



生物多样性 ›› 2013, Vol. 21 ›› Issue (5): 601-609. DOI: 10.3724/SP.J.1003.2013.09098 cstr: 32101.14.SP.J.1003.2013.09098
王德元1, 彭婕1, 陈雅静1, 吕国胜1, 张小平1,2, 邵剑文1,3,*( )
)
收稿日期:2013-04-18
接受日期:2013-07-01
出版日期:2013-09-20
发布日期:2013-10-08
通讯作者:
邵剑文
基金资助:
Deyun Wang1, Jie Peng1, Yajing Chen1, Guosheng Lü1, Xiaoping Zhang1,2, Jianwen Shao1,3,*( )
)
Received:2013-04-18
Accepted:2013-07-01
Online:2013-09-20
Published:2013-10-08
Contact:
Shao Jianwen
摘要:
毛茛叶报春(Primula ranunculoides)是我国特有的珍稀濒危花卉, 具较好的园艺开发潜力, 其野生种群已十分稀少。本文利用9对微卫星引物对7个自然种群共222个个体的遗传多样性和遗传结构进行了研究。结果表明, 与近缘种相比, 毛茛叶报春具较低的遗传多样性, 种群间分化明显。种群的平均观察杂合度为0.286, 期望杂合度为0.330, 种群遗传多样性与植株密度有显著的正相关性, 而与种群大小和面积无显著相关性。种群间的平均基因流Nm = 0.730。AMOVA分析表明毛茛叶报春有48.08%的变异存在于种群间, 51.92%出现在种群内。毛茛叶报春种下的遗传结构与地形关系密切, 山脉间的两湖平原(江汉平原和洞庭湖平原)及武宁和修水低洼农耕区等异质生境是其主要的基因流屏障。建议将毛茛叶报春划分为九岭山、幕阜山、七姊妹山和银炉4个单元进行遗传多样性管理。
王德元, 彭婕, 陈雅静, 吕国胜, 张小平, 邵剑文 (2013) 毛茛叶报春的遗传多样性及遗传结构. 生物多样性, 21, 601-609. DOI: 10.3724/SP.J.1003.2013.09098.
Deyun Wang,Jie Peng,Yajing Chen,Guosheng Lü,Xiaoping Zhang,Jianwen Shao (2013) Genetic diversity and genetic structure of the rare and endangered species, Primula ranunculoides. Biodiversity Science, 21, 601-609. DOI: 10.3724/SP.J.1003.2013.09098.
| 种群 Population | 地点 Locality | 生境 Habitat | 地理位置 Geographical location | 海拔 Altitude (m) | 面积 Area (m2) | 花朵类型 Flower type | 种群大小 Population size |
|---|---|---|---|---|---|---|---|
| 杨家坪A YJPA | 江西省修水县杨家坪 Yangjiaping, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside under forests | 28°47'45"N, 114°43'56"E | 300-375 | 720 | 两型花 Distyle | ~2,000 |
| 杨家坪B YJPB | 江西省修水县杨家坪 Yangjiaping, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside, under forest | 28°47'04"N, 114°44'13"E | 350-360 | 1,060 | 两型花 Distyle | ~6,000 |
| 毛竹山 MZS | 江西省修水县毛竹山 Maozhushan, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside, under forest | 28°50'31"N, 114°52'00"E | 390-410 | 1,650 | 两型花 Distyle | ~1,500 |
| 三界A SJA | 湖北省通山县三界 Sanjie, Tongshan County, Hubei | 林下溪边或路边 Brook-side or roadside, under forest | 29°24'16"N, 114°27'15"E | 255-285 | 545 | 两型花 Distyle | ~1,000 |
| 三界B SJB | 湖北省通山县三界 Sanjie, Tongshan County, Hubei | 林下溪边或路边 Brook-side or roadside, under forest | 29°22'57"N, 114°27'08"E | 265-280 | 385 | 两型花 Distyle | ~700 |
| 七姊妹山 JPT | 湖北省宣恩县七姊妹山 Qizimei Mount, Xuanen County, Hubei | 林下溪边 Brook-side, under forest | 30°02'09"N, 109°43'57"E | 1,310-1,365 | 1,200 | 同型花 Monostyle | ~600 |
| 银炉 YL | 江西省武宁县银炉 Yinlu, Wuning County, Jiangxi | 溪边石壁 Rock near brook-side | 28°59'31''N, 114°49'39''E | 420 | 50 | 两型花 Distyle | ~200 |
表1 本研究中毛茛叶报春种群取样信息
Table 1 Locality and information of studied populations of Primula ranunculoides
| 种群 Population | 地点 Locality | 生境 Habitat | 地理位置 Geographical location | 海拔 Altitude (m) | 面积 Area (m2) | 花朵类型 Flower type | 种群大小 Population size |
|---|---|---|---|---|---|---|---|
| 杨家坪A YJPA | 江西省修水县杨家坪 Yangjiaping, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside under forests | 28°47'45"N, 114°43'56"E | 300-375 | 720 | 两型花 Distyle | ~2,000 |
| 杨家坪B YJPB | 江西省修水县杨家坪 Yangjiaping, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside, under forest | 28°47'04"N, 114°44'13"E | 350-360 | 1,060 | 两型花 Distyle | ~6,000 |
| 毛竹山 MZS | 江西省修水县毛竹山 Maozhushan, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside, under forest | 28°50'31"N, 114°52'00"E | 390-410 | 1,650 | 两型花 Distyle | ~1,500 |
| 三界A SJA | 湖北省通山县三界 Sanjie, Tongshan County, Hubei | 林下溪边或路边 Brook-side or roadside, under forest | 29°24'16"N, 114°27'15"E | 255-285 | 545 | 两型花 Distyle | ~1,000 |
| 三界B SJB | 湖北省通山县三界 Sanjie, Tongshan County, Hubei | 林下溪边或路边 Brook-side or roadside, under forest | 29°22'57"N, 114°27'08"E | 265-280 | 385 | 两型花 Distyle | ~700 |
| 七姊妹山 JPT | 湖北省宣恩县七姊妹山 Qizimei Mount, Xuanen County, Hubei | 林下溪边 Brook-side, under forest | 30°02'09"N, 109°43'57"E | 1,310-1,365 | 1,200 | 同型花 Monostyle | ~600 |
| 银炉 YL | 江西省武宁县银炉 Yinlu, Wuning County, Jiangxi | 溪边石壁 Rock near brook-side | 28°59'31''N, 114°49'39''E | 420 | 50 | 两型花 Distyle | ~200 |
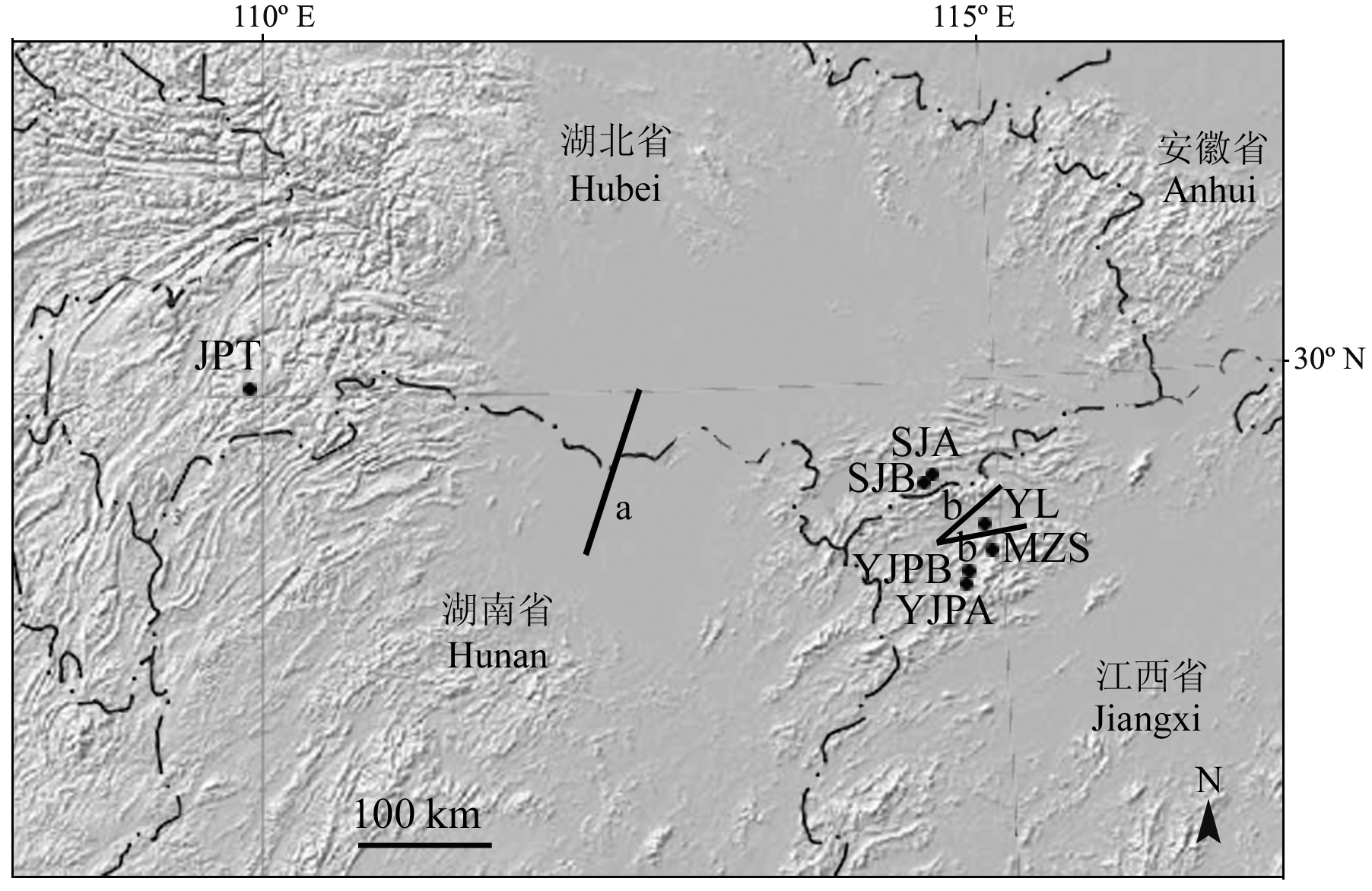
图1 采样点分布图及BARRIER软件分析的第一(a)和第二(b)基因流屏障(种群代号同表1)
Fig. 1 Geographic location of the studied populations and the first two boundaries (a to b) detected using BARRIER. Population codes see Table 1.
| 位点 Locus | 序列 Repeat motif | 引物 Primer (5'-3') | 退火温度(℃) Annealing temperature | 荧光标记 Fluorescent label | 等位基因数 No. of alleles |
|---|---|---|---|---|---|
| Pm1 | (TC)10(CT)3 | F:ATCTTTGAGGTCCTTTTA | 50 | FAM | 7 |
| R: ATCGCCCAATGGAGTGAA | |||||
| Pm2 | (AG)14 | F: CGCCTACAGTGTTTGGGA | 55 | FAM | 2 |
| R: CTATCTCACCTGCGTTCT | |||||
| Pm7 | (AG)3GG(AG)8 | F: TTGTTCACCGACGCATAC | 54 | HEX | 18 |
| R: TTACACGCACCAAATCAT | |||||
| Pm9 | (TTTC)2(TC)6 | F: AGACTCACGAGGAATACG | 50 | HEX | 6 |
| R: AGAAAAGGAGGAGACAAA | |||||
| Pm12 | (TC)10(CT)3 | F: TAAAACTCCTGGAGGGGTAC | 51 | TAMRA | 10 |
| R: ATCGCCCAATGGAGTGAA | |||||
| Pm13 | (GATAGG)2GAT(AG)7 | F: GAGGACAGGCACCACAGA | 54 | TAMRA | 6 |
| R: TCCCCAACTTCATGCTCTT | |||||
| Pm14 | (AG)3(GA)12 | F: TCGCCCAATGGAGTGAAC | 50 | TAMRA | 9 |
| R: TCTTTGAGGTCCTTTTAT | |||||
| Pm16 | (GAGGGA)3(GA)3 | F: AACCACTCGTCGTCCTAA | 51 | FAM | 5 |
| R: CGATAGATTGCCTTACCC | |||||
| Pm17 | (GA)9 | F: TAAATCAAGGTAGCAACT | 51 | HEX | 9 |
| R: TACCTACCATTACTCCC |
表2 本研究使用的微卫星位点及引物序列
Table 2 The sequences of microsatellite loci and primers in this study
| 位点 Locus | 序列 Repeat motif | 引物 Primer (5'-3') | 退火温度(℃) Annealing temperature | 荧光标记 Fluorescent label | 等位基因数 No. of alleles |
|---|---|---|---|---|---|
| Pm1 | (TC)10(CT)3 | F:ATCTTTGAGGTCCTTTTA | 50 | FAM | 7 |
| R: ATCGCCCAATGGAGTGAA | |||||
| Pm2 | (AG)14 | F: CGCCTACAGTGTTTGGGA | 55 | FAM | 2 |
| R: CTATCTCACCTGCGTTCT | |||||
| Pm7 | (AG)3GG(AG)8 | F: TTGTTCACCGACGCATAC | 54 | HEX | 18 |
| R: TTACACGCACCAAATCAT | |||||
| Pm9 | (TTTC)2(TC)6 | F: AGACTCACGAGGAATACG | 50 | HEX | 6 |
| R: AGAAAAGGAGGAGACAAA | |||||
| Pm12 | (TC)10(CT)3 | F: TAAAACTCCTGGAGGGGTAC | 51 | TAMRA | 10 |
| R: ATCGCCCAATGGAGTGAA | |||||
| Pm13 | (GATAGG)2GAT(AG)7 | F: GAGGACAGGCACCACAGA | 54 | TAMRA | 6 |
| R: TCCCCAACTTCATGCTCTT | |||||
| Pm14 | (AG)3(GA)12 | F: TCGCCCAATGGAGTGAAC | 50 | TAMRA | 9 |
| R: TCTTTGAGGTCCTTTTAT | |||||
| Pm16 | (GAGGGA)3(GA)3 | F: AACCACTCGTCGTCCTAA | 51 | FAM | 5 |
| R: CGATAGATTGCCTTACCC | |||||
| Pm17 | (GA)9 | F: TAAATCAAGGTAGCAACT | 51 | HEX | 9 |
| R: TACCTACCATTACTCCC |
| 种群 Population | 取样数 Sample size | 有效种群大小 Effective population size | 期望杂合度(He) Expected heterozygosity | 观察杂合度(Ho) Observed heterozygosity | 稀有等位基因 Rare alleles | 近交系数(Fis) Inbreeding coefficient |
|---|---|---|---|---|---|---|
| YJPA | 32 | 166 | 0.430 | 0.424 | 2 | 0.014 ns |
| YJPB | 31 | 231 | 0.412 | 0.399 | 4 | 0.032** |
| MZS | 30 | 105 | 0.297 | 0.155 | 2 | 0.484*** |
| SJA | 33 | 169 | 0.348 | 0.330 | 4 | 0.055*** |
| SJB | 31 | 149 | 0.281 | 0.230 | 1 | 0.184*** |
| JPT | 33 | 127 | 0.203 | 0.132 | 2 | 0.350*** |
| YL | 32 | 153 | 0.340 | 0.328 | 2 | 0.037** |
| 平均 Mean | 31.7 | 157.1 | 0.330 | 0.286 | 2.4 |
表3 毛茛叶报春各个种群的遗传参数
Table 3 Genetic characteristics of Primula ranunculoides populations
| 种群 Population | 取样数 Sample size | 有效种群大小 Effective population size | 期望杂合度(He) Expected heterozygosity | 观察杂合度(Ho) Observed heterozygosity | 稀有等位基因 Rare alleles | 近交系数(Fis) Inbreeding coefficient |
|---|---|---|---|---|---|---|
| YJPA | 32 | 166 | 0.430 | 0.424 | 2 | 0.014 ns |
| YJPB | 31 | 231 | 0.412 | 0.399 | 4 | 0.032** |
| MZS | 30 | 105 | 0.297 | 0.155 | 2 | 0.484*** |
| SJA | 33 | 169 | 0.348 | 0.330 | 4 | 0.055*** |
| SJB | 31 | 149 | 0.281 | 0.230 | 1 | 0.184*** |
| JPT | 33 | 127 | 0.203 | 0.132 | 2 | 0.350*** |
| YL | 32 | 153 | 0.340 | 0.328 | 2 | 0.037** |
| 平均 Mean | 31.7 | 157.1 | 0.330 | 0.286 | 2.4 |
| YJPA | YJPB | MZS | SJA | SJB | JPT | YL | |
|---|---|---|---|---|---|---|---|
| YJPA | - | 0.091 | 0.208 | 0.420 | 0.528 | 0.875 | 0.702 |
| YJPB | 0.096 | - | 0.167 | 0.311 | 0.363 | 0.793 | 0.588 |
| MZS | 0.256 | 0.222 | - | 0.336 | 0.378 | 0.827 | 0.742 |
| SJA | 0.301 | 0.262 | 0.399 | - | 0.030 | 0.921 | 0.473 |
| SJB | 0.385 | 0.332 | 0.476 | 0.047 | - | 0.929 | 0.467 |
| JPT | 0.510 | 0.526 | 0.645 | 0.569 | 0.599 | - | 0.318 |
| YL | 0.445 | 0.424 | 0.536 | 0.392 | 0.428 | 0.367 | - |
表4 毛茛叶报春种群间的遗传分化系数(Fst, 对角线下)和遗传距离(D, 对角线上)
Table 4 Pairwise population differentiation (below diagonal) and genetic distances (above diagonal) among Primula ranunculoides populations
| YJPA | YJPB | MZS | SJA | SJB | JPT | YL | |
|---|---|---|---|---|---|---|---|
| YJPA | - | 0.091 | 0.208 | 0.420 | 0.528 | 0.875 | 0.702 |
| YJPB | 0.096 | - | 0.167 | 0.311 | 0.363 | 0.793 | 0.588 |
| MZS | 0.256 | 0.222 | - | 0.336 | 0.378 | 0.827 | 0.742 |
| SJA | 0.301 | 0.262 | 0.399 | - | 0.030 | 0.921 | 0.473 |
| SJB | 0.385 | 0.332 | 0.476 | 0.047 | - | 0.929 | 0.467 |
| JPT | 0.510 | 0.526 | 0.645 | 0.569 | 0.599 | - | 0.318 |
| YL | 0.445 | 0.424 | 0.536 | 0.392 | 0.428 | 0.367 | - |
| 变异来源 Source | 自由度 d.f. | 方差和 Sum of squares | 变异组分 Variance components | 占总变异比例% Percentage of variation | 显著性检验 Significance tests |
|---|---|---|---|---|---|
| 种群间 Among populations | 6 | 225.635 | 0.6596 | 48.08 | P<0.001 |
| 种群内 Within populations | 215 | 310.851 | 0.7124 | 51.92 | P<0.001 |
| 总和 Total | 222 | 536.486 | 1.3720 | 100% |
表5 毛茛叶报春分子变异的AMOVA分析结果
Table 5 Analysis of molecular variance (AMOVA) within/among Primula ranunculoides populations
| 变异来源 Source | 自由度 d.f. | 方差和 Sum of squares | 变异组分 Variance components | 占总变异比例% Percentage of variation | 显著性检验 Significance tests |
|---|---|---|---|---|---|
| 种群间 Among populations | 6 | 225.635 | 0.6596 | 48.08 | P<0.001 |
| 种群内 Within populations | 215 | 310.851 | 0.7124 | 51.92 | P<0.001 |
| 总和 Total | 222 | 536.486 | 1.3720 | 100% |
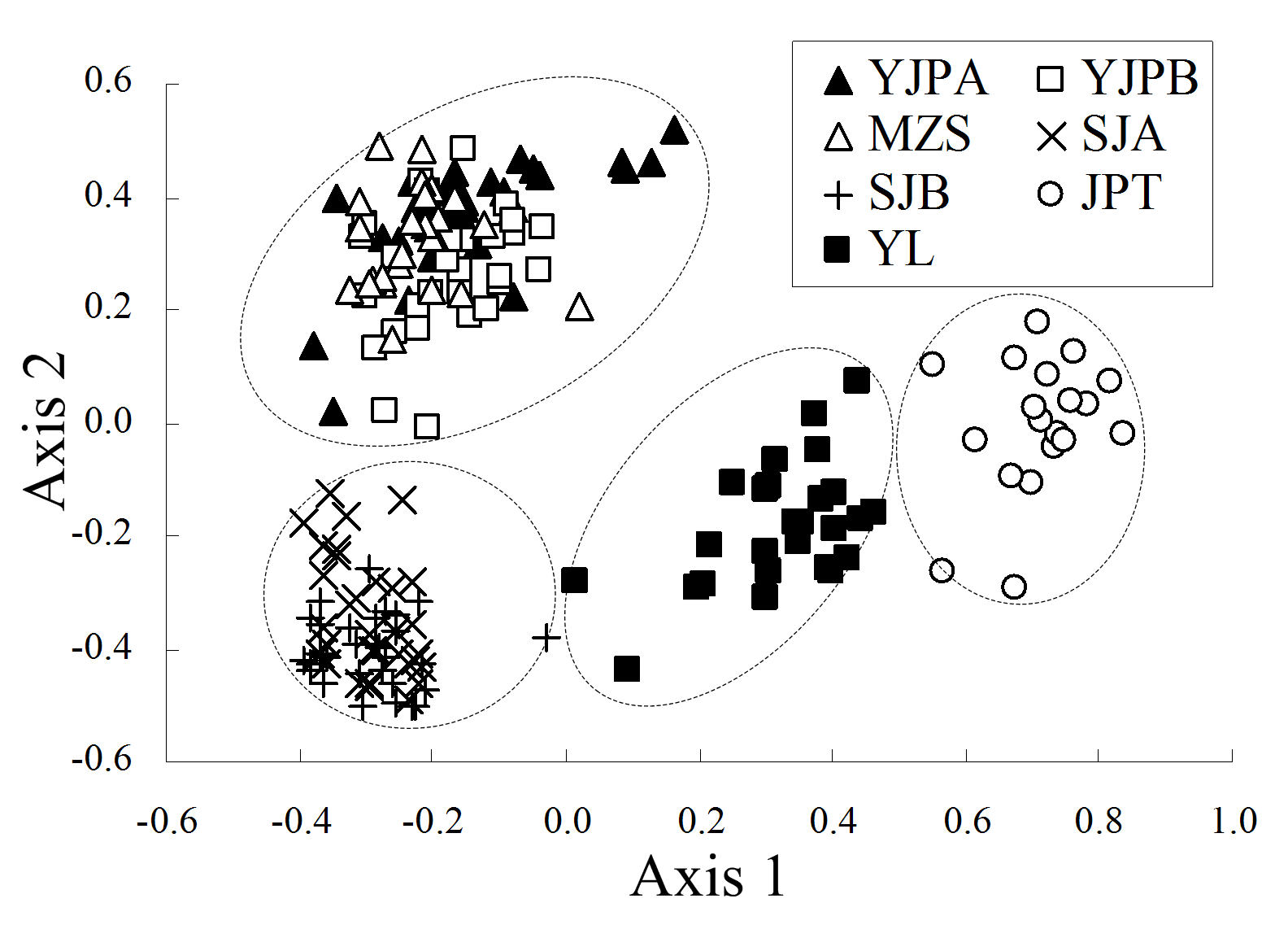
图2 毛茛叶报春7个种群的主成分分析结果。Axis1和Axis2分别代表40.0%和26.9%的总变异。种群代号同表1。
Fig. 2 The result of Principal coordinates (PCO) analysis of Primula ranunculoides populations. The first and second axis extracted 40.0% and 26.9% of the total genetic variance, respectively. Population codes see Table 1.
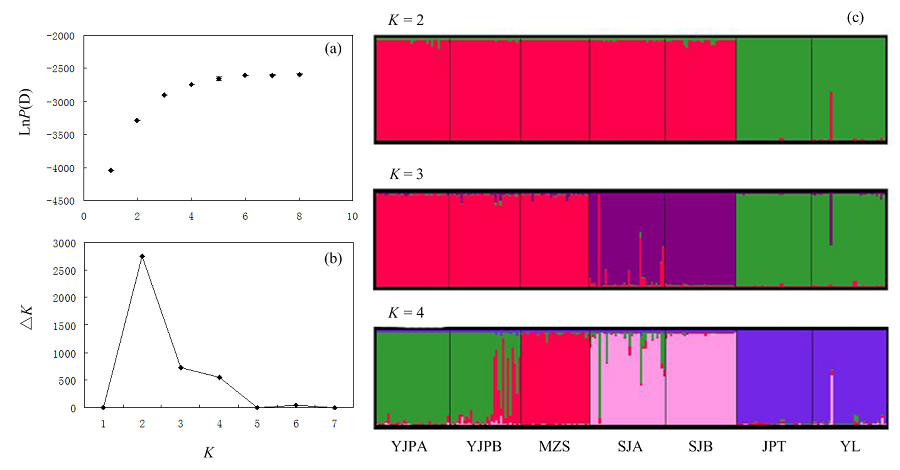
图3 STRUCTURE对遗传结构的分析结果。(a)不同K值运算的lnP(D)平均值; (b)依据Evanno等(2005)计算的△K值; (c) K = 2-4时的个体分配柱形图。
Fig. 3 The results of genetic structure analyses by STRUCTURE soft. (a) Plot of mean posterior probability lnP(D) values of each K; (b) The corresponding △K statistics calculated according to Evanno et al. (2005); (c) Histogram of the structure analysis for the model with K = 2-4. Population codes see Table 1.
| 1 | Beerli P (2008) MIGRATE Version 3.0―a Maximum Likelihood and Bayesian Estimator of Gene Flow Using the Coalescent. Distributed over the Internet at. |
| 2 | Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, Logiciel sous WindowsTM pour la Génétique des Populations. Laboratoire Génome, Populations, Intera- ctions, Université de Montpellier II, Montpellier. |
| 3 | Chen FH (1948) A new Chinese Primula.Notes from the Royal Botanic Garden Edinburgh, 20, 120. |
| 4 | Hu CM (胡启明) (1990) Primulaceae. In: Flora Reipublicae Popularis Sinicae (中国植物志) (ed. Wu ZY (吴征镒)), Tomus 59(2), pp. 1-277. Science Press, Beijing. (in Chinese) |
| 5 | Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation.Annual Review of Ecology and Systematics, 24, 217-242. |
| 6 | Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study.Molecular Ecology, 14, 2611-2620. |
| 7 | Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis.Evolutionary Bioinformatics Online, 1, 47-50. |
| 8 | Foxe JP, Stift M, Tedder A, Haudry A, Wright SI, Mable BK (2010) Reconstructing origins of loss of self-incompatibility and selfing in north American Arabidopsis lyrata: a population genetic context.Evolution, 64, 3495-3510. |
| 9 | Guillernsut P, Maréchal-Drouard L (1992) Isolation of plant DNA: a fast, inexpensive, and reliable method.Plant Molecular Biology Reporter, 10, 60-65. |
| 10 | Guillot G, Mortier F, Estoup A (2005) GENELAND: a computer package for landscape genetics.Molecular Ecology Notes, 5, 712-715. |
| 11 | Hao G, Hu CM, Lee NS (2002) Circumscriptions and phylogenetic relationships of Primula sect. Auganthus and Ranunculoides: evidence from nrDNA ITS sequences.Acta Botanica Sinica, 44, 72-75. |
| 12 | Honjo M, Kitamoto N, Ueno S, Tsumura Y, Washitani I, Ohsawa R (2009) Management units of the endangered herb Primula sieboldii based on microsatellite variation among and within populations throughout Japan.Conservation Genetics, 10, 257-267. |
| 13 | Hu CM, Kelso S (1996) Primulaceae. In: Flora of China (eds Wu ZY, Raven PH), Vol. 15, pp. 99-185. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis. |
| 14 | Huang Y, Wang XQ, Yang CY, Long CL (2010) Development of 11 polymorphic microsatellite loci from Primula amthystina Franchet. (Primulaceae).Hortscience, 45, 148-149. |
| 15 | Ishihama F, Ueno S, Tsumura Y, Washitani I (2006) Effects of density and floral morph on pollen flow and seed reproduction of an endangered heterostylous herb, Primula sieboldii.Journal of Ecology, 94, 846-855. |
| 16 | Jacquemyn H, Olivier H, Peter G, Isabel RU (2004) Genetic structure of the forest herb Primula elatior in a changing landscape.Molecular Ecology, 13, 211-219. |
| 17 | Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure.Bioinformatics, 23, 1801-1806. |
| 18 | Kaeuffer R, Réale D, Coltman DW, Pontier D (2007) Detecting population structure using Structure software: effect of background linkage disequilibrium.Heredity, 99, 374-380. |
| 19 | Lienert J, Fischer M (2003) Habitat fragmentation affects the common wetland specialist Primula farinosa in north-east Switzerland.Journal of Ecology, 91, 587-599. |
| 20 | Manni F, Guérard E (2004) BARRIER Version 2.2. Manual of the User. Population Genetics Team, Museum of Mankind, Paris. |
| 21 | Mantel N (1967) The detection of disease clustering and a generalized regression approach.Cancer Research, 27, 209-220. |
| 22 | Miller MP (1997) Tools for Population Genetic Analysis (TFPGA), Version 1.3. Department of Biological Sciences. Northern Arizona University, Arizona. |
| 23 | Ness RW, Wright SI, Barrett SCH (2010) Mating-system variation, demographic history and patterns of nucleotide diversity in the tristylous plant Eichhornia paniculata.Genetics, 184, 381-392. |
| 24 | Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants.Perspectives in Plant Ecology Evolution and Systematics, 3, 93-114. |
| 25 | Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research.Molecular Ecology Notes, 6, 288-295. |
| 26 | Peng YQ, Shao JW, Wu HL, Zhang XP, Zhu GP (2009) Isolation and characterization of fifteen polymorphic microsatellite loci from Primula merrilliana (Primulaceae), an endemic from China.Conservation Genetics, 10, 1441-1443. |
| 27 | Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data.Genetics, 155, 945-959. |
| 28 | Richards AJ (2003) Primula, 2nd edn. Timber Press, Portland. |
| 29 | Rosenberg NA (2004) Distruct: a program for the graphical display of population structure.Molecular Ecology Notes, 4, 137-138. |
| 30 | Rousset F (2008) Genepop’007: a complete re-implementation of the genepop software for Windows and Linux.Molecular Ecology Resources, 8, 103-106. |
| 31 | Shao JW, Chen WL, Peng YQ, Zhu GP, Zhang XP (2009) Genetic diversity within and among populations of the endan gered and endemic species Primula merrilliana in China. Biochemical Systematics and Ecology, 37, 699-706. |
| 32 | Shao JW, Wu YF, Kan XZ, Liang TJ, Zhang XP (2012) Reappraisal of Primula ranunculoides (Primulaceae), an endangered species endemic to China, based on morphological, molecular genetic and reproductive characters.Botanical Journal of the Linnean Society, 169, 338-349. |
| 33 | Shao JW (邵剑文), Zhang XP (张小平), Zhang ZX (张中信), Zhu GP (朱国萍) (2008a) Identification of effective pollinators of Primula merrilliana and effects of flower density and population size on pollination efficiency.Journal of Systematics and Evolution, 46, 537-544. (in Chinese with English abstract) |
| 34 | Shao JW, Zhang ZX, Zhu GP, Zhang XP (2008b) Effects of population size on reproductive success of the endangered and endemic species Primula merrilliana. Journal of Integrative Plant Biology, 50, 1151-1160. |
| 35 | Slatkin M (1985) Gene flow in natural populations.Annual Review of Ecology and Systematics, 16, 393-430. |
| 36 | van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data.Molecular Ecology Notes, 4, 535-538. |
| 37 | van Rossum F, De Sousa SC, Triest L (2004) Genetic consequences of habitat fragmentation in an agricultural landscape on the common Primula veris, and comparison with its rare congener, P. vulgaris.Conservation Genetics, 5, 231-245. |
| 38 | van Rossum F, Triest L (2003) Spatial genetic structure and reproductive success in fragmented and continuous populations of Primula vulgaris.Folia Geobotanica, 38, 239-254. |
| 39 | van Rossum F, Triest L (2006) Fine-scale genetic structure of the common Primula elatior (Primulaceae) at an early stage of population fragmentation.American Journal of Botany, 93, 1281-1288. |
| 40 | Xue DW, Ge XJ, Hao G, Zhang CQ (2004) High genetic diversity in a rare, narrowly endemic primrose species: Primula interjacens by ISSR analysis.Acta Botanica Sinica, 46, 1163-1169. |
| 41 | Yan HF, Ge XJ, Hu CM, Hao G (2010) Isolation and characterization of microsatellite loci for the ornamental plant Primula obconica Hance (Primulaceae).Hortscience, 45, 314-315. |
| 42 | Yuan N, Comes HP, Mao YR, Qi XS, Qiu YX (2012) Genetic effects of recent habitat fragmentation in the Thousand-Island Lake region of southeast China on the distylous herb Hedyotis chrysotricha (Rubiaceae).American Journal of Botany, 99, 1715-1725. |
| 43 | Zhang DX, Hewitt GM (2003) Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects.Molecular Ecology, 12, 563-584. |
| [1] | 周志华, 金效华, 罗颖, 李迪强, 岳建兵, 刘芳, 何拓, 李希, 董晖, 罗鹏. 中国林草部门落实《昆明-蒙特利尔全球生物多样性框架》的机制、成效分析及建议[J]. 生物多样性, 2025, 33(3): 24487-. |
| [2] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [3] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [4] | 钟超, 廖亚琴, 刘伟杰, 隋昊志, 陈清华. 广东沿海海草床的现状、面临的威胁与保护建议[J]. 生物多样性, 2024, 32(2): 23201-. |
| [5] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [6] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [7] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [8] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [9] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [10] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [11] | 席辉辉, 王祎晴, 潘跃芝, 许恬, 湛青青, 刘健, 冯秀彦, 龚洵. 中国苏铁属植物资源和保护[J]. 生物多样性, 2022, 30(7): 21495-. |
| [12] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [13] | 牛晓锋, 王晓梅, 张研, 赵志鹏, 樊恩源. 鲟鱼分子鉴定方法的整合应用[J]. 生物多样性, 2022, 30(6): 22034-. |
| [14] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [15] | 崔静, 徐明芳, 章群, 李瑶, 曾晓舒, 李莎. 基于3种线粒体标记探讨中日沿海角木叶鲽遗传多样性差异[J]. 生物多样性, 2022, 30(5): 21485-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn