Li WN, Otecko NO, Geng ZY, Qu XY, Wu YP, Yang XR, Jin JQ, Han JL, Zhong FS, Zhang XQ, Zhang YP ( 2018) Was chicken domesticated in northern China? New evidence from mitochondrial genomes. Science Bulletin, 63, 743-746.
DOI URL |
| [13] | Jia XX, Tang XJ, Fan YF, Lu JX, Huang SH, Ge QL, Gao YS, Han W ( 2017) Genetic diversity of local chicken breeds in East China based on mitochondrial DNA D-loop region. Biodiversity Science, 25, 540-548. (in Chinese with English abstract) |
| [ 贾晓旭, 唐修君, 樊艳凤, 陆俊贤, 黄胜海, 葛庆联, 高玉时, 韩威 ( 2017) 华东地区地方鸡品种mtDNA控制区遗传多样性. 生物多样性, 25, 540-548.] | |
| [14] | Kameshpandian P, Thomas S, Nagarajan M ( 2016) Genetic diversity and relationship of Indian Muscovy duck populations. Mitochondrial DNA Part A, 29, 1-5. |
| [15] | Li SZ ( 2005) Bencao Gangmu, 2nd edn. People’s Medical Publishing House, Beijing. (in Chinese) |
| [ 李时珍 ( 2005) 本草纲目(第2版). 人民卫生出版社, 北京.] | |
| [16] | Lin MM, Wang GQ, He ZG, Ma P, Liu Y ( 2017) Genetic diversity of Tianshui black-bone chicken based on DNA D-loop sequences. China Poultry, 39(1), 57-59. (in Chinese) |
| [ 林萌萌, 王国琪, 何振刚, 马平, 刘玉 ( 2017) 天水乌鸡线粒体DNA D-loop序列遗传多样性分析. 中国家禽, 39(1), 57-59.] | |
| [17] | Liu HT ( 2006) The formation, utilization and development of Dehua black-bone chicken. Fujian Journal of Animal Husbandry and Veterinary Medicine, 2(S1), 50-52. (in Chinese) |
| [ 刘鸿涛 ( 2006) 德化黑鸡的形成、利用现状与发展思路. 福建畜牧兽医, 2(S1), 50-52.] | |
| [18] | Liu YP ( 2002) Genetic Diversity of Chinese Black-bone Chickens and Origin of Domestic Chickens. PhD dissertation, Sichuan Agricultural University, Ya’an, Sichuan. (in Chinese with English abstract) |
| [ 刘益平 ( 2002) 中国乌骨鸡遗传多样性及家鸡起源研究. 博士学位论文, 四川农业大学, 四川雅安.] | |
| [19] |
Liu YP, Wu GS, Yao YG, Miao YW, Luikart GD, Baig M, Pereira AB, Ding ZL, Palanichamy MG, Zhang YP ( 2006) Multiple maternal origins of chickens: Out of the Asian jungles. Molecular Phylogenetics and Evolution, 38, 12-19.
DOI URL |
| [20] | Miao YW, Sun LM, Tong JJ, Li DL, Yuan YY, Yang MC, Hao WF, Huo JL, Yang R, Yu SL ( 2013) Maternal genetic analysis of Puer Maojiao and Nanjian Lver black-bone chicken using mtDNA D-loop sequences. Acta Ecologiae Animalis Domastici, 34(7), 10-14. (in Chinese with English abstract) |
| [ 苗永旺, 孙利民, 童晶晶, 李大林, 袁跃云, 杨满灿, 郝伟峰, 霍金龙, 杨润, 余仕亮 ( 2013) 普洱毛脚乌鸡与南涧绿耳乌鸡线粒体DNA母系遗传分析. 家畜生态学报, 34(7), 10-14.] | |
| [21] |
Peters J, Lebrasseur O, Deng H, Larson D ( 2016) Holocene cultural history of red jungle fowl (Gallus gallus) and its domestic descendant in East Asia. Quaternary Science Reviews, 142, 102-119.
DOI URL |
| [22] | Ren G, Ma H, Ma C, Wang W, Chen W, Ma L ( 2016) Genetic diversity and population structure of Portunus sanguinolentus (Herbst, 1783) revealed by mtDNA COI sequences. Mitochondrial DNA Part A: DNA Mapping, Sequencing, and Analysis, 28, 740-746. |
| [23] | Rousset F ( 1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics , 145, 1219-1228. |
| [24] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A ( 2017) DnaSP 6: DNA sequence polymorphism analysis of large datasets. Molecular Biology Evolution, 34, 3299-3302.
DOI URL |
| [25] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S ( 2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology & Evolution, 30, 2725-2729. |
| [26] |
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG ( 1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882.
DOI URL |
| [27] | Tu YJ, Gao YS, Su YJ, Wang KH, Tong HB ( 2011) Genetic diversity and phylogenetic analysis of COI gene in some indigenous chicken breeds. Journal of Anhui Agricultural University, 38, 39-42. (in Chinese with English abstract) |
| [ 屠云洁, 高玉时, 苏一军, 王克华, 童海兵 ( 2011) 我国部分地方鸡种COI基因多态性及其分子系统进化研究. 安徽农业大学学报, 38, 39-42.] | |
| [28] | Wang L, Li Q, Kong LF, Yu H ( 2018) Population genetic structure and demographic history of Barbatia virescens along Chinese coast based on mitochondrial COI sequences. Oceanologia et Limnologia Sinica, 49, 87-95. (in Chinese with English abstract) |
| [ 王玲, 李琪, 孔令锋, 于红 ( 2018) 基于COI基因的中国沿海青蚶野生群体遗传结构及种群动态研究. 海洋与湖沼, 49, 87-95.] | |
| [29] | Wei L, Liu SG, Shi XW ( 2008) Genetic diversity of the Xuefeng black bone chicken based on microsatellite markers. Biodiversity Science, 16, 503-508. (in Chinese with English abstract) |
| [ 魏麟, 刘胜贵, 史宪伟 ( 2008) 雪峰乌骨鸡自然群体遗传多样性的微卫星分析. 生物多样性, 16, 503-508.] | |
| [30] | Wu HH, Xie RQ ( 1999) The origin, history and usage of black-bone chickens. Guide to Chinese Poultry, 16(11), 56. (in Chinese) |
| [ 吾豪华, 谢若泉 ( 1999) 乌鸡产地、历史及作用浅议. 中国家禽导刊, 16(11), 56.] | |
| [31] | Xiang H, Gao J, Yu B, Xiang H, Gao JQ, Yu BQ, Zhou H, Cai DW, Zhang YW, Chen XY, Wang X, Hofreiter M, Zhao XB ( 2014) Early Holocene chicken domestication in northern China. Proceedings of the National Academy of Sciences, USA, 111, 17564-17569. |
| [32] | Xu WJ, Zhu WQ, Shu JT, Song C, Chen KW ( 2014) Study on genetic diversity and phylogenetic evolution in Chinese main black-bone chicken. Chinese Journal of Animal Science, 50(23), 10-14. (in Chinese with English abstract) |
| [ 徐文娟, 朱文奇, 束婧婷, 宋迟, 陈宽维 ( 2014) 我国主要乌骨鸡品种遗传多样性和系统进化研究. 中国畜牧杂志, 50(23), 10-14.] | |
| [33] | Yuan J ( 2010) Zooarchaeological study on the domestic animals in ancient China. Quaternary Science, 30, 298-306. (in Chinese with English abstract) |
| [ 袁靖 ( 2010) 中国古代家养动物的动物考古学研究. 第四纪研究, 30, 298-306.] | |
| [34] |
Zhang T, Du W, Lu H, Wang L ( 2018) Genetic diversity of mitochondrial DNA of Chinese black-bone chicken. Brazilian Journal of Poultry Science, 20, 565-572.
DOI URL |
| [1] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [2] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [3] | 武星彤, 陈璐, 王敏求, 张原, 林雪莹, 李鑫玉, 周宏, 文亚峰. 丹霞梧桐群体遗传结构及其遗传分化[J]. 生物多样性, 2018, 26(11): 1168-1179. |
| [4] | 刘青青, 董志军. 基于线粒体COI基因分析钩手水母的群体遗传结构[J]. 生物多样性, 2018, 26(11): 1204-1211. |
| [5] | 盛芳, 陈淑英, 田嘉, 李鹏, 秦雪, 罗淑萍, 李疆. 新疆准噶尔山楂不同居群的遗传多样性[J]. 生物多样性, 2017, 25(5): 518-530. |
| [6] | 刘若愚, 孙忠民, 姚建亭, 胡自民, 段德麟. 中国近海重要生态建群红藻真江蓠的群体遗传多样性[J]. 生物多样性, 2016, 24(7): 781-790. |
| [7] | 于少帅, 徐启聪, 林彩丽, 王圣洁, 田国忠. 植原体遗传多样性研究现状与展望[J]. 生物多样性, 2016, 24(2): 205-215. |
| [8] | 向妮, 肖炎农, 段灿星, 王晓鸣, 朱振东. 利用SSR标记分析茄镰孢豌豆专化型的遗传多样性[J]. 生物多样性, 2012, 20(6): 693-702. |
| [9] | 周蓉, 李佳琦, 李铀, 刘迺发, 房峰杰, 施丽敏, 王莹. 基于线粒体DNA的大石鸡种群遗传变异[J]. 生物多样性, 2012, 20(4): 451-459. |
| [10] | 陈碧云, 胡琼, Christina Dixelius, 李国庆, 伍晓明. 利用SRAP分析核盘菌遗传多样性[J]. 生物多样性, 2010, 18(5): 509-515. |
| [11] | 张俊红, 黄华宏, 童再康, 程龙军, 梁跃龙, 陈奕良. 光皮桦6个南方天然群体的遗传多样性[J]. 生物多样性, 2010, 18(3): 233-240. |
| [12] | 张根, 席贻龙, 薛颖昊, 胡忻, 项贤领, 温新利. 基于rDNA ITS序列探讨粉煤灰污染对萼花臂尾轮虫种群遗传多样性的影响[J]. 生物多样性, 2010, 18(3): 241-250. |
| [13] | 傅洪拓, 乔慧, 姚建华, 龚永生, 吴滟, 蒋速飞, 熊贻伟. 基于SRAP分子标记的海南沼虾种群遗传多样性[J]. 生物多样性, 2010, 18(2): 145-149. |
| [14] | 王长忠, 李忠, 梁宏伟, 呼光富, 吴勤超, 邹桂伟, 罗相忠. 长江下游地区4个克氏原螯虾群体的遗传多样性分析[J]. 生物多样性, 2009, 17(5): 518-523. |
| [15] | 王峥峰, 葛学军. 不仅仅是遗传多样性: 植物保护遗传学进展[J]. 生物多样性, 2009, 17(4): 330-339. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn