



生物多样性 ›› 2008, Vol. 16 ›› Issue (6): 555-561. DOI: 10.3724/SP.J.1003.2008.08132 cstr: 32101.14.SP.J.1003.2008.08132
潘兆娥, 孙君灵, 王西文( ), 贾银华, 周忠丽, 庞保印, 杜雄明(
), 贾银华, 周忠丽, 庞保印, 杜雄明( )
)
收稿日期:2008-06-11
接受日期:2008-11-07
出版日期:2008-11-20
发布日期:2008-11-20
通讯作者:
杜雄明
基金资助:
Zhao-E Pan, Junling Sun, Yinhua Jia, Zhongli Zhou, Baoyin Pang, Xiongming Du( )
)
Received:2008-06-11
Accepted:2008-11-07
Online:2008-11-20
Published:2008-11-20
Contact:
Xiongming Du
About author:* E-mail: duxm@cricaas.com.cn; dxm630723@163.com摘要:
棉花品种间遗传多样性的评价和遗传图谱构建都需要大量多态性的SSR标记。本研究通过构建棉花参比种质(panel), 高效快速地筛选出棉花多态性核心引物。本实验选用12份表型性状和遗传差异较大的棉花种质作为参比种质, 对来自Cotton Microsatellite Database (CMD)网站上 (
潘兆娥, 孙君灵, 王西文, 贾银华, 周忠丽, 庞保印, 杜雄明 (2008) 基于棉花参比种质的SSR多态性核心引物筛选. 生物多样性, 16, 555-561. DOI: 10.3724/SP.J.1003.2008.08132.
Zhao-E Pan, Junling Sun, Yinhua Jia, Zhongli Zhou, Baoyin Pang, Xiongming Du (2008) Screening of SSR core primers with polymorphism on a cotton panel. Biodiversity Science, 16, 555-561. DOI: 10.3724/SP.J.1003.2008.08132.
| 参比种质 Accession of panel | 染色体组Genome | 种质类型 Biological status of accession | 特征及描述 Characters and description | |||||
|---|---|---|---|---|---|---|---|---|
| 1 常抗棉 Changkang mian | (AD)1 | 陆地棉 Gossypium hirsutum | 抗黄萎病 Verticillium wilt resistance | |||||
| 2 南丹巴地大花 Nandan badidahua | (AD)1 | 陆地棉 G. hirsutum | 产量低, 品质差 Low yield and poor fiber quality | |||||
| 3 徐142无絮 Xu 142 lintless-fuzless | (AD)1 | 陆地棉 G. hirsutum | 无绒无絮 Lintless and fuzless | |||||
| 4 TM-1 | (AD)1 | 陆地棉 G. hirsutum | 通用陆地棉标准系 Common standard lines of G. hirsutum | |||||
| 5 棕2-63 Zong2-63 | (AD)1 | 陆地棉 G. hirsutum | 棕色纤维 Brown fiber | |||||
| 6 海7124 Hai7124 | (AD)2 | 海岛棉 G. barbadense | 中国海岛棉标准系 Standard lines of G. barbadense in China | |||||
| 7 3-79 | (AD)2 | 海岛棉 G. barbadense | 通用海岛棉标准系 Common standard lines of G. barbadense | |||||
| 8 雷蒙地棉 Gossypium raimondii | D5 | 野生棉 Wild species | D5染色体组 D5 genome | |||||
| 9 达尔文氏棉 G. darwinii | (AD)4 | 野生棉 Wild species | 四倍体野生棉(AD)4Wild species of tetraploid with (AD)4genome | |||||
| 10 石系亚1号 Shixiya No.1 | A2 | 中国亚洲棉标准系 Standard lines of G. arboretum in China | ||||||
| 11 DPL972 | A2 | 亚洲棉 G. arboretum | 光子 Naked seed | |||||
| 12 高台草棉 Gaotai caomian | A1 | 草棉 G. herbaceum | 极早熟, 耐高温、干旱 Very early mature, tolerance to high temperature and draught | |||||
表1 12份参比种质及相关信息
Table 1 Twelve panel accessions and their relative information
| 参比种质 Accession of panel | 染色体组Genome | 种质类型 Biological status of accession | 特征及描述 Characters and description | |||||
|---|---|---|---|---|---|---|---|---|
| 1 常抗棉 Changkang mian | (AD)1 | 陆地棉 Gossypium hirsutum | 抗黄萎病 Verticillium wilt resistance | |||||
| 2 南丹巴地大花 Nandan badidahua | (AD)1 | 陆地棉 G. hirsutum | 产量低, 品质差 Low yield and poor fiber quality | |||||
| 3 徐142无絮 Xu 142 lintless-fuzless | (AD)1 | 陆地棉 G. hirsutum | 无绒无絮 Lintless and fuzless | |||||
| 4 TM-1 | (AD)1 | 陆地棉 G. hirsutum | 通用陆地棉标准系 Common standard lines of G. hirsutum | |||||
| 5 棕2-63 Zong2-63 | (AD)1 | 陆地棉 G. hirsutum | 棕色纤维 Brown fiber | |||||
| 6 海7124 Hai7124 | (AD)2 | 海岛棉 G. barbadense | 中国海岛棉标准系 Standard lines of G. barbadense in China | |||||
| 7 3-79 | (AD)2 | 海岛棉 G. barbadense | 通用海岛棉标准系 Common standard lines of G. barbadense | |||||
| 8 雷蒙地棉 Gossypium raimondii | D5 | 野生棉 Wild species | D5染色体组 D5 genome | |||||
| 9 达尔文氏棉 G. darwinii | (AD)4 | 野生棉 Wild species | 四倍体野生棉(AD)4Wild species of tetraploid with (AD)4genome | |||||
| 10 石系亚1号 Shixiya No.1 | A2 | 中国亚洲棉标准系 Standard lines of G. arboretum in China | ||||||
| 11 DPL972 | A2 | 亚洲棉 G. arboretum | 光子 Naked seed | |||||
| 12 高台草棉 Gaotai caomian | A1 | 草棉 G. herbaceum | 极早熟, 耐高温、干旱 Very early mature, tolerance to high temperature and draught | |||||
| 引物类别 Type of primer | 引物数目 No. of primer | 核心多态性引物数 No. of core polymorphic primers | 陆地棉间多态性引物数 No. of polymorphic primers among upland lines | 定位在染色体或连锁群上的多态性引物数 No. of polymorphic primers located on chromosome or linkage group | 引物序列贡献者 Primer sequence contributors |
|---|---|---|---|---|---|
| BNL | 690 | 46 | 46 | 43 | Ben Burr, Michael Blewitt and Eileen Matz (USA) |
| CIR | 392 | 11 | 7 | 11 | Trung-Bieu Nguyen, Marc Giband, Ange-Marie Risterucci, Jean-Marc Lacape (France) |
| CM | 53 | 5 | 4 | 0 | James P. Connell, Muhammad J. Iqbal, Tim Huizinga, Avutu S. Reddy, Suyata Pammi (USA) |
| MGHES | 84 | 10 | 9 | 5 | Samina N. Qureshi, Sukumar Saha, Ramesh V. Kantety (USA) |
| MUCS | 624 | 5 | 3 | 4 | Mauricio Ulloa, Young-Hoon Park, Magdy S. Alabady, Brad Sickler, Thea A. Wilkins (USA) |
| MUSS | 554 | 7 | 6 | 2 | Mauricio Ulloa, Young-Hoon Park, Magdy S. Alabady, Brad Sickler, Thea A. Wilkins (USA) |
| NAU | 2,033 | 129 | 118 | 106 | Tianzhen Zhang, Wangzhen Guo (China) |
| HAU | 20 | 0 | 0 | 0 | Xianlong Zhang, Zhongxu Lin (China) |
| DPL | 200 | 27 | 23 | 26 | David Fang (USA) |
| GH | 200 | 21 | 16 | 0 | Steven M. Hoffman, Daniel Grum, Alan E. Pepper, John Z. Yu, Russel J. Kohel, Jin-Hua Xiao (USA) |
| JESPR | 309 | 17 | 14 | 12 | O. Umesh K. Reddy, Alan E. Pepper, Ibrokhim Abdurakhmonov et al. (USA) |
| TMB | 750 | 36 | 28 | 19 | John Yu, Russell J. Kohel, Jianmin Dong (USA) |
| CSHES | 5 | 5 | 3 | 0 | Tianzhen Zhang, Wangzhen Guo (China) |
| TOTAL | 5,914 | 319 | 277 | 228 |
表2 本实验所用的CMD(http://www.cottonmarker.org/)引物及在参比种质中的多态性信息
Table 2 CMD (http://www.cottonmarker.org/) primers in the experiment and their polymorphism information in the panel
| 引物类别 Type of primer | 引物数目 No. of primer | 核心多态性引物数 No. of core polymorphic primers | 陆地棉间多态性引物数 No. of polymorphic primers among upland lines | 定位在染色体或连锁群上的多态性引物数 No. of polymorphic primers located on chromosome or linkage group | 引物序列贡献者 Primer sequence contributors |
|---|---|---|---|---|---|
| BNL | 690 | 46 | 46 | 43 | Ben Burr, Michael Blewitt and Eileen Matz (USA) |
| CIR | 392 | 11 | 7 | 11 | Trung-Bieu Nguyen, Marc Giband, Ange-Marie Risterucci, Jean-Marc Lacape (France) |
| CM | 53 | 5 | 4 | 0 | James P. Connell, Muhammad J. Iqbal, Tim Huizinga, Avutu S. Reddy, Suyata Pammi (USA) |
| MGHES | 84 | 10 | 9 | 5 | Samina N. Qureshi, Sukumar Saha, Ramesh V. Kantety (USA) |
| MUCS | 624 | 5 | 3 | 4 | Mauricio Ulloa, Young-Hoon Park, Magdy S. Alabady, Brad Sickler, Thea A. Wilkins (USA) |
| MUSS | 554 | 7 | 6 | 2 | Mauricio Ulloa, Young-Hoon Park, Magdy S. Alabady, Brad Sickler, Thea A. Wilkins (USA) |
| NAU | 2,033 | 129 | 118 | 106 | Tianzhen Zhang, Wangzhen Guo (China) |
| HAU | 20 | 0 | 0 | 0 | Xianlong Zhang, Zhongxu Lin (China) |
| DPL | 200 | 27 | 23 | 26 | David Fang (USA) |
| GH | 200 | 21 | 16 | 0 | Steven M. Hoffman, Daniel Grum, Alan E. Pepper, John Z. Yu, Russel J. Kohel, Jin-Hua Xiao (USA) |
| JESPR | 309 | 17 | 14 | 12 | O. Umesh K. Reddy, Alan E. Pepper, Ibrokhim Abdurakhmonov et al. (USA) |
| TMB | 750 | 36 | 28 | 19 | John Yu, Russell J. Kohel, Jianmin Dong (USA) |
| CSHES | 5 | 5 | 3 | 0 | Tianzhen Zhang, Wangzhen Guo (China) |
| TOTAL | 5,914 | 319 | 277 | 228 |
| 引物名称 Primer | 所属染色体或 连锁群 Chromosome or linkage group | 退火温度 Annealing temperature (Tm) | 参比种质 Panel | 陆地棉 | 其他 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Panel等位基因 多态信息含量 Allelic polymorphism information content (PIC) | Panel基因型 多样性(H') Genotype polymorphism | Panel有效等 位基因数(Ne) Effective number of alleles | Panel等位 基因数 No. of allelic loci | Panel多态性 等位基因 No. of polymorphic allelic | 等位 基因数 No. of allelic loci in upland lines | 多态性 等位基因数 No. of polymorphic allelic loci in upland lines | 棉种 等位基因数 No. of allelic loci among other species | 棉种多态性 等位基因数 No. of polymorphic allelic loci among other species | |||
| BNL1015 | 57 | 0.837 | 1.945 | 6.125 | 8 | 7 | 7 | 6 | 6 | 5 | |
| BNL1026 | 54 | 0.861 | 2.117 | 7.209 | 10 | 10 | 7 | 4 | 7 | 7 | |
| BNL1034 | 11,17,21 | 54 | 0.826 | 1.900 | 5.740 | 8 | 8 | 5 | 4 | 8 | 8 |
| BNL1040 | 13,18 | 56 | 0.889 | 2.349 | 9.000 | 13 | 13 | 8 | 6 | 10 | 10 |
| BNL1053 | 5,21,LGA03 | 58 | 0.847 | 2.036 | 6.553 | 10 | 10 | 7 | 5 | 6 | 6 |
| BNL119 | 20 | 56 | 0.789 | 1.700 | 4.737 | 7 | 7 | 5 | 4 | 5 | 5 |
| BNL1227 | 12,26 | 58 | 0.844 | 2.088 | 6.422 | 11 | 11 | 6 | 5 | 7 | 7 |
| BNL1317 | 9,23 | 53 | 0.845 | 1.960 | 6.451 | 8 | 8 | 6 | 5 | 5 | 5 |
| BNL1421 | 13,18 | 55 | 0.894 | 2.389 | 9.397 | 14 | 14 | 7 | 3 | 14 | 14 |
| BNL1440 | 6,8,25 | 55 | 0.811 | 1.806 | 5.297 | 7 | 7 | 5 | 4 | 5 | 5 |
| BNL1495 | 13,18 | 53 | 0.806 | 1.694 | 5.149 | 5 | 5 | 4 | 4 | 5 | 5 |
| BNL1513 | 24 | 56 | 0.842 | 1.998 | 6.330 | 9 | 9 | 8 | 7 | 6 | 6 |
| BNL1517 | 25 | 56 | 0.882 | 2.315 | 8.442 | 12 | 12 | 8 | 7 | 6 | 6 |
| BNL1580 | 3,21 | 57 | 0.843 | 1.952 | 6.373 | 8 | 8 | 4 | 4 | 4 | 4 |
| BNL1604 | 7,16 | 57 | 0.813 | 1.777 | 5.342 | 7 | 7 | 3 | 2 | 5 | 5 |
| BNL1669 | 9,26 | 55 | 0.782 | 1.604 | 4.588 | 6 | 5 | 5 | 2 | 5 | 4 |
| BNL1694 | 7,16 | 55 | 0.879 | 2.200 | 8.291 | 10 | 10 | 6 | 5 | 7 | 7 |
| BNL193 | 12,18 | 57 | 0.818 | 1.946 | 5.497 | 9 | 9 | 6 | 6 | 4 | 4 |
| BNL226 | 3,14 | 57 | 0.871 | 2.161 | 7.734 | 10 | 10 | 5 | 2 | 8 | 8 |
| BNL243 | 18 | 57 | 0.864 | 2.175 | 7.364 | 11 | 11 | 6 | 3 | 7 | 7 |
| BNL2449 | 10,13 | 55 | 0.883 | 2.260 | 8.583 | 12 | 12 | 9 | 7 | 11 | 11 |
| BNL2485 | 56 | 0.810 | 1.754 | 5.263 | 7 | 7 | 5 | 2 | 6 | 6 | |
| BNL252 | 9,19,24 | 55 | 0.857 | 2.138 | 7.014 | 11 | 11 | 7 | 7 | 9 | 9 |
| BNL2590 | 9,23 | 55 | 0.878 | 2.191 | 8.198 | 10 | 10 | 6 | 4 | 6 | 6 |
| BNL2646 | 7,9,15 | 55 | 0.715 | 1.480 | 3.512 | 6 | 6 | 4 | 3 | 3 | 3 |
| BNL3033 | 15 | 55 | 0.749 | 1.580 | 3.986 | 6 | 5 | 5 | 4 | 3 | 2 |
| BNL3048 | 17 | 55 | 0.848 | 2.094 | 6.596 | 11 | 11 | 6 | 4 | 10 | 10 |
| BNL3140 | 23 | 55 | 0.797 | 1.761 | 4.923 | 7 | 7 | 6 | 4 | 4 | 4 |
| BNL3261 | 12 | 56 | 0.860 | 2.087 | 7.161 | 10 | 8 | 8 | 3 | 7 | 5 |
| BNL3383 | 23,26,LGD09 | 56 | 0.893 | 2.341 | 9.332 | 12 | 12 | 12 | 8 | 8 | 8 |
| BNL3474 | 8,9,24,26 | 53 | 0.852 | 2.065 | 6.744 | 10 | 10 | 6 | 4 | 8 | 8 |
| BNL3502 | 14 | 55 | 0.837 | 1.878 | 6.128 | 7 | 7 | 4 | 4 | 4 | 4 |
| BNL358 | 22 | 53 | 0.917 | 2.569 | 11.985 | 15 | 15 | 9 | 1 | 12 | 12 |
| BNL3590 | 2,8,17 | 57 | 0.864 | 2.143 | 7.360 | 11 | 10 | 8 | 4 | 8 | 7 |
| BNL3650 | 6,LGA06 | 53 | 0.881 | 2.224 | 8.430 | 11 | 11 | 8 | 4 | 8 | 7 |
| BNL3806 | 25,LGD06,LGA06 | 55 | 0.789 | 1.757 | 4.741 | 7 | 6 | 4 | 2 | 4 | 3 |
| BNL3976 | 5,21 | 57 | 0.771 | 1.601 | 4.359 | 6 | 5 | 6 | 4 | 4 | 3 |
| BNL4030 | 5,22 | 57 | 0.792 | 1.659 | 4.804 | 6 | 6 | 4 | 2 | 6 | 6 |
| BNL448 | 22 | 57 | 0.863 | 2.107 | 7.283 | 10 | 8 | 8 | 1 | 8 | 6 |
| BNL511 | 9 | 57 | 0.893 | 2.317 | 9.308 | 12 | 12 | 9 | 8 | 10 | 10 |
| BNL530 | 4 | 57 | 0.904 | 2.433 | 10.464 | 13 | 13 | 9 | 4 | 13 | 13 |
| BNL542 | 5,21 | 53 | 0.874 | 2.163 | 7.949 | 10 | 10 | 5 | 4 | 9 | 9 |
| BNL827 | 25 | 57 | 0.899 | 2.341 | 9.936 | 11 | 11 | 10 | 8 | 9 | 9 |
| BNL830 | 15 | 53 | 0.714 | 1.375 | 3.495 | 5 | 5 | 4 | 2 | 3 | 3 |
| BNL834 | 17 | 55 | 0.893 | 2.348 | 9.321 | 12 | 12 | 8 | 4 | 6 | 6 |
| BNL946 | 20 | 56 | 0.896 | 2.377 | 9.624 | 12 | 12 | 12 | 7 | 5 | 4 |
| CIR139 | 19 | 51 | 0.647 | 1.203 | 2.829 | 4 | 3 | 2 | 0 | 3 | 2 |
| CIR167 | 6,26 | 50 | 0.838 | 1.956 | 6.171 | 8 | 8 | 3 | 0 | 7 | 7 |
| CIR246 | 14 | 51 | 0.850 | 2.004 | 6.651 | 9 | 9 | 6 | 5 | 8 | 8 |
| CIR249 | 4 | 51 | 0.859 | 2.012 | 7.111 | 8 | 8 | 6 | 6 | 7 | 7 |
| CIR307 | 15,LGD01 | 51 | 0.774 | 1.590 | 4.430 | 6 | 6 | 4 | 2 | 5 | 5 |
| CIR32 | 26 | 51 | 0.722 | 1.410 | 3.596 | 5 | 5 | 3 | 0 | 4 | 4 |
| CIR328 | 5 | 51 | 0.827 | 2.011 | 5.780 | 10 | 10 | 6 | 5 | 9 | 9 |
| CIR332 | 3,LGA03 | 51 | 0.893 | 2.345 | 9.368 | 12 | 12 | 6 | 2 | 12 | 12 |
| CIR353 | 9 | 53 | 0.874 | 2.235 | 7.954 | 12 | 12 | 8 | 5 | 8 | 8 |
| CIR62 | 5,19 | 51 | 0.824 | 1.934 | 5.694 | 9 | 8 | 5 | 2 | 6 | 5 |
| CIR78 | 26 | 50 | 0.808 | 1.768 | 5.213 | 7 | 5 | 4 | 0 | 6 | 4 |
| CM162 | 55 | 0.895 | 2.423 | 9.564 | 14 | 14 | 9 | 5 | 9 | 9 | |
| CM45 | 56 | 0.746 | 1.564 | 3.930 | 6 | 6 | 4 | 3 | 4 | 4 | |
| CM51 | 58 | 0.816 | 1.777 | 5.442 | 7 | 7 | 4 | 0 | 6 | 6 | |
| CM61 | 57 | 0.792 | 1.667 | 4.813 | 6 | 6 | 3 | 3 | 3 | 3 | |
| CM67 | 57 | 0.819 | 1.752 | 5.538 | 6 | 6 | 3 | 2 | 5 | 5 | |
| CSHES29 | 55 | 0.724 | 1.494 | 3.629 | 7 | 6 | 3 | 0 | 7 | 6 | |
| CSHES32 | 53 | 0.719 | 1.391 | 3.564 | 5 | 4 | 3 | 0 | 4 | 4 | |
| CSHES51 | 57 | 0.840 | 1.888 | 6.259 | 7 | 7 | 4 | 3 | 4 | 4 | |
| CSHES62 | 55 | 0.753 | 1.654 | 4.041 | 7 | 6 | 7 | 6 | 3 | 2 | |
| CSHES82 | 53 | 0.726 | 1.410 | 3.645 | 5 | 5 | 4 | 3 | 4 | 4 | |
| DPL209 | 11 | 59 | 0.897 | 2.362 | 9.672 | 13 | 13 | 12 | 9 | 9 | 9 |
| DPL216 | 2 | 58 | 0.801 | 1.685 | 5.026 | 6 | 4 | 0 | 6 | 6 | |
| DPL220 | 8 | 58 | 0.881 | 2.382 | 8.399 | 14 | 13 | 10 | 8 | 9 | 8 |
| DPL225 | 20 | 58 | 0.853 | 1.982 | 6.782 | 8 | 8 | 6 | 2 | 6 | 6 |
| DPL234 | 58 | 0.872 | 2.160 | 7.803 | 10 | 10 | 7 | 7 | 8 | 8 | |
| DPL238 | 6 | 59 | 0.858 | 2.094 | 7.043 | 10 | 10 | 5 | 2 | 9 | 9 |
| DPL249 | 18 | 58 | 0.852 | 1.975 | 6.768 | 8 | 8 | 5 | 4 | 6 | 6 |
| DPL253 | 11 | 58 | 0.759 | 1.558 | 4.149 | 6 | 4 | 5 | 1 | 3 | 1 |
| DPL262 | 23 | 58 | 0.811 | 1.832 | 5.303 | 8 | 8 | 5 | 2 | 7 | 7 |
| DPL283 | 16 | 58 | 0.728 | 1.642 | 3.674 | 8 | 7 | 2 | 0 | 7 | 6 |
| DPL300 | 15 | 58 | 0.773 | 1.616 | 4.412 | 6 | 6 | 5 | 4 | 4 | 4 |
| DPL325 | 11 | 58 | 0.790 | 1.659 | 4.765 | 6 | 6 | 3 | 2 | 4 | 4 |
| DPL354 | 17 | 59 | 0.864 | 2.200 | 7.339 | 11 | 11 | 7 | 6 | 6 | 6 |
| DPL431 | 10 | 57 | 0.735 | 1.433 | 3.769 | 5 | 5 | 4 | 4 | 2 | 2 |
| DPL46 | 2 | 58 | 0.858 | 2.114 | 7.053 | 10 | 10 | 6 | 2 | 9 | 9 |
| DPL461 | 24 | 58 | 0.826 | 1.885 | 5.762 | 8 | 8 | 4 | 4 | 4 | 4 |
| DPL511 | 16 | 58 | 0.867 | 2.162 | 7.529 | 11 | 11 | 7 | 4 | 7 | 7 |
| DPL513 | 1 | 59 | 0.816 | 1.881 | 5.444 | 9 | 9 | 6 | 4 | 5 | 4 |
| DPL519 | 25 | 58 | 0.722 | 1.570 | 3.600 | 7 | 7 | 2 | 0 | 6 | 6 |
| DPL528 | 21 | 58 | 0.905 | 2.416 | 10.492 | 12 | 12 | 8 | 6 | 10 | 10 |
| DPL570 | 11 | 58 | 0.851 | 2.032 | 6.714 | 9 | 9 | 6 | 2 | 9 | 9 |
| DPL681 | 6 | 57 | 0.727 | 1.559 | 3.658 | 7 | 7 | 4 | 3 | 3 | 2 |
| DPL69 | 26 | 60 | 0.692 | 1.125 | 3.245 | 4 | 4 | 3 | 2 | 3 | 3 |
| DPL71 | 19 | 58 | 0.707 | 1.484 | 3.413 | 6 | 6 | 4 | 3 | 4 | 4 |
| DPL73 | 14 | 59 | 0.844 | 1.984 | 6.408 | 9 | 8 | 6 | 0 | 7 | 6 |
| DPL799 | 25 | 59 | 0.807 | 1.915 | 5.172 | 9 | 9 | 6 | 5 | 4 | 4 |
| DPL910 | 18 | 56 | 0.870 | 2.250 | 7.704 | 12 | 12 | 6 | 6 | 8 | 8 |
| GH110 | 56 | 0.881 | 2.251 | 8.395 | 11 | 11 | 7 | 7 | 4 | 4 | |
| GH111 | 56 | 0.877 | 2.284 | 8.133 | 13 | 13 | 7 | 6 | 12 | 12 | |
| GH112 | 58 | 0.873 | 2.217 | 7.903 | 11 | 11 | 6 | 5 | 11 | 11 | |
| GH132 | 58 | 0.852 | 1.995 | 6.744 | 8 | 8 | 4 | 0 | 6 | 6 | |
| GH158 | 58 | 0.837 | 2.008 | 6.146 | 9 | 9 | 5 | 2 | 9 | 9 | |
| GH220 | 58 | 0.675 | 1.333 | 3.077 | 5 | 5 | 3 | 2 | 4 | 4 | |
| GH243 | 60 | 0.764 | 1.608 | 4.233 | 6 | 5 | 3 | 0 | 5 | 4 | |
| GH247 | 60 | 0.886 | 2.233 | 8.779 | 10 | 10 | 9 | 7 | 8 | 8 | |
| GH268 | 60 | 0.790 | 1.651 | 4.766 | 6 | 6 | 2 | 0 | 5 | 5 | |
| GH273 | 57 | 0.648 | 1.072 | 2.844 | 3 | 3 | 3 | 3 | 3 | 3 | |
| GH277 | 58 | 0.659 | 1.313 | 2.935 | 5 | 5 | 5 | 5 | 2 | 1 | |
| GH32 | 60 | 0.798 | 1.698 | 4.962 | 6 | 6 | 3 | 0 | 4 | 4 | |
| GH34 | 58 | 0.780 | 1.685 | 4.546 | 7 | 7 | 5 | 3 | 5 | ||
| GH447 | 58 | 0.819 | 1.849 | 5.528 | 8 | 8 | 7 | 6 | 5 | 5 | |
| GH532 | 56 | 0.870 | 2.206 | 7.718 | 11 | 10 | 9 | 7 | 7 | 6 | |
| GH56 | 55 | 0.823 | 1.986 | 5.635 | 10 | 9 | 7 | 4 | 4 | 3 | |
| GH59 | 55 | 0.901 | 2.394 | 10.117 | 12 | 12 | 6 | 1 | 10 | 10 | |
| GH60 | 57 | 0.776 | 1.550 | 4.455 | 5 | 5 | 2 | 0 | 5 | 5 | |
| GH70 | 55 | 0.870 | 2.107 | 7.716 | 9 | 9 | 5 | 2 | 9 | 8 | |
| GH72 | 57 | 0.828 | 1.964 | 5.797 | 9 | 9 | 6 | 4 | 5 | 5 | |
| GH75 | 56 | 0.855 | 2.077 | 6.914 | 10 | 10 | 7 | 7 | 6 | 6 | |
| JESPR101 | 2,8 | 56 | 0.817 | 1.837 | 5.454 | 8 | 7 | 4 | 3 | 6 | 5 |
| JESPR102 | 15,16 | 55 | 0.758 | 1.466 | 4.129 | 5 | 5 | 4 | 3 | 5 | 5 |
| JESPR114 | 23,LGD09 | 58 | 0.842 | 2.070 | 6.333 | 11 | 11 | 9 | 8 | 5 | 5 |
| JESPR133 | 55 | 0.882 | 2.089 | 8.471 | 10 | 10 | 7 | 3 | 8 | 8 | |
| JESPR138 | 55 | 0.716 | 1.427 | 3.522 | 5 | 4 | 2 | 0 | 5 | 4 | |
| JESPR156 | 14,21 | 58 | 0.780 | 2.324 | 4.540 | 13 | 13 | 10 | 7 | 8 | 8 |
| JESPR158 | 21,LGA03 | 57 | 0.891 | 2.298 | 9.195 | 11 | 11 | 7 | 4 | 5 | 5 |
| JESPR204 | 5,13,18,19 | 57 | 0.887 | 2.311 | 8.828 | 12 | 12 | 9 | 6 | 6 | 6 |
| JESPR208 | 9,23 | 54 | 0.885 | 2.264 | 8.699 | 11 | 11 | 6 | 3 | 10 | 10 |
| JESPR222 | 58 | 0.810 | 1.794 | 5.261 | 7 | 7 | 3 | 3 | 7 | 7 | |
| JESPR227 | 25 | 57 | 0.801 | 1.868 | 5.031 | 9 | 9 | 5 | 2 | 7 | 7 |
| JESPR231 | 3,14 | 57 | 0.867 | 2.092 | 7.506 | 9 | 9 | 6 | 4 | 5 | 5 |
| JESPR247 | 6 | 55 | 0.768 | 1.661 | 4.319 | 8 | 8 | 5 | 2 | 6 | 6 |
| JESPR274 | 9,23,24 | 61 | 0.753 | 1.480 | 4.050 | 5 | 5 | 3 | 1 | 4 | 4 |
| JESPR42 | 57 | 0.810 | 1.772 | 5.261 | 7 | 7 | 3 | 2 | 5 | 5 | |
| JESPR45 | 57 | 0.610 | 1.110 | 2.564 | 4 | 4 | 2 | 0 | 3 | 3 | |
| JESPR50 | 5,LGA04,LGD06 | 56 | 0.873 | 2.113 | 7.881 | 9 | 9 | 4 | 0 | 8 | 8 |
| MGHES08 | 57 | 0.817 | 1.779 | 5.473 | 6 | 5 | 6 | 1 | 6 | 6 | |
| MGHES16 | 21,LGA03 | 55 | 0.812 | 1.857 | 5.315 | 9 | 7 | 5 | 1 | 8 | 6 |
| MGHES18 | 58 | 0.846 | 1.522 | 6.480 | 7 | 7 | 4 | 1 | 6 | 6 | |
| MGHES38 | LGA03 | 54 | 0.743 | 2.281 | 3.890 | 13 | 13 | 6 | 2 | 10 | 10 |
| MGHES40 | 56 | 0.766 | 1.700 | 4.269 | 7 | 6 | 7 | 6 | 3 | 2 | |
| MGHES41 | 55 | 0.794 | 2.081 | 4.861 | 8 | 8 | 3 | 2 | 8 | 8 | |
| MGHES46 | 56 | 0.842 | 2.008 | 6.345 | 10 | 10 | 5 | 0 | 8 | 8 | |
| MGHES58 | 7 | 57 | 0.870 | 2.255 | 7.697 | 13 | 13 | 5 | 2 | 11 | 11 |
| MGHES70 | 9 | 57 | 0.907 | 2.429 | 10.777 | 12 | 12 | 10 | 9 | 8 | 8 |
| MGHES73 | 9 | 60 | 0.819 | 1.893 | 5.528 | 8 | 8 | 4 | 3 | 8 | 8 |
| MUCS101 | 4 | 57 | 0.762 | 1.572 | 4.197 | 6 | 6 | 6 | 5 | 4 | 3 |
| MUCS127 | 3 | 55 | 0.717 | 2.101 | 3.531 | 10 | 9 | 10 | 9 | 5 | 4 |
| MUCS347 | 21 | 57 | 0.834 | 1.982 | 6.025 | 9 | 9 | 3 | 0 | 8 | 7 |
| MUCS466 | 58 | 0.779 | 1.552 | 4.521 | 5 | 4 | 4 | 0 | 5 | 4 | |
| MUCS530 | 11 | 58 | 0.735 | 1.442 | 3.769 | 5 | 4 | 5 | 2 | 5 | 4 |
| MUSS130 | 60 | 0.856 | 1.996 | 6.928 | 8 | 7 | 6 | 0 | 8 | 7 | |
| MUSS138 | 57 | 0.859 | 2.051 | 7.067 | 9 | 9 | 7 | 3 | 8 | 8 | |
| MUSS422 | 10 | 56 | 0.860 | 2.064 | 7.141 | 9 | 9 | 5 | 4 | 9 | 9 |
| MUSS440 | 15 | 58 | 0.900 | 2.375 | 9.979 | 12 | 12 | 9 | 9 | 8 | 7 |
| MUSS46 | 58 | 0.771 | 1.572 | 4.360 | 6 | 4 | 5 | 1 | 4 | 2 | |
| MUSS59 | 57 | 0.859 | 2.005 | 7.084 | 8 | 8 | 7 | 5 | 8 | 8 | |
| MUSS65 | 57 | 0.837 | 1.936 | 6.149 | 8 | 6 | 8 | 6 | 6 | 4 | |
| NAU1014 | 9,11 | 53 | 0.886 | 2.301 | 8.744 | 12 | 12 | 5 | 2 | 12 | 12 |
| NAU1015 | 5 | 53 | 0.852 | 2.141 | 6.748 | 12 | 12 | 3 | 0 | 12 | 12 |
| NAU1017 | 8,24 | 55 | 0.826 | 1.834 | 5.762 | 7 | 70 | 4 | 3 | 7 | 7 |
| NAU1027 | 6,19 | 53 | 0.845 | 2.149 | 6.450 | 11 | 11 | 5 | 4 | 11 | 11 |
| NAU1028 | 17,23 | 55 | 0.895 | 2.387 | 9.483 | 13 | 13 | 7 | 4 | 12 | 12 |
| NAU1042 | 5,19 | 55 | 0.783 | 1.709 | 4.605 | 7 | 7 | 4 | 3 | 7 | 7 |
| NAU1043 | 7,LGA07 | 55 | 0.881 | 2.193 | 8.400 | 10 | 10 | 8 | 6 | 10 | 9 |
| NAU1048 | 7,11 | 53 | 0.832 | 1.898 | 5.938 | 8 | 7 | 6 | 4 | 8 | 6 |
| NAU1053 | 7 | 53 | 0.850 | 1.997 | 6.651 | 9 | 9 | 6 | 2 | 9 | 9 |
| NAU1070 | 3,9 | 55 | 0.897 | 2.368 | 9.665 | 12 | 12 | 8 | 7 | 11 | 11 |
| NAU1085 | 55 | 0.882 | 2.227 | 8.495 | 11 | 11 | 7 | 6 | 10 | 10 | |
| NAU1093 | LGA06 | 57 | 0.896 | 2.371 | 9.656 | 12 | 12 | 8 | 5 | 9 | 9 |
| NAU1102 | LGD05 | 53 | 0.893 | 2.299 | 9.333 | 11 | 11 | 9 | 2 | 10 | 10 |
| NAU1103 | 57 | 0.815 | 1.941 | 5.402 | 9 | 8 | 9 | 8 | 3 | 2 | |
| NAU1125 | 19 | 53 | 0.820 | 1.846 | 5.558 | 8 | 8 | 5 | 4 | 7 | 7 |
| NAU1155 | 55 | 0.763 | 1.586 | 4.212 | 7 | 7 | 7 | 6 | 3 | 1 | |
| NAU1167 | 3,8,17,LGA03 | 55 | 0.880 | 2.154 | 8.345 | 9 | 9 | 9 | 9 | 8 | 8 |
| NAU1169 | 3,10,20 | 53 | 0.856 | 2.041 | 6.942 | 9 | 9 | 7 | 5 | 7 | 7 |
| NAU1186 | 55 | 0.803 | 1.728 | 5.085 | 7 | 7 | 7 | 6 | 4 | 4 | |
| NAU1187 | LGD05 | 55 | 0.794 | 1.684 | 4.861 | 6 | 6 | 5 | 4 | 4 | 4 |
| NAU1190 | 3,16,LGA03 | 53 | 0.863 | 2.066 | 7.316 | 9 | 9 | 5 | 3 | 7 | 7 |
| NAU1196 | 55 | 0.859 | 2.075 | 7.078 | 9 | 9 | 7 | 6 | 5 | 5 | |
| NAU1200 | LGA05 | 55 | 0.848 | 1.984 | 6.568 | 8 | 8 | 7 | 7 | 6 | 6 |
| NAU1201 | 13,14 | 55 | 0.710 | 1.398 | 3.447 | 5 | 5 | 2 | 2 | 5 | 5 |
| NAU1222 | 7,20,23 | 55 | 0.667 | 1.242 | 3.000 | 4 | 4 | 1 | 0 | 4 | 4 |
| NAU1225 | 7 | 53 | 0.832 | 1.921 | 5.965 | 8 | 8 | 5 | 4 | 5 | 5 |
| NAU1230 | LGD05 | 55 | 0.812 | 1.793 | 5.321 | 7 | 7 | 5 | 4 | 4 | 4 |
| NAU1233 | 10 | 55 | 0.875 | 2.374 | 8.004 | 14 | 13 | 10 | 9 | 8 | 7 |
| NAU1248 | 3,20 | 55 | 0.815 | 1.782 | 5.411 | 7 | 7 | 4 | 0 | 6 | 6 |
| NAU1255 | LGD05 | 55 | 0.829 | 1.852 | 5.841 | 7 | 7 | 5 | 4 | 4 | 4 |
| NAU1269 | LGD05 | 55 | 0.864 | 2.082 | 7.364 | 9 | 9 | 6 | 6 | 5 | 5 |
| NAU1273 | 53 | 0.830 | 1.781 | 5.880 | 6 | 6 | 6 | 2 | 5 | 4 | |
| NAU1302 | 3,5,24 | 51 | 0.904 | 2.459 | 10.421 | 13 | 12 | 10 | 8 | 8 | 7 |
| NAU1322 | 24 | 55 | 0.891 | 2.419 | 9.188 | 14 | 14 | 11 | 11 | 8 | 8 |
| NAU1336 | 8,19 | 57 | 0.830 | 1.847 | 5.882 | 7 | 7 | 4 | 0 | 7 | 7 |
| NAU1356 | 52 | 0.874 | 2.203 | 7.934 | 12 | 11 | 7 | 0 | 11 | 8 | |
| NAU1362 | 7 | 55 | 0.901 | 2.459 | 10.140 | 14 | 14 | 8 | 5 | 7 | 7 |
| NAU1366 | 3,5,21 | 55 | 0.902 | 2.458 | 10.240 | 14 | 14 | 11 | 7 | 11 | 11 |
| NAU1369 | 8,9,24 | 53 | 0.770 | 1.678 | 4.356 | 7 | 7 | 5 | 4 | 4 | 4 |
| NAU1379 | 55 | 0.860 | 2.051 | 7.165 | 9 | 9 | 7 | 5 | 8 | 8 | |
| NAU2026 | 12,LGD04 | 55 | 0.881 | 2.186 | 8.395 | 10 | 10 | 8 | 5 | 8 | 8 |
| NAU2083 | 1 | 57 | 0.837 | 1.977 | 6.125 | 9 | 9 | 7 | 6 | 3 | 3 |
| NAU2108 | 7 | 56 | 0.842 | 1.926 | 6.341 | 8 | 7 | 5 | 0 | 8 | 7 |
| NAU2120 | 22 | 58 | 0.845 | 2.001 | 6.472 | 9 | 9 | 5 | 2 | 8 | 8 |
| NAU2126 | 22 | 59 | 0.840 | 2.026 | 6.231 | 10 | 10 | 7 | 5 | 7 | 7 |
| NAU2139 | 19 | 55 | 0.857 | 2.059 | 7.000 | 9 | 9 | 6 | 4 | 7 | 7 |
| NAU2152 | 11,16,17 | 53 | 0.880 | 2.341 | 8.308 | 14 | 14 | 5 | 2 | 12 | 12 |
| NAU2156 | 6 | 55 | 0.853 | 2.118 | 6.818 | 11 | 11 | 5 | 4 | 9 | 9 |
| NAU2162 | 22 | 55 | 0.885 | 2.246 | 8.668 | 11 | 11 | 8 | 4 | 7 | 7 |
| NAU2173 | 14 | 53 | 0.736 | 1.434 | 3.789 | 5 | 5 | 3 | 1 | 5 | 5 |
| NAU2190 | 14 | 53 | 0.823 | 2.267 | 5.657 | 9 | 9 | 6 | 2 | 5 | 5 |
| NAU2251 | 12,26 | 55 | 0.844 | 1.944 | 6.400 | 8 | 8 | 7 | 6 | 4 | 4 |
| NAU2257 | 11 | 55 | 0.874 | 2.198 | 7.963 | 11 | 11 | 7 | 5 | 8 | 8 |
| NAU2272 | 5,14 | 53 | 0.846 | 2.076 | 6.504 | 10 | 10 | 7 | 5 | 7 | 7 |
| NAU2274 | 19 | 55 | 0.764 | 1.690 | 4.245 | 8 | 8 | 7 | 6 | 5 | 5 |
| NAU2277 | 2 | 57 | 0.847 | 1.961 | 6.541 | 8 | 8 | 5 | 5 | 8 | 8 |
| NAU2308 | 7 | 55 | 0.813 | 1.729 | 5.355 | 6 | 6 | 3 | 0 | 6 | 6 |
| NAU2336 | 14 | 55 | 0.813 | 1.872 | 5.333 | 8 | 8 | 5 | 4 | 6 | 6 |
| NAU2342 | 55 | 0.817 | 1.794 | 5.452 | 7 | 7 | 6 | 5 | 6 | 6 | |
| NAU2343 | 56 | 0.778 | 1.809 | 4.500 | 9 | 8 | 8 | 7 | 4 | 3 | |
| NAU2361 | 21 | 56 | 0.710 | 1.400 | 3.448 | 5 | 5 | 3 | 2 | 3 | 3 |
| NAU2398 | 55 | 0.892 | 2.327 | 9.256 | 12 | 12 | 4 | 0 | 9 | 9 | |
| NAU2494 | 5 | 55 | 0.828 | 1.975 | 5.823 | 10 | 10 | 6 | 3 | 6 | 6 |
| NAU2508 | 53 | 0.785 | 1.573 | 4.655 | 5 | 5 | 5 | 3 | 5 | 5 | |
| NAU2600 | 53 | 0.868 | 2.151 | 7.564 | 10 | 10 | 5 | 4 | 8 | 8 | |
| NAU2604 | 19 | 55 | 0.701 | 1.285 | 3.346 | 4 | 3 | 4 | 1 | 2 | 1 |
| NAU2631 | 24 | 53 | 0.838 | 1.937 | 6.158 | 8 | 8 | 7 | 6 | 6 | 6 |
| NAU2654 | 4 | 53 | 0.885 | 2.295 | 8.674 | 12 | 11 | 9 | 7 | 10 | 9 |
| NAU2679 | 6,25,LGD06 | 57 | 0.908 | 2.498 | 10.813 | 14 | 14 | 6 | 4 | 9 | 9 |
| NAU2741 | 1,19 | 52 | 0.722 | 1.418 | 3.593 | 5 | 4 | 4 | 3 | 4 | 3 |
| NAU2776 | 10,20 | 55 | 0.719 | 1.317 | 3.558 | 4 | 4 | 4 | 3 | 3 | 3 |
| NAU2782 | 22 | 55 | 0.908 | 2.490 | 10.912 | 14 | 14 | 10 | 3 | 7 | 7 |
| NAU2816 | 19 | 55 | 0.812 | 1.721 | 5.319 | 6 | 6 | 5 | 1 | 5 | 5 |
| NAU2820 | 7,16 | 55 | 0.849 | 2.038 | 6.627 | 10 | 10 | 5 | 1 | 10 | 10 |
| NAU2836 | 3,17 | 55 | 0.844 | 1.994 | 6.391 | 9 | 9 | 6 | 3 | 5 | 5 |
| NAU2898 | 17 | 55 | 0.811 | 1.944 | 5.294 | 9 | 9 | 6 | 5 | 4 | 4 |
| NAU3013 | 10 | 57 | 0.799 | 1.733 | 4.971 | 7 | 7 | 4 | 1 | 5 | 5 |
| NAU3018 | 13 | 57 | 0.791 | 1.694 | 4.787 | 7 | 6 | 5 | 2 | 6 | 5 |
| NAU3096 | 5,19 | 53 | 0.768 | 1.611 | 4.304 | 6 | 5 | 6 | 4 | 4 | 3 |
| NAU3100 | 23 | 55 | 0.847 | 2.034 | 6.542 | 10 | 9 | 8 | 4 | 5 | 4 |
| NAU3102 | 52 | 0.507 | 0.953 | 2.027 | 4 | 4 | 4 | 4 | 1 | 0 | |
| NAU3127 | 4 | 57 | 0.739 | 1.448 | 3.835 | 5 | 5 | 3 | 2 | 5 | 5 |
| NAU3212 | 5 | 53 | 0.904 | 2.438 | 10.365 | 13 | 13 | 5 | 3 | 10 | 9 |
| NAU3225 | 14 | 53 | 0.853 | 2.093 | 6.818 | 10 | 10 | 5 | 3 | 6 | 6 |
| NAU3254 | 1 | 53 | 0.828 | 1.891 | 5.806 | 9 | 8 | 9 | 6 | 5 | 4 |
| NAU3305 | 26 | 56 | 0.860 | 2.130 | 7.143 | 10 | 10 | 6 | 5 | 5 | 5 |
| NAU3308 | 14 | 53 | 0.786 | 1.657 | 4.672 | 6 | 5 | 5 | 2 | 4 | 3 |
| NAU3325 | 5 | 56 | 0.785 | 1.732 | 4.661 | 7 | 6 | 5 | 4 | 4 | 3 |
| NAU3414 | 9,23 | 53 | 0.760 | 1.575 | 4.173 | 6 | 4 | 5 | 3 | 4 | 2 |
| NAU3419 | 2 | 57 | 0.852 | 2.021 | 6.737 | 9 | 9 | 6 | 4 | 8 | 8 |
| NAU3427 | 6 | 55 | 0.897 | 2.334 | 9.727 | 11 | 11 | 8 | 4 | 7 | 7 |
| NAU3467 | 10 | 55 | 0.739 | 1.469 | 3.835 | 6 | 5 | 4 | 1 | 5 | 4 |
| NAU3468 | 13 | 53 | 0.860 | 2.095 | 7.153 | 10 | 10 | 9 | 6 | 8 | 8 |
| NAU3499 | 8,19,24 | 53 | 0.862 | 2.187 | 7.251 | 11 | 10 | 7 | 5 | 6 | 5 |
| NAU3942 | 22 | 55 | 0.840 | 1.922 | 6.256 | 8 | 7 | 6 | 2 | 8 | 7 |
| NAU4042 | 19 | 53 | 0.864 | 2.137 | 7.333 | 10 | 10 | 9 | 8 | 9 | 9 |
| NAU4047 | 12 | 55 | 0.540 | 0.898 | 2.174 | 3 | 2 | 3 | 2 | 2 | 1 |
| NAU4057 | 5 | 53 | 0.890 | 2.343 | 9.109 | 13 | 13 | 11 | 8 | 8 | 8 |
| NAU4855 | 21 | 55 | 0.907 | 2.518 | 10.762 | 15 | 15 | 7 | 3 | 12 | 12 |
| NAU5013 | 20 | 60 | 0.813 | 1.810 | 5.359 | 7 | 6 | 7 | 5 | 5 | 4 |
| NAU5046 | 22 | 57 | 0.877 | 2.235 | 8.100 | 11 | 11 | 7 | 6 | 6 | 6 |
| NAU5064 | 11 | 57 | 0.832 | 1.902 | 5.954 | 8 | 8 | 7 | 4 | 6 | 6 |
| NAU5099 | 22 | 55 | 0.880 | 2.159 | 8.333 | 9 | 9 | 5 | 3 | 5 | 5 |
| NAU5107 | 1 | 55 | 0.857 | 2.018 | 7.010 | 8 | 8 | 7 | 5 | 6 | 6 |
| NAU5120 | 16 | 53 | 0.873 | 2.152 | 7.881 | 10 | 10 | 8 | 3 | 10 | 10 |
| NAU5128 | 8, | 53 | 0.847 | 2.052 | 6.553 | 10 | 9 | 8 | 6 | 6 | 5 |
| NAU5129 | 8 | 55 | 0.883 | 2.237 | 8.533 | 11 | 11 | 6 | 5 | 5 | 5 |
| NAU5163 | 1 | 55 | 0.804 | 1.742 | 5.115 | 7 | 7 | 6 | 5 | 7 | 7 |
| NAU5368 | 8 | 55 | 0.855 | 2.069 | 6.881 | 10 | 10 | 6 | 5 | 6 | 6 |
| NAU5373 | 6,25 | 55 | 0.824 | 1.817 | 5.686 | 7 | 7 | 5 | 4 | 5 | 5 |
| NAU5379 | 6,24 | 55 | 0.741 | 1.458 | 3.861 | 5 | 5 | 4 | 4 | 3 | 3 |
| NAU5416 | 53 | 0.768 | 1.639 | 4.310 | 6 | 6 | 4 | 3 | 3 | 3 | |
| NAU5426 | 53 | 0.847 | 2.022 | 6.519 | 9 | 9 | 7 | 6 | 3 | 3 | |
| NAU5433 | 6 | 55 | 0.889 | 2.364 | 9.000 | 13 | 13 | 9 | 7 | 5 | 5 |
| NAU5434 | 6 | 57 | 0.883 | 2.311 | 8.528 | 12 | 11 | 9 | 7 | 5 | 4 |
| NAU5463 | 53 | 0.780 | 1.789 | 4.545 | 8 | 7 | 8 | 7 | 1 | 0 | |
| NAU5494 | 9 | 55 | 0.831 | 1.923 | 5.918 | 8 | 8 | 5 | 3 | 7 | 7 |
| NAU749 | 55 | 0.744 | 1.534 | 3.913 | 6 | 5 | 5 | 4 | 5 | 4 | |
| NAU868 | 55 | 0.857 | 2.079 | 7.010 | 9 | 9 | 8 | 6 | 4 | 4 | |
| NAU874 | LGA06 | 55 | 0.824 | 1.828 | 5.684 | 7 | 7 | 5 | 3 | 5 | 5 |
| NAU876 | 55 | 0.798 | 1.604 | 4.944 | 5 | 5 | 5 | 0 | 5 | 5 | |
| NAU879 | 10,LGA05 | 55 | 0.856 | 2.077 | 6.964 | 10 | 10 | 7 | 6 | 9 | 9 |
| NAU895 | 2 | 53 | 0.719 | 1.428 | 3.564 | 5 | 4 | 2 | 0 | 5 | 4 |
| NAU899 | 53 | 0.770 | 1.548 | 4.349 | 6 | 6 | 6 | 2 | 4 | 4 | |
| NAU905 | 6,25 | 53 | 0.865 | 2.161 | 7.420 | 11 | 11 | 5 | 3 | 10 | 10 |
| NAU934 | 5 | 51 | 0.895 | 2.357 | 9.483 | 12 | 12 | 6 | 1 | 12 | 12 |
| NAU965 | 53 | 0.900 | 2.378 | 9.975 | 12 | 12 | 6 | 4 | 11 | 11 | |
| NAU984 | 21 | 53 | 0.786 | 1.766 | 4.667 | 8 | 8 | 2 | 0 | 8 | 8 |
| NAU990 | 53 | 0.730 | 1.496 | 3.705 | 6 | 5 | 3 | 2 | 5 | 3 | |
| TMB0283 | 1 | 55 | 0.806 | 1.760 | 5.160 | 7 | 7 | 3 | 1 | 6 | 6 |
| TMB0301 | 15 | 55 | 0.752 | 1.564 | 4.038 | 6 | 6 | 2 | 0 | 5 | 5 |
| TMB0312 | 55 | 0.676 | 1.307 | 3.085 | 5 | 5 | 4 | 3 | 3 | 3 | |
| TMB0313 | 25 | 55 | 0.758 | 1.598 | 4.128 | 6 | 5 | 5 | 4 | 3 | 2 |
| TMB0325 | 10 | 60 | 0.802 | 1.730 | 5.055 | 7 | 5 | 4 | 0 | 6 | 4 |
| TMB0327 | 12 | 55 | 0.836 | 1.932 | 6.095 | 8 | 8 | 6 | 5 | 7 | 7 |
| TMB0375 | 15 | 57 | 0.822 | 1.759 | 5.622 | 6 | 6 | 5 | 0 | 4 | 4 |
| TMB0409 | 57 | 0.842 | 1.983 | 6.323 | 9 | 9 | 5 | 5 | 9 | 9 | |
| TMB0436 | 6,25 | 55 | 0.824 | 1.819 | 5.686 | 7 | 7 | 4 | 4 | 7 | 7 |
| TMB0508 | 25 | 60 | 0.761 | 1.566 | 4.181 | 6 | 6 | 3 | 0 | 5 | 5 |
| TMB10 | 55 | 0.877 | 2.177 | 8.114 | 10 | 10 | 6 | 0 | 9 | 9 | |
| TMB1120 | 5 | 57 | 0.869 | 2.169 | 7.649 | 11 | 11 | 7 | 4 | 9 | 9 |
| TMB1152 | 57 | 0.884 | 2.255 | 8.638 | 11 | 11 | 9 | 9 | 11 | 11 | |
| TMB1181 | 57 | 0.823 | 1.883 | 5.642 | 8 | 8 | 5 | 5 | 8 | 8 | |
| TMB1224 | 55 | 0.880 | 2.237 | 8.363 | 11 | 11 | 4 | 0 | 10 | 10 | |
| TMB1232 | 11,21 | 56 | 0.868 | 2.133 | 7.603 | 10 | 10 | 7 | 4 | 6 | 6 |
| TMB1268 | 17 | 57 | 0.886 | 2.330 | 8.791 | 12 | 12 | 7 | 6 | 7 | 7 |
| TMB1288 | 10 | 55 | 0.819 | 1.831 | 5.519 | 8 | 8 | 6 | 2 | 7 | 7 |
| TMB1296 | 5,19 | 55 | 0.884 | 2.266 | 8.647 | 11 | 11 | 7 | 3 | 10 | 10 |
| TMB1409 | 56 | 0.874 | 2.229 | 7.941 | 11 | 11 | 5 | 4 | 9 | 9 | |
| TMB1638 | 18 | 53 | 0.753 | 1.544 | 4.042 | 6 | 6 | 4 | 1 | 4 | 4 |
| TMB1667 | 11 | 60 | 0.811 | 1.877 | 5.289 | 9 | 7 | 5 | 2 | 8 | 6 |
| TMB1706 | 16 | 53 | 0.873 | 2.158 | 7.855 | 10 | 10 | 5 | 0 | 8 | 8 |
| TMB1746 | 55 | 0.892 | 2.336 | 9.251 | 12 | 12 | 5 | 3 | 11 | 11 | |
| TMB176 | 57 | 0.834 | 1.995 | 6.012 | 10 | 9 | 9 | 8 | 5 | 4 | |
| TMB1791 | 57 | 0.827 | 1.854 | 5.789 | 8 | 8 | 6 | 4 | 7 | 7 | |
| TMB1939 | 57 | 0.913 | 2.570 | 11.512 | 15 | 15 | 6 | 4 | 14 | 14 | |
| TMB1989 | 56 | 0.813 | 1.793 | 5.342 | 7 | 7 | 5 | 4 | 6 | 6 | |
| TMB2038 | 57 | 0.876 | 2.200 | 8.092 | 11 | 11 | 8 | 6 | 7 | 7 | |
| TMB2762 | 18 | 55 | 0.810 | 1.817 | 5.258 | 7 | 6 | 7 | 6 | 3 | 2 |
| TMB2789 | 12 | 60 | 0.793 | 1.678 | 4.837 | 6 | 5 | 4 | 0 | 6 | 5 |
| TMB2945 | 57 | 0.738 | 1.418 | 3.814 | 5 | 5 | 4 | 1 | 4 | 4 | |
| TMB515 | 60 | 0.811 | 1.813 | 5.298 | 8 | 8 | 6 | 4 | 6 | 6 | |
| TMB537 | 57 | 0.791 | 1.671 | 4.781 | 6 | 5 | 5 | 3 | 4 | 3 | |
| TMB682 | 55 | 0.663 | 1.232 | 2.969 | 4 | 3 | 3 | 1 | 2 | 1 | |
| TMB858 | 9 | 57 | 0.828 | 1.935 | 5.817 | 9 | 9 | 6 | 4 | 8 | 7 |
| Total | 262.600 | 615.582 | 2011.469 | 2776 | 2755 | 1849 | 1153 | 2068 | 1967 | ||
| Average | 0.823 | 1.930 | 6.306 | 8.702 | 8.636 | 5.796 | 3.614 | 6.483 | 6.166 | ||
附录I 基于棉花参比种质筛选的SSR核心引物及相关多态性信息
Appendix I SSR core primers screened from 5,914 primers used for identification of cotton cultivars on a cotton panel and relative polymorphism information
| 引物名称 Primer | 所属染色体或 连锁群 Chromosome or linkage group | 退火温度 Annealing temperature (Tm) | 参比种质 Panel | 陆地棉 | 其他 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Panel等位基因 多态信息含量 Allelic polymorphism information content (PIC) | Panel基因型 多样性(H') Genotype polymorphism | Panel有效等 位基因数(Ne) Effective number of alleles | Panel等位 基因数 No. of allelic loci | Panel多态性 等位基因 No. of polymorphic allelic | 等位 基因数 No. of allelic loci in upland lines | 多态性 等位基因数 No. of polymorphic allelic loci in upland lines | 棉种 等位基因数 No. of allelic loci among other species | 棉种多态性 等位基因数 No. of polymorphic allelic loci among other species | |||
| BNL1015 | 57 | 0.837 | 1.945 | 6.125 | 8 | 7 | 7 | 6 | 6 | 5 | |
| BNL1026 | 54 | 0.861 | 2.117 | 7.209 | 10 | 10 | 7 | 4 | 7 | 7 | |
| BNL1034 | 11,17,21 | 54 | 0.826 | 1.900 | 5.740 | 8 | 8 | 5 | 4 | 8 | 8 |
| BNL1040 | 13,18 | 56 | 0.889 | 2.349 | 9.000 | 13 | 13 | 8 | 6 | 10 | 10 |
| BNL1053 | 5,21,LGA03 | 58 | 0.847 | 2.036 | 6.553 | 10 | 10 | 7 | 5 | 6 | 6 |
| BNL119 | 20 | 56 | 0.789 | 1.700 | 4.737 | 7 | 7 | 5 | 4 | 5 | 5 |
| BNL1227 | 12,26 | 58 | 0.844 | 2.088 | 6.422 | 11 | 11 | 6 | 5 | 7 | 7 |
| BNL1317 | 9,23 | 53 | 0.845 | 1.960 | 6.451 | 8 | 8 | 6 | 5 | 5 | 5 |
| BNL1421 | 13,18 | 55 | 0.894 | 2.389 | 9.397 | 14 | 14 | 7 | 3 | 14 | 14 |
| BNL1440 | 6,8,25 | 55 | 0.811 | 1.806 | 5.297 | 7 | 7 | 5 | 4 | 5 | 5 |
| BNL1495 | 13,18 | 53 | 0.806 | 1.694 | 5.149 | 5 | 5 | 4 | 4 | 5 | 5 |
| BNL1513 | 24 | 56 | 0.842 | 1.998 | 6.330 | 9 | 9 | 8 | 7 | 6 | 6 |
| BNL1517 | 25 | 56 | 0.882 | 2.315 | 8.442 | 12 | 12 | 8 | 7 | 6 | 6 |
| BNL1580 | 3,21 | 57 | 0.843 | 1.952 | 6.373 | 8 | 8 | 4 | 4 | 4 | 4 |
| BNL1604 | 7,16 | 57 | 0.813 | 1.777 | 5.342 | 7 | 7 | 3 | 2 | 5 | 5 |
| BNL1669 | 9,26 | 55 | 0.782 | 1.604 | 4.588 | 6 | 5 | 5 | 2 | 5 | 4 |
| BNL1694 | 7,16 | 55 | 0.879 | 2.200 | 8.291 | 10 | 10 | 6 | 5 | 7 | 7 |
| BNL193 | 12,18 | 57 | 0.818 | 1.946 | 5.497 | 9 | 9 | 6 | 6 | 4 | 4 |
| BNL226 | 3,14 | 57 | 0.871 | 2.161 | 7.734 | 10 | 10 | 5 | 2 | 8 | 8 |
| BNL243 | 18 | 57 | 0.864 | 2.175 | 7.364 | 11 | 11 | 6 | 3 | 7 | 7 |
| BNL2449 | 10,13 | 55 | 0.883 | 2.260 | 8.583 | 12 | 12 | 9 | 7 | 11 | 11 |
| BNL2485 | 56 | 0.810 | 1.754 | 5.263 | 7 | 7 | 5 | 2 | 6 | 6 | |
| BNL252 | 9,19,24 | 55 | 0.857 | 2.138 | 7.014 | 11 | 11 | 7 | 7 | 9 | 9 |
| BNL2590 | 9,23 | 55 | 0.878 | 2.191 | 8.198 | 10 | 10 | 6 | 4 | 6 | 6 |
| BNL2646 | 7,9,15 | 55 | 0.715 | 1.480 | 3.512 | 6 | 6 | 4 | 3 | 3 | 3 |
| BNL3033 | 15 | 55 | 0.749 | 1.580 | 3.986 | 6 | 5 | 5 | 4 | 3 | 2 |
| BNL3048 | 17 | 55 | 0.848 | 2.094 | 6.596 | 11 | 11 | 6 | 4 | 10 | 10 |
| BNL3140 | 23 | 55 | 0.797 | 1.761 | 4.923 | 7 | 7 | 6 | 4 | 4 | 4 |
| BNL3261 | 12 | 56 | 0.860 | 2.087 | 7.161 | 10 | 8 | 8 | 3 | 7 | 5 |
| BNL3383 | 23,26,LGD09 | 56 | 0.893 | 2.341 | 9.332 | 12 | 12 | 12 | 8 | 8 | 8 |
| BNL3474 | 8,9,24,26 | 53 | 0.852 | 2.065 | 6.744 | 10 | 10 | 6 | 4 | 8 | 8 |
| BNL3502 | 14 | 55 | 0.837 | 1.878 | 6.128 | 7 | 7 | 4 | 4 | 4 | 4 |
| BNL358 | 22 | 53 | 0.917 | 2.569 | 11.985 | 15 | 15 | 9 | 1 | 12 | 12 |
| BNL3590 | 2,8,17 | 57 | 0.864 | 2.143 | 7.360 | 11 | 10 | 8 | 4 | 8 | 7 |
| BNL3650 | 6,LGA06 | 53 | 0.881 | 2.224 | 8.430 | 11 | 11 | 8 | 4 | 8 | 7 |
| BNL3806 | 25,LGD06,LGA06 | 55 | 0.789 | 1.757 | 4.741 | 7 | 6 | 4 | 2 | 4 | 3 |
| BNL3976 | 5,21 | 57 | 0.771 | 1.601 | 4.359 | 6 | 5 | 6 | 4 | 4 | 3 |
| BNL4030 | 5,22 | 57 | 0.792 | 1.659 | 4.804 | 6 | 6 | 4 | 2 | 6 | 6 |
| BNL448 | 22 | 57 | 0.863 | 2.107 | 7.283 | 10 | 8 | 8 | 1 | 8 | 6 |
| BNL511 | 9 | 57 | 0.893 | 2.317 | 9.308 | 12 | 12 | 9 | 8 | 10 | 10 |
| BNL530 | 4 | 57 | 0.904 | 2.433 | 10.464 | 13 | 13 | 9 | 4 | 13 | 13 |
| BNL542 | 5,21 | 53 | 0.874 | 2.163 | 7.949 | 10 | 10 | 5 | 4 | 9 | 9 |
| BNL827 | 25 | 57 | 0.899 | 2.341 | 9.936 | 11 | 11 | 10 | 8 | 9 | 9 |
| BNL830 | 15 | 53 | 0.714 | 1.375 | 3.495 | 5 | 5 | 4 | 2 | 3 | 3 |
| BNL834 | 17 | 55 | 0.893 | 2.348 | 9.321 | 12 | 12 | 8 | 4 | 6 | 6 |
| BNL946 | 20 | 56 | 0.896 | 2.377 | 9.624 | 12 | 12 | 12 | 7 | 5 | 4 |
| CIR139 | 19 | 51 | 0.647 | 1.203 | 2.829 | 4 | 3 | 2 | 0 | 3 | 2 |
| CIR167 | 6,26 | 50 | 0.838 | 1.956 | 6.171 | 8 | 8 | 3 | 0 | 7 | 7 |
| CIR246 | 14 | 51 | 0.850 | 2.004 | 6.651 | 9 | 9 | 6 | 5 | 8 | 8 |
| CIR249 | 4 | 51 | 0.859 | 2.012 | 7.111 | 8 | 8 | 6 | 6 | 7 | 7 |
| CIR307 | 15,LGD01 | 51 | 0.774 | 1.590 | 4.430 | 6 | 6 | 4 | 2 | 5 | 5 |
| CIR32 | 26 | 51 | 0.722 | 1.410 | 3.596 | 5 | 5 | 3 | 0 | 4 | 4 |
| CIR328 | 5 | 51 | 0.827 | 2.011 | 5.780 | 10 | 10 | 6 | 5 | 9 | 9 |
| CIR332 | 3,LGA03 | 51 | 0.893 | 2.345 | 9.368 | 12 | 12 | 6 | 2 | 12 | 12 |
| CIR353 | 9 | 53 | 0.874 | 2.235 | 7.954 | 12 | 12 | 8 | 5 | 8 | 8 |
| CIR62 | 5,19 | 51 | 0.824 | 1.934 | 5.694 | 9 | 8 | 5 | 2 | 6 | 5 |
| CIR78 | 26 | 50 | 0.808 | 1.768 | 5.213 | 7 | 5 | 4 | 0 | 6 | 4 |
| CM162 | 55 | 0.895 | 2.423 | 9.564 | 14 | 14 | 9 | 5 | 9 | 9 | |
| CM45 | 56 | 0.746 | 1.564 | 3.930 | 6 | 6 | 4 | 3 | 4 | 4 | |
| CM51 | 58 | 0.816 | 1.777 | 5.442 | 7 | 7 | 4 | 0 | 6 | 6 | |
| CM61 | 57 | 0.792 | 1.667 | 4.813 | 6 | 6 | 3 | 3 | 3 | 3 | |
| CM67 | 57 | 0.819 | 1.752 | 5.538 | 6 | 6 | 3 | 2 | 5 | 5 | |
| CSHES29 | 55 | 0.724 | 1.494 | 3.629 | 7 | 6 | 3 | 0 | 7 | 6 | |
| CSHES32 | 53 | 0.719 | 1.391 | 3.564 | 5 | 4 | 3 | 0 | 4 | 4 | |
| CSHES51 | 57 | 0.840 | 1.888 | 6.259 | 7 | 7 | 4 | 3 | 4 | 4 | |
| CSHES62 | 55 | 0.753 | 1.654 | 4.041 | 7 | 6 | 7 | 6 | 3 | 2 | |
| CSHES82 | 53 | 0.726 | 1.410 | 3.645 | 5 | 5 | 4 | 3 | 4 | 4 | |
| DPL209 | 11 | 59 | 0.897 | 2.362 | 9.672 | 13 | 13 | 12 | 9 | 9 | 9 |
| DPL216 | 2 | 58 | 0.801 | 1.685 | 5.026 | 6 | 4 | 0 | 6 | 6 | |
| DPL220 | 8 | 58 | 0.881 | 2.382 | 8.399 | 14 | 13 | 10 | 8 | 9 | 8 |
| DPL225 | 20 | 58 | 0.853 | 1.982 | 6.782 | 8 | 8 | 6 | 2 | 6 | 6 |
| DPL234 | 58 | 0.872 | 2.160 | 7.803 | 10 | 10 | 7 | 7 | 8 | 8 | |
| DPL238 | 6 | 59 | 0.858 | 2.094 | 7.043 | 10 | 10 | 5 | 2 | 9 | 9 |
| DPL249 | 18 | 58 | 0.852 | 1.975 | 6.768 | 8 | 8 | 5 | 4 | 6 | 6 |
| DPL253 | 11 | 58 | 0.759 | 1.558 | 4.149 | 6 | 4 | 5 | 1 | 3 | 1 |
| DPL262 | 23 | 58 | 0.811 | 1.832 | 5.303 | 8 | 8 | 5 | 2 | 7 | 7 |
| DPL283 | 16 | 58 | 0.728 | 1.642 | 3.674 | 8 | 7 | 2 | 0 | 7 | 6 |
| DPL300 | 15 | 58 | 0.773 | 1.616 | 4.412 | 6 | 6 | 5 | 4 | 4 | 4 |
| DPL325 | 11 | 58 | 0.790 | 1.659 | 4.765 | 6 | 6 | 3 | 2 | 4 | 4 |
| DPL354 | 17 | 59 | 0.864 | 2.200 | 7.339 | 11 | 11 | 7 | 6 | 6 | 6 |
| DPL431 | 10 | 57 | 0.735 | 1.433 | 3.769 | 5 | 5 | 4 | 4 | 2 | 2 |
| DPL46 | 2 | 58 | 0.858 | 2.114 | 7.053 | 10 | 10 | 6 | 2 | 9 | 9 |
| DPL461 | 24 | 58 | 0.826 | 1.885 | 5.762 | 8 | 8 | 4 | 4 | 4 | 4 |
| DPL511 | 16 | 58 | 0.867 | 2.162 | 7.529 | 11 | 11 | 7 | 4 | 7 | 7 |
| DPL513 | 1 | 59 | 0.816 | 1.881 | 5.444 | 9 | 9 | 6 | 4 | 5 | 4 |
| DPL519 | 25 | 58 | 0.722 | 1.570 | 3.600 | 7 | 7 | 2 | 0 | 6 | 6 |
| DPL528 | 21 | 58 | 0.905 | 2.416 | 10.492 | 12 | 12 | 8 | 6 | 10 | 10 |
| DPL570 | 11 | 58 | 0.851 | 2.032 | 6.714 | 9 | 9 | 6 | 2 | 9 | 9 |
| DPL681 | 6 | 57 | 0.727 | 1.559 | 3.658 | 7 | 7 | 4 | 3 | 3 | 2 |
| DPL69 | 26 | 60 | 0.692 | 1.125 | 3.245 | 4 | 4 | 3 | 2 | 3 | 3 |
| DPL71 | 19 | 58 | 0.707 | 1.484 | 3.413 | 6 | 6 | 4 | 3 | 4 | 4 |
| DPL73 | 14 | 59 | 0.844 | 1.984 | 6.408 | 9 | 8 | 6 | 0 | 7 | 6 |
| DPL799 | 25 | 59 | 0.807 | 1.915 | 5.172 | 9 | 9 | 6 | 5 | 4 | 4 |
| DPL910 | 18 | 56 | 0.870 | 2.250 | 7.704 | 12 | 12 | 6 | 6 | 8 | 8 |
| GH110 | 56 | 0.881 | 2.251 | 8.395 | 11 | 11 | 7 | 7 | 4 | 4 | |
| GH111 | 56 | 0.877 | 2.284 | 8.133 | 13 | 13 | 7 | 6 | 12 | 12 | |
| GH112 | 58 | 0.873 | 2.217 | 7.903 | 11 | 11 | 6 | 5 | 11 | 11 | |
| GH132 | 58 | 0.852 | 1.995 | 6.744 | 8 | 8 | 4 | 0 | 6 | 6 | |
| GH158 | 58 | 0.837 | 2.008 | 6.146 | 9 | 9 | 5 | 2 | 9 | 9 | |
| GH220 | 58 | 0.675 | 1.333 | 3.077 | 5 | 5 | 3 | 2 | 4 | 4 | |
| GH243 | 60 | 0.764 | 1.608 | 4.233 | 6 | 5 | 3 | 0 | 5 | 4 | |
| GH247 | 60 | 0.886 | 2.233 | 8.779 | 10 | 10 | 9 | 7 | 8 | 8 | |
| GH268 | 60 | 0.790 | 1.651 | 4.766 | 6 | 6 | 2 | 0 | 5 | 5 | |
| GH273 | 57 | 0.648 | 1.072 | 2.844 | 3 | 3 | 3 | 3 | 3 | 3 | |
| GH277 | 58 | 0.659 | 1.313 | 2.935 | 5 | 5 | 5 | 5 | 2 | 1 | |
| GH32 | 60 | 0.798 | 1.698 | 4.962 | 6 | 6 | 3 | 0 | 4 | 4 | |
| GH34 | 58 | 0.780 | 1.685 | 4.546 | 7 | 7 | 5 | 3 | 5 | ||
| GH447 | 58 | 0.819 | 1.849 | 5.528 | 8 | 8 | 7 | 6 | 5 | 5 | |
| GH532 | 56 | 0.870 | 2.206 | 7.718 | 11 | 10 | 9 | 7 | 7 | 6 | |
| GH56 | 55 | 0.823 | 1.986 | 5.635 | 10 | 9 | 7 | 4 | 4 | 3 | |
| GH59 | 55 | 0.901 | 2.394 | 10.117 | 12 | 12 | 6 | 1 | 10 | 10 | |
| GH60 | 57 | 0.776 | 1.550 | 4.455 | 5 | 5 | 2 | 0 | 5 | 5 | |
| GH70 | 55 | 0.870 | 2.107 | 7.716 | 9 | 9 | 5 | 2 | 9 | 8 | |
| GH72 | 57 | 0.828 | 1.964 | 5.797 | 9 | 9 | 6 | 4 | 5 | 5 | |
| GH75 | 56 | 0.855 | 2.077 | 6.914 | 10 | 10 | 7 | 7 | 6 | 6 | |
| JESPR101 | 2,8 | 56 | 0.817 | 1.837 | 5.454 | 8 | 7 | 4 | 3 | 6 | 5 |
| JESPR102 | 15,16 | 55 | 0.758 | 1.466 | 4.129 | 5 | 5 | 4 | 3 | 5 | 5 |
| JESPR114 | 23,LGD09 | 58 | 0.842 | 2.070 | 6.333 | 11 | 11 | 9 | 8 | 5 | 5 |
| JESPR133 | 55 | 0.882 | 2.089 | 8.471 | 10 | 10 | 7 | 3 | 8 | 8 | |
| JESPR138 | 55 | 0.716 | 1.427 | 3.522 | 5 | 4 | 2 | 0 | 5 | 4 | |
| JESPR156 | 14,21 | 58 | 0.780 | 2.324 | 4.540 | 13 | 13 | 10 | 7 | 8 | 8 |
| JESPR158 | 21,LGA03 | 57 | 0.891 | 2.298 | 9.195 | 11 | 11 | 7 | 4 | 5 | 5 |
| JESPR204 | 5,13,18,19 | 57 | 0.887 | 2.311 | 8.828 | 12 | 12 | 9 | 6 | 6 | 6 |
| JESPR208 | 9,23 | 54 | 0.885 | 2.264 | 8.699 | 11 | 11 | 6 | 3 | 10 | 10 |
| JESPR222 | 58 | 0.810 | 1.794 | 5.261 | 7 | 7 | 3 | 3 | 7 | 7 | |
| JESPR227 | 25 | 57 | 0.801 | 1.868 | 5.031 | 9 | 9 | 5 | 2 | 7 | 7 |
| JESPR231 | 3,14 | 57 | 0.867 | 2.092 | 7.506 | 9 | 9 | 6 | 4 | 5 | 5 |
| JESPR247 | 6 | 55 | 0.768 | 1.661 | 4.319 | 8 | 8 | 5 | 2 | 6 | 6 |
| JESPR274 | 9,23,24 | 61 | 0.753 | 1.480 | 4.050 | 5 | 5 | 3 | 1 | 4 | 4 |
| JESPR42 | 57 | 0.810 | 1.772 | 5.261 | 7 | 7 | 3 | 2 | 5 | 5 | |
| JESPR45 | 57 | 0.610 | 1.110 | 2.564 | 4 | 4 | 2 | 0 | 3 | 3 | |
| JESPR50 | 5,LGA04,LGD06 | 56 | 0.873 | 2.113 | 7.881 | 9 | 9 | 4 | 0 | 8 | 8 |
| MGHES08 | 57 | 0.817 | 1.779 | 5.473 | 6 | 5 | 6 | 1 | 6 | 6 | |
| MGHES16 | 21,LGA03 | 55 | 0.812 | 1.857 | 5.315 | 9 | 7 | 5 | 1 | 8 | 6 |
| MGHES18 | 58 | 0.846 | 1.522 | 6.480 | 7 | 7 | 4 | 1 | 6 | 6 | |
| MGHES38 | LGA03 | 54 | 0.743 | 2.281 | 3.890 | 13 | 13 | 6 | 2 | 10 | 10 |
| MGHES40 | 56 | 0.766 | 1.700 | 4.269 | 7 | 6 | 7 | 6 | 3 | 2 | |
| MGHES41 | 55 | 0.794 | 2.081 | 4.861 | 8 | 8 | 3 | 2 | 8 | 8 | |
| MGHES46 | 56 | 0.842 | 2.008 | 6.345 | 10 | 10 | 5 | 0 | 8 | 8 | |
| MGHES58 | 7 | 57 | 0.870 | 2.255 | 7.697 | 13 | 13 | 5 | 2 | 11 | 11 |
| MGHES70 | 9 | 57 | 0.907 | 2.429 | 10.777 | 12 | 12 | 10 | 9 | 8 | 8 |
| MGHES73 | 9 | 60 | 0.819 | 1.893 | 5.528 | 8 | 8 | 4 | 3 | 8 | 8 |
| MUCS101 | 4 | 57 | 0.762 | 1.572 | 4.197 | 6 | 6 | 6 | 5 | 4 | 3 |
| MUCS127 | 3 | 55 | 0.717 | 2.101 | 3.531 | 10 | 9 | 10 | 9 | 5 | 4 |
| MUCS347 | 21 | 57 | 0.834 | 1.982 | 6.025 | 9 | 9 | 3 | 0 | 8 | 7 |
| MUCS466 | 58 | 0.779 | 1.552 | 4.521 | 5 | 4 | 4 | 0 | 5 | 4 | |
| MUCS530 | 11 | 58 | 0.735 | 1.442 | 3.769 | 5 | 4 | 5 | 2 | 5 | 4 |
| MUSS130 | 60 | 0.856 | 1.996 | 6.928 | 8 | 7 | 6 | 0 | 8 | 7 | |
| MUSS138 | 57 | 0.859 | 2.051 | 7.067 | 9 | 9 | 7 | 3 | 8 | 8 | |
| MUSS422 | 10 | 56 | 0.860 | 2.064 | 7.141 | 9 | 9 | 5 | 4 | 9 | 9 |
| MUSS440 | 15 | 58 | 0.900 | 2.375 | 9.979 | 12 | 12 | 9 | 9 | 8 | 7 |
| MUSS46 | 58 | 0.771 | 1.572 | 4.360 | 6 | 4 | 5 | 1 | 4 | 2 | |
| MUSS59 | 57 | 0.859 | 2.005 | 7.084 | 8 | 8 | 7 | 5 | 8 | 8 | |
| MUSS65 | 57 | 0.837 | 1.936 | 6.149 | 8 | 6 | 8 | 6 | 6 | 4 | |
| NAU1014 | 9,11 | 53 | 0.886 | 2.301 | 8.744 | 12 | 12 | 5 | 2 | 12 | 12 |
| NAU1015 | 5 | 53 | 0.852 | 2.141 | 6.748 | 12 | 12 | 3 | 0 | 12 | 12 |
| NAU1017 | 8,24 | 55 | 0.826 | 1.834 | 5.762 | 7 | 70 | 4 | 3 | 7 | 7 |
| NAU1027 | 6,19 | 53 | 0.845 | 2.149 | 6.450 | 11 | 11 | 5 | 4 | 11 | 11 |
| NAU1028 | 17,23 | 55 | 0.895 | 2.387 | 9.483 | 13 | 13 | 7 | 4 | 12 | 12 |
| NAU1042 | 5,19 | 55 | 0.783 | 1.709 | 4.605 | 7 | 7 | 4 | 3 | 7 | 7 |
| NAU1043 | 7,LGA07 | 55 | 0.881 | 2.193 | 8.400 | 10 | 10 | 8 | 6 | 10 | 9 |
| NAU1048 | 7,11 | 53 | 0.832 | 1.898 | 5.938 | 8 | 7 | 6 | 4 | 8 | 6 |
| NAU1053 | 7 | 53 | 0.850 | 1.997 | 6.651 | 9 | 9 | 6 | 2 | 9 | 9 |
| NAU1070 | 3,9 | 55 | 0.897 | 2.368 | 9.665 | 12 | 12 | 8 | 7 | 11 | 11 |
| NAU1085 | 55 | 0.882 | 2.227 | 8.495 | 11 | 11 | 7 | 6 | 10 | 10 | |
| NAU1093 | LGA06 | 57 | 0.896 | 2.371 | 9.656 | 12 | 12 | 8 | 5 | 9 | 9 |
| NAU1102 | LGD05 | 53 | 0.893 | 2.299 | 9.333 | 11 | 11 | 9 | 2 | 10 | 10 |
| NAU1103 | 57 | 0.815 | 1.941 | 5.402 | 9 | 8 | 9 | 8 | 3 | 2 | |
| NAU1125 | 19 | 53 | 0.820 | 1.846 | 5.558 | 8 | 8 | 5 | 4 | 7 | 7 |
| NAU1155 | 55 | 0.763 | 1.586 | 4.212 | 7 | 7 | 7 | 6 | 3 | 1 | |
| NAU1167 | 3,8,17,LGA03 | 55 | 0.880 | 2.154 | 8.345 | 9 | 9 | 9 | 9 | 8 | 8 |
| NAU1169 | 3,10,20 | 53 | 0.856 | 2.041 | 6.942 | 9 | 9 | 7 | 5 | 7 | 7 |
| NAU1186 | 55 | 0.803 | 1.728 | 5.085 | 7 | 7 | 7 | 6 | 4 | 4 | |
| NAU1187 | LGD05 | 55 | 0.794 | 1.684 | 4.861 | 6 | 6 | 5 | 4 | 4 | 4 |
| NAU1190 | 3,16,LGA03 | 53 | 0.863 | 2.066 | 7.316 | 9 | 9 | 5 | 3 | 7 | 7 |
| NAU1196 | 55 | 0.859 | 2.075 | 7.078 | 9 | 9 | 7 | 6 | 5 | 5 | |
| NAU1200 | LGA05 | 55 | 0.848 | 1.984 | 6.568 | 8 | 8 | 7 | 7 | 6 | 6 |
| NAU1201 | 13,14 | 55 | 0.710 | 1.398 | 3.447 | 5 | 5 | 2 | 2 | 5 | 5 |
| NAU1222 | 7,20,23 | 55 | 0.667 | 1.242 | 3.000 | 4 | 4 | 1 | 0 | 4 | 4 |
| NAU1225 | 7 | 53 | 0.832 | 1.921 | 5.965 | 8 | 8 | 5 | 4 | 5 | 5 |
| NAU1230 | LGD05 | 55 | 0.812 | 1.793 | 5.321 | 7 | 7 | 5 | 4 | 4 | 4 |
| NAU1233 | 10 | 55 | 0.875 | 2.374 | 8.004 | 14 | 13 | 10 | 9 | 8 | 7 |
| NAU1248 | 3,20 | 55 | 0.815 | 1.782 | 5.411 | 7 | 7 | 4 | 0 | 6 | 6 |
| NAU1255 | LGD05 | 55 | 0.829 | 1.852 | 5.841 | 7 | 7 | 5 | 4 | 4 | 4 |
| NAU1269 | LGD05 | 55 | 0.864 | 2.082 | 7.364 | 9 | 9 | 6 | 6 | 5 | 5 |
| NAU1273 | 53 | 0.830 | 1.781 | 5.880 | 6 | 6 | 6 | 2 | 5 | 4 | |
| NAU1302 | 3,5,24 | 51 | 0.904 | 2.459 | 10.421 | 13 | 12 | 10 | 8 | 8 | 7 |
| NAU1322 | 24 | 55 | 0.891 | 2.419 | 9.188 | 14 | 14 | 11 | 11 | 8 | 8 |
| NAU1336 | 8,19 | 57 | 0.830 | 1.847 | 5.882 | 7 | 7 | 4 | 0 | 7 | 7 |
| NAU1356 | 52 | 0.874 | 2.203 | 7.934 | 12 | 11 | 7 | 0 | 11 | 8 | |
| NAU1362 | 7 | 55 | 0.901 | 2.459 | 10.140 | 14 | 14 | 8 | 5 | 7 | 7 |
| NAU1366 | 3,5,21 | 55 | 0.902 | 2.458 | 10.240 | 14 | 14 | 11 | 7 | 11 | 11 |
| NAU1369 | 8,9,24 | 53 | 0.770 | 1.678 | 4.356 | 7 | 7 | 5 | 4 | 4 | 4 |
| NAU1379 | 55 | 0.860 | 2.051 | 7.165 | 9 | 9 | 7 | 5 | 8 | 8 | |
| NAU2026 | 12,LGD04 | 55 | 0.881 | 2.186 | 8.395 | 10 | 10 | 8 | 5 | 8 | 8 |
| NAU2083 | 1 | 57 | 0.837 | 1.977 | 6.125 | 9 | 9 | 7 | 6 | 3 | 3 |
| NAU2108 | 7 | 56 | 0.842 | 1.926 | 6.341 | 8 | 7 | 5 | 0 | 8 | 7 |
| NAU2120 | 22 | 58 | 0.845 | 2.001 | 6.472 | 9 | 9 | 5 | 2 | 8 | 8 |
| NAU2126 | 22 | 59 | 0.840 | 2.026 | 6.231 | 10 | 10 | 7 | 5 | 7 | 7 |
| NAU2139 | 19 | 55 | 0.857 | 2.059 | 7.000 | 9 | 9 | 6 | 4 | 7 | 7 |
| NAU2152 | 11,16,17 | 53 | 0.880 | 2.341 | 8.308 | 14 | 14 | 5 | 2 | 12 | 12 |
| NAU2156 | 6 | 55 | 0.853 | 2.118 | 6.818 | 11 | 11 | 5 | 4 | 9 | 9 |
| NAU2162 | 22 | 55 | 0.885 | 2.246 | 8.668 | 11 | 11 | 8 | 4 | 7 | 7 |
| NAU2173 | 14 | 53 | 0.736 | 1.434 | 3.789 | 5 | 5 | 3 | 1 | 5 | 5 |
| NAU2190 | 14 | 53 | 0.823 | 2.267 | 5.657 | 9 | 9 | 6 | 2 | 5 | 5 |
| NAU2251 | 12,26 | 55 | 0.844 | 1.944 | 6.400 | 8 | 8 | 7 | 6 | 4 | 4 |
| NAU2257 | 11 | 55 | 0.874 | 2.198 | 7.963 | 11 | 11 | 7 | 5 | 8 | 8 |
| NAU2272 | 5,14 | 53 | 0.846 | 2.076 | 6.504 | 10 | 10 | 7 | 5 | 7 | 7 |
| NAU2274 | 19 | 55 | 0.764 | 1.690 | 4.245 | 8 | 8 | 7 | 6 | 5 | 5 |
| NAU2277 | 2 | 57 | 0.847 | 1.961 | 6.541 | 8 | 8 | 5 | 5 | 8 | 8 |
| NAU2308 | 7 | 55 | 0.813 | 1.729 | 5.355 | 6 | 6 | 3 | 0 | 6 | 6 |
| NAU2336 | 14 | 55 | 0.813 | 1.872 | 5.333 | 8 | 8 | 5 | 4 | 6 | 6 |
| NAU2342 | 55 | 0.817 | 1.794 | 5.452 | 7 | 7 | 6 | 5 | 6 | 6 | |
| NAU2343 | 56 | 0.778 | 1.809 | 4.500 | 9 | 8 | 8 | 7 | 4 | 3 | |
| NAU2361 | 21 | 56 | 0.710 | 1.400 | 3.448 | 5 | 5 | 3 | 2 | 3 | 3 |
| NAU2398 | 55 | 0.892 | 2.327 | 9.256 | 12 | 12 | 4 | 0 | 9 | 9 | |
| NAU2494 | 5 | 55 | 0.828 | 1.975 | 5.823 | 10 | 10 | 6 | 3 | 6 | 6 |
| NAU2508 | 53 | 0.785 | 1.573 | 4.655 | 5 | 5 | 5 | 3 | 5 | 5 | |
| NAU2600 | 53 | 0.868 | 2.151 | 7.564 | 10 | 10 | 5 | 4 | 8 | 8 | |
| NAU2604 | 19 | 55 | 0.701 | 1.285 | 3.346 | 4 | 3 | 4 | 1 | 2 | 1 |
| NAU2631 | 24 | 53 | 0.838 | 1.937 | 6.158 | 8 | 8 | 7 | 6 | 6 | 6 |
| NAU2654 | 4 | 53 | 0.885 | 2.295 | 8.674 | 12 | 11 | 9 | 7 | 10 | 9 |
| NAU2679 | 6,25,LGD06 | 57 | 0.908 | 2.498 | 10.813 | 14 | 14 | 6 | 4 | 9 | 9 |
| NAU2741 | 1,19 | 52 | 0.722 | 1.418 | 3.593 | 5 | 4 | 4 | 3 | 4 | 3 |
| NAU2776 | 10,20 | 55 | 0.719 | 1.317 | 3.558 | 4 | 4 | 4 | 3 | 3 | 3 |
| NAU2782 | 22 | 55 | 0.908 | 2.490 | 10.912 | 14 | 14 | 10 | 3 | 7 | 7 |
| NAU2816 | 19 | 55 | 0.812 | 1.721 | 5.319 | 6 | 6 | 5 | 1 | 5 | 5 |
| NAU2820 | 7,16 | 55 | 0.849 | 2.038 | 6.627 | 10 | 10 | 5 | 1 | 10 | 10 |
| NAU2836 | 3,17 | 55 | 0.844 | 1.994 | 6.391 | 9 | 9 | 6 | 3 | 5 | 5 |
| NAU2898 | 17 | 55 | 0.811 | 1.944 | 5.294 | 9 | 9 | 6 | 5 | 4 | 4 |
| NAU3013 | 10 | 57 | 0.799 | 1.733 | 4.971 | 7 | 7 | 4 | 1 | 5 | 5 |
| NAU3018 | 13 | 57 | 0.791 | 1.694 | 4.787 | 7 | 6 | 5 | 2 | 6 | 5 |
| NAU3096 | 5,19 | 53 | 0.768 | 1.611 | 4.304 | 6 | 5 | 6 | 4 | 4 | 3 |
| NAU3100 | 23 | 55 | 0.847 | 2.034 | 6.542 | 10 | 9 | 8 | 4 | 5 | 4 |
| NAU3102 | 52 | 0.507 | 0.953 | 2.027 | 4 | 4 | 4 | 4 | 1 | 0 | |
| NAU3127 | 4 | 57 | 0.739 | 1.448 | 3.835 | 5 | 5 | 3 | 2 | 5 | 5 |
| NAU3212 | 5 | 53 | 0.904 | 2.438 | 10.365 | 13 | 13 | 5 | 3 | 10 | 9 |
| NAU3225 | 14 | 53 | 0.853 | 2.093 | 6.818 | 10 | 10 | 5 | 3 | 6 | 6 |
| NAU3254 | 1 | 53 | 0.828 | 1.891 | 5.806 | 9 | 8 | 9 | 6 | 5 | 4 |
| NAU3305 | 26 | 56 | 0.860 | 2.130 | 7.143 | 10 | 10 | 6 | 5 | 5 | 5 |
| NAU3308 | 14 | 53 | 0.786 | 1.657 | 4.672 | 6 | 5 | 5 | 2 | 4 | 3 |
| NAU3325 | 5 | 56 | 0.785 | 1.732 | 4.661 | 7 | 6 | 5 | 4 | 4 | 3 |
| NAU3414 | 9,23 | 53 | 0.760 | 1.575 | 4.173 | 6 | 4 | 5 | 3 | 4 | 2 |
| NAU3419 | 2 | 57 | 0.852 | 2.021 | 6.737 | 9 | 9 | 6 | 4 | 8 | 8 |
| NAU3427 | 6 | 55 | 0.897 | 2.334 | 9.727 | 11 | 11 | 8 | 4 | 7 | 7 |
| NAU3467 | 10 | 55 | 0.739 | 1.469 | 3.835 | 6 | 5 | 4 | 1 | 5 | 4 |
| NAU3468 | 13 | 53 | 0.860 | 2.095 | 7.153 | 10 | 10 | 9 | 6 | 8 | 8 |
| NAU3499 | 8,19,24 | 53 | 0.862 | 2.187 | 7.251 | 11 | 10 | 7 | 5 | 6 | 5 |
| NAU3942 | 22 | 55 | 0.840 | 1.922 | 6.256 | 8 | 7 | 6 | 2 | 8 | 7 |
| NAU4042 | 19 | 53 | 0.864 | 2.137 | 7.333 | 10 | 10 | 9 | 8 | 9 | 9 |
| NAU4047 | 12 | 55 | 0.540 | 0.898 | 2.174 | 3 | 2 | 3 | 2 | 2 | 1 |
| NAU4057 | 5 | 53 | 0.890 | 2.343 | 9.109 | 13 | 13 | 11 | 8 | 8 | 8 |
| NAU4855 | 21 | 55 | 0.907 | 2.518 | 10.762 | 15 | 15 | 7 | 3 | 12 | 12 |
| NAU5013 | 20 | 60 | 0.813 | 1.810 | 5.359 | 7 | 6 | 7 | 5 | 5 | 4 |
| NAU5046 | 22 | 57 | 0.877 | 2.235 | 8.100 | 11 | 11 | 7 | 6 | 6 | 6 |
| NAU5064 | 11 | 57 | 0.832 | 1.902 | 5.954 | 8 | 8 | 7 | 4 | 6 | 6 |
| NAU5099 | 22 | 55 | 0.880 | 2.159 | 8.333 | 9 | 9 | 5 | 3 | 5 | 5 |
| NAU5107 | 1 | 55 | 0.857 | 2.018 | 7.010 | 8 | 8 | 7 | 5 | 6 | 6 |
| NAU5120 | 16 | 53 | 0.873 | 2.152 | 7.881 | 10 | 10 | 8 | 3 | 10 | 10 |
| NAU5128 | 8, | 53 | 0.847 | 2.052 | 6.553 | 10 | 9 | 8 | 6 | 6 | 5 |
| NAU5129 | 8 | 55 | 0.883 | 2.237 | 8.533 | 11 | 11 | 6 | 5 | 5 | 5 |
| NAU5163 | 1 | 55 | 0.804 | 1.742 | 5.115 | 7 | 7 | 6 | 5 | 7 | 7 |
| NAU5368 | 8 | 55 | 0.855 | 2.069 | 6.881 | 10 | 10 | 6 | 5 | 6 | 6 |
| NAU5373 | 6,25 | 55 | 0.824 | 1.817 | 5.686 | 7 | 7 | 5 | 4 | 5 | 5 |
| NAU5379 | 6,24 | 55 | 0.741 | 1.458 | 3.861 | 5 | 5 | 4 | 4 | 3 | 3 |
| NAU5416 | 53 | 0.768 | 1.639 | 4.310 | 6 | 6 | 4 | 3 | 3 | 3 | |
| NAU5426 | 53 | 0.847 | 2.022 | 6.519 | 9 | 9 | 7 | 6 | 3 | 3 | |
| NAU5433 | 6 | 55 | 0.889 | 2.364 | 9.000 | 13 | 13 | 9 | 7 | 5 | 5 |
| NAU5434 | 6 | 57 | 0.883 | 2.311 | 8.528 | 12 | 11 | 9 | 7 | 5 | 4 |
| NAU5463 | 53 | 0.780 | 1.789 | 4.545 | 8 | 7 | 8 | 7 | 1 | 0 | |
| NAU5494 | 9 | 55 | 0.831 | 1.923 | 5.918 | 8 | 8 | 5 | 3 | 7 | 7 |
| NAU749 | 55 | 0.744 | 1.534 | 3.913 | 6 | 5 | 5 | 4 | 5 | 4 | |
| NAU868 | 55 | 0.857 | 2.079 | 7.010 | 9 | 9 | 8 | 6 | 4 | 4 | |
| NAU874 | LGA06 | 55 | 0.824 | 1.828 | 5.684 | 7 | 7 | 5 | 3 | 5 | 5 |
| NAU876 | 55 | 0.798 | 1.604 | 4.944 | 5 | 5 | 5 | 0 | 5 | 5 | |
| NAU879 | 10,LGA05 | 55 | 0.856 | 2.077 | 6.964 | 10 | 10 | 7 | 6 | 9 | 9 |
| NAU895 | 2 | 53 | 0.719 | 1.428 | 3.564 | 5 | 4 | 2 | 0 | 5 | 4 |
| NAU899 | 53 | 0.770 | 1.548 | 4.349 | 6 | 6 | 6 | 2 | 4 | 4 | |
| NAU905 | 6,25 | 53 | 0.865 | 2.161 | 7.420 | 11 | 11 | 5 | 3 | 10 | 10 |
| NAU934 | 5 | 51 | 0.895 | 2.357 | 9.483 | 12 | 12 | 6 | 1 | 12 | 12 |
| NAU965 | 53 | 0.900 | 2.378 | 9.975 | 12 | 12 | 6 | 4 | 11 | 11 | |
| NAU984 | 21 | 53 | 0.786 | 1.766 | 4.667 | 8 | 8 | 2 | 0 | 8 | 8 |
| NAU990 | 53 | 0.730 | 1.496 | 3.705 | 6 | 5 | 3 | 2 | 5 | 3 | |
| TMB0283 | 1 | 55 | 0.806 | 1.760 | 5.160 | 7 | 7 | 3 | 1 | 6 | 6 |
| TMB0301 | 15 | 55 | 0.752 | 1.564 | 4.038 | 6 | 6 | 2 | 0 | 5 | 5 |
| TMB0312 | 55 | 0.676 | 1.307 | 3.085 | 5 | 5 | 4 | 3 | 3 | 3 | |
| TMB0313 | 25 | 55 | 0.758 | 1.598 | 4.128 | 6 | 5 | 5 | 4 | 3 | 2 |
| TMB0325 | 10 | 60 | 0.802 | 1.730 | 5.055 | 7 | 5 | 4 | 0 | 6 | 4 |
| TMB0327 | 12 | 55 | 0.836 | 1.932 | 6.095 | 8 | 8 | 6 | 5 | 7 | 7 |
| TMB0375 | 15 | 57 | 0.822 | 1.759 | 5.622 | 6 | 6 | 5 | 0 | 4 | 4 |
| TMB0409 | 57 | 0.842 | 1.983 | 6.323 | 9 | 9 | 5 | 5 | 9 | 9 | |
| TMB0436 | 6,25 | 55 | 0.824 | 1.819 | 5.686 | 7 | 7 | 4 | 4 | 7 | 7 |
| TMB0508 | 25 | 60 | 0.761 | 1.566 | 4.181 | 6 | 6 | 3 | 0 | 5 | 5 |
| TMB10 | 55 | 0.877 | 2.177 | 8.114 | 10 | 10 | 6 | 0 | 9 | 9 | |
| TMB1120 | 5 | 57 | 0.869 | 2.169 | 7.649 | 11 | 11 | 7 | 4 | 9 | 9 |
| TMB1152 | 57 | 0.884 | 2.255 | 8.638 | 11 | 11 | 9 | 9 | 11 | 11 | |
| TMB1181 | 57 | 0.823 | 1.883 | 5.642 | 8 | 8 | 5 | 5 | 8 | 8 | |
| TMB1224 | 55 | 0.880 | 2.237 | 8.363 | 11 | 11 | 4 | 0 | 10 | 10 | |
| TMB1232 | 11,21 | 56 | 0.868 | 2.133 | 7.603 | 10 | 10 | 7 | 4 | 6 | 6 |
| TMB1268 | 17 | 57 | 0.886 | 2.330 | 8.791 | 12 | 12 | 7 | 6 | 7 | 7 |
| TMB1288 | 10 | 55 | 0.819 | 1.831 | 5.519 | 8 | 8 | 6 | 2 | 7 | 7 |
| TMB1296 | 5,19 | 55 | 0.884 | 2.266 | 8.647 | 11 | 11 | 7 | 3 | 10 | 10 |
| TMB1409 | 56 | 0.874 | 2.229 | 7.941 | 11 | 11 | 5 | 4 | 9 | 9 | |
| TMB1638 | 18 | 53 | 0.753 | 1.544 | 4.042 | 6 | 6 | 4 | 1 | 4 | 4 |
| TMB1667 | 11 | 60 | 0.811 | 1.877 | 5.289 | 9 | 7 | 5 | 2 | 8 | 6 |
| TMB1706 | 16 | 53 | 0.873 | 2.158 | 7.855 | 10 | 10 | 5 | 0 | 8 | 8 |
| TMB1746 | 55 | 0.892 | 2.336 | 9.251 | 12 | 12 | 5 | 3 | 11 | 11 | |
| TMB176 | 57 | 0.834 | 1.995 | 6.012 | 10 | 9 | 9 | 8 | 5 | 4 | |
| TMB1791 | 57 | 0.827 | 1.854 | 5.789 | 8 | 8 | 6 | 4 | 7 | 7 | |
| TMB1939 | 57 | 0.913 | 2.570 | 11.512 | 15 | 15 | 6 | 4 | 14 | 14 | |
| TMB1989 | 56 | 0.813 | 1.793 | 5.342 | 7 | 7 | 5 | 4 | 6 | 6 | |
| TMB2038 | 57 | 0.876 | 2.200 | 8.092 | 11 | 11 | 8 | 6 | 7 | 7 | |
| TMB2762 | 18 | 55 | 0.810 | 1.817 | 5.258 | 7 | 6 | 7 | 6 | 3 | 2 |
| TMB2789 | 12 | 60 | 0.793 | 1.678 | 4.837 | 6 | 5 | 4 | 0 | 6 | 5 |
| TMB2945 | 57 | 0.738 | 1.418 | 3.814 | 5 | 5 | 4 | 1 | 4 | 4 | |
| TMB515 | 60 | 0.811 | 1.813 | 5.298 | 8 | 8 | 6 | 4 | 6 | 6 | |
| TMB537 | 57 | 0.791 | 1.671 | 4.781 | 6 | 5 | 5 | 3 | 4 | 3 | |
| TMB682 | 55 | 0.663 | 1.232 | 2.969 | 4 | 3 | 3 | 1 | 2 | 1 | |
| TMB858 | 9 | 57 | 0.828 | 1.935 | 5.817 | 9 | 9 | 6 | 4 | 8 | 7 |
| Total | 262.600 | 615.582 | 2011.469 | 2776 | 2755 | 1849 | 1153 | 2068 | 1967 | ||
| Average | 0.823 | 1.930 | 6.306 | 8.702 | 8.636 | 5.796 | 3.614 | 6.483 | 6.166 | ||
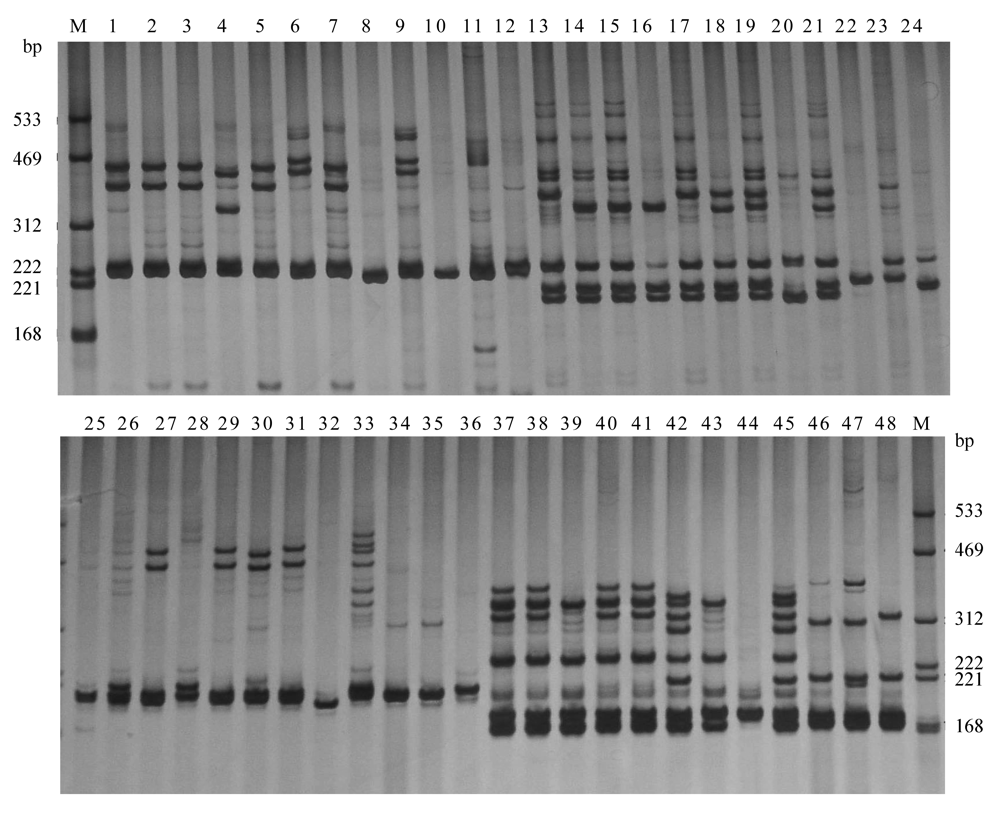
图1 引物NAU1369 、NAU1379、CIR328和CIR332在参比种质中的扩增结果(种质顺序左边起依次为: 常抗棉, 南丹巴地大花, 徐142无絮, TM-1, 棕2-63, 海7124, 3-79, 雷蒙地棉, 达尔文氏棉, 石系亚1号, DPL972, 高台草棉); M: 为分子量标准, 泳道1-12、13-24、25-36、37-48分别是引物NAU 1369、NAU1379、CIR328和CIR332在12个panel材料中的扩增结果。
Fig. 1 The amplified results of primer NAU1369, NAU1379, CIR328 and CIR332 in the panel. The panel lines from left to right were Changkang mian, Nandan badidahua, Xu 142 lintless-fuzzless, TM-1, Zong2-63, Hai7124, 3-79, G. raimondii, G. darwinii, Shixiya No.1, DPL972, and Gaotai caomian. The lane M represents the marker, the lanes 1-12, 13-24, 25-36, 37-48 represent the amplified results of primer NAU 1369, NAU1379, CIR328 and CIR332 in the panel with 12 accessions respectively.
| 多态性引物名称 Polymorphic primer | 引物序列 Primer sequence (5’→3’) | 染色体位置 Chromosome location | 基因组分布 Genome distribution | |||||
|---|---|---|---|---|---|---|---|---|
| MGHES18 | GCCATCAATTGGTGAAGCAT ATGCCTCGGTGAGAAAATTG | |||||||
| TMB0283 | GGCTCCAAAATTGAAACGTG GTGGACATTGGCATTCATTG | 1 | A | |||||
| TMB0436 | TGTGGCACAACCTTCCAAT CGTGTTCTCCATTTGATTCAT | 6, 25 | A, D | |||||
| GH111 | GTTGCAACCTTGGAAACCA GGGTTGCCGTTAGACCAG | |||||||
| NAU905 | TGGCTGAACTTTGCAATTTA AAGCAAGGGAGGTAATCCTT | 6, 25 | A, D | |||||
| NAU1085 | AGTCGCCCCTTCTCTAATTT TGTAAACCGAACTCGTTGTG | |||||||
| NAU2251 | TTCTCCAGTAACCAACAAAGG AAAATATCATCCCCGTCAAA | 12, 26 | A, D | |||||
| BNL252 | TGAAGAGCTCGTTGTTGCAC CGAAAGAGACAAGCAATGCA | 9, 24 | A, D | |||||
| BNL530 | CGTAGGATGGAAACGAAAGC GCCACACTTTTCCCTCTCAA | 4 | A | |||||
| BNL1227 | CATCAAGATCTATCTCTCTCTATACCG TTTACCCTCCGATCTCAACG | 12, 26 | A, D | |||||
| BNL1034 | TTGCTTTCAATGGAAAACCC CGTCGCAAAGTTGAGAATCA | 11, 17, 21 | A, D | |||||
| DPL209 | GAAGGAACCTCGTGATTATTTGAG GACCGGTAGACAGAGATGAGAAAT | 11 | A | |||||
| DPL225 | TAAAGTGGATACAATGACGACGAC CTTCAGCCATACATGCTGTTTATC | 20 | D | |||||
表3 在陆地棉、海岛棉及亚洲棉三大栽培棉种内都有多态性的通用SSR引物
Table 3 The common SSR primers with polymorphism in three cultivated species including upland cotton, sea island cotton and Asiatic cotton
| 多态性引物名称 Polymorphic primer | 引物序列 Primer sequence (5’→3’) | 染色体位置 Chromosome location | 基因组分布 Genome distribution | |||||
|---|---|---|---|---|---|---|---|---|
| MGHES18 | GCCATCAATTGGTGAAGCAT ATGCCTCGGTGAGAAAATTG | |||||||
| TMB0283 | GGCTCCAAAATTGAAACGTG GTGGACATTGGCATTCATTG | 1 | A | |||||
| TMB0436 | TGTGGCACAACCTTCCAAT CGTGTTCTCCATTTGATTCAT | 6, 25 | A, D | |||||
| GH111 | GTTGCAACCTTGGAAACCA GGGTTGCCGTTAGACCAG | |||||||
| NAU905 | TGGCTGAACTTTGCAATTTA AAGCAAGGGAGGTAATCCTT | 6, 25 | A, D | |||||
| NAU1085 | AGTCGCCCCTTCTCTAATTT TGTAAACCGAACTCGTTGTG | |||||||
| NAU2251 | TTCTCCAGTAACCAACAAAGG AAAATATCATCCCCGTCAAA | 12, 26 | A, D | |||||
| BNL252 | TGAAGAGCTCGTTGTTGCAC CGAAAGAGACAAGCAATGCA | 9, 24 | A, D | |||||
| BNL530 | CGTAGGATGGAAACGAAAGC GCCACACTTTTCCCTCTCAA | 4 | A | |||||
| BNL1227 | CATCAAGATCTATCTCTCTCTATACCG TTTACCCTCCGATCTCAACG | 12, 26 | A, D | |||||
| BNL1034 | TTGCTTTCAATGGAAAACCC CGTCGCAAAGTTGAGAATCA | 11, 17, 21 | A, D | |||||
| DPL209 | GAAGGAACCTCGTGATTATTTGAG GACCGGTAGACAGAGATGAGAAAT | 11 | A | |||||
| DPL225 | TAAAGTGGATACAATGACGACGAC CTTCAGCCATACATGCTGTTTATC | 20 | D | |||||
| [1] | Bertini C, Schuster I, Sediyama T, Barros E, Moreira M (2006) Characterization and genetic diversity analysis of cotton cultivars using microsatellites. Genetics and Molecular Biology, 29,321-329. |
| [2] | Bie S (别墅), Kong FL (孔繁玲), Zhou YY (周有耀), Zhang GM (张光梅), Zhang QY (张群远), Wang XG (王孝纲) (2001) Genetic diversity analysis of representative elite cotton varieties in three main cotton regions in China by RAPD and its relation with agronomic characteristics. Scientia Agricultura Sinica (中国农业科学), 34,597-603. (in Chinese with English abstract) |
| [3] | Botstein D, White R L, Skolnick M (1981) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32,314-331. |
| [4] | Chen G, Du XM (2006) Genetic diversity of source germplasm of upland cotton in China as determined by SSR marker analysis. Acta Genetica Sinica (遗传学报), 33,733-745. |
| [5] | Chen XM (陈新民), He ZH (何中虎), Shi JR (史建荣), Xia LQ (夏兰芹), Ward R, Zhou Y (周阳), Jiang GL (蒋国梁) (2003) Genetic diversity of high quality winter wheat varieties (lines) based on SSR markers. Acta Agronomica Sinica (作物学报), 29,13-19. (in Chinese with English abstract) |
| [6] | Dong YC (董玉琛), Cao YS (曹永生), Zhang XY (张学勇), Liu SC (刘三才), Wang LF (王兰芬), You GX (游光霞), Pang BS (庞斌双), Li LH (李立会), Jia JZ (贾继增) (2003) Establishment of candidate core collections in Chinese common wheat germplasm. Journal of Plant Genetic Resources (植物遗传资源学报), 4,l-8. (in Chinese with English abstract) |
| [7] | Du JY (杜金友), Li Y (黎裕), Wang TY (王天宇), Shi YS (石云素), Song YC (宋燕春), Wang HB (王海波) (2003) Studies of genetic diversity in maize inbred lines based on SSRs and AFLPs markers. Acta Agriculturae Boreali-Sinica (华北农学报), 18(1),59-63. (in Chinese with English abstract) |
| [8] | Guo WZ (郭旺珍), Wang K (王凯), Zhang TZ (张天真) (2003) A and D genome evolution in Gossypium revealed using SSR molecular markers. Acta Genetica Sinica (遗传学报), 30,183-188. (in Chinese with English abstract) |
| [9] |
Guo WZ, Cai CP, Wang CB, Han ZG, Song XL, Wang K, Niu XW, Wang C, Lu KY, Shi B, Zhang TZ (2007) A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics, 176,527-541.
URL PMID |
| [10] | Hai L (海林), Wang KJ (王克晶), Yang K (杨凯) (2002) Genetic diversity of semi-wild soybean using SSR markers. Acta Botanica Boreali-Occidentalia Sinica (西北植物学报), 22,751-757. (in Chinese with English abstract) |
| [11] | Huang ZK (黄滋康) (2007) Cotton Variety and Pedigree in China (中国棉花品种及其系谱). China Agriculture Press, Beijing. (in Chinese) |
| [12] | Khan SA, Hussain D, Askari E, Stewart J McD, Malik KA, Zafar Y (2000) Molecular phylogeny of Gossypium species by DNA fingerprinting. Theoretical and Applied Genetics, 101,931-938. |
| [13] | Li ZC (李自超), Zhang HL (张洪亮), Cao YS (曹永生), Qiu ZE (裘宗恩), Wei XH (魏兴华), Tang SX (汤圣祥), Yu P (余萍), Wang XK (王象坤) (2003) Studies on the sampling strategy for primary core collection of Chinese ingenious rice. Acta Agronomica Sinica (作物学报), 29,20-24. (in Chinese with English abstract) |
| [14] | Ma X (马轩), Du XM (杜雄明), Sun JL (孙君灵) (2003) SSR fingerprinting analysis on 18 colored cotton lines. Journal of Plant Genetic Resources (植物遗传资源学报), 4,305-310. (in Chinese with English abstract) |
| [15] | Pan ZE (潘兆娥), Du XM (杜雄明), Sun JL (孙君灵), Zhou ZL (周忠丽), Pang BY (庞保印) (2004) SSR marker analysis of upland cotton transferred into hormone gene. Molecular Plant Breeding (分子植物育种), 2,513-519. (in Chinese with English abstract) |
| [16] | Pang CY, Du XM, Ma ZY (2006) Evaluation of the introgressed lines and screening for elite germplasm in Gossypium. Chinese Science Bulletin, 51,304-312. |
| [17] | Shannon CE, Weaver W (1949) The Mathematical Theory of Communication, pp. 3-24. The University of Illinois, Urbana, Chicago. |
| [18] | Shen FF (沈法富), Yu YJ (于元杰), Liu FZ (刘凤珍), Yin CY (尹承佾) (1996) Isolation of nuclear DNA from cotton and its RAPD analysis. Acta Gossypii Sinica (棉花学报), 8,246-249. (in Chinese) |
| [19] | Song GL (宋国立), Cui RX (崔荣霞), Wang KB (王坤波), Guo LP (郭立平), Li SH (黎绍惠), Wang CY (王春英), Zhang XD (张香娣) (1998) A rapid improved CTAB method for extraction of cotton genomic DNA. Acta Gossypii Sinica (棉花学报), 10,273-275. (in Chinese with English abstract) |
| [20] | Wang LX (王丽侠), Li YH (李英慧), Li W (李伟), Zhu L (朱莉), Guan Y (关媛), Ning XC (宁学成), Guan RX (关荣霞), Liu ZX (刘章雄), Chang RZ (常汝镇), Qiu LJ (邱丽娟) (2004) Establishment of a core collection of Changjiang spring sowing soybean. Biodiversity Science (生物多样性), 12,578-585. (in Chinese with English abstract) |
| [21] | Wu YT (武耀廷), Zhang TZ (张天真), Guo WZ (郭旺珍), Yin JM (殷剑美) (2001) Detecting polymorphism among upland cotton (Gossypium hirsutum L.) cultivars and their roles in seed purity of hybrids with SSR markers. Cotton Science (棉花学报), 13,131-133. (in Chinese with English abstract) |
| [22] | Xie ZH (解志红), Sun Y (孙毅), Wu RH (吴日恒), Wang JX (王景雪), Liu HM (刘惠民) (2002) Current status, prospect on molecular marker in cotton. China Cotton (中国棉花), 29(5),5-8. (in Chinese) |
| [23] | Zhang JF, Stewart J McD (2000) Economical and rapid method for extracting cotton genomic DNA. Journal of Cotton Science, 4,193-201. |
| [24] | Zhu ZD (朱振东), Jia JZ (贾继增) (2003) Microsatellite marker development and applications in wheat genetics and breeding. Hereditas (Beijing) (遗传), 25,355-360. (in Chinese with English abstract) |
| [25] | Zuo KJ (左开井), Sun JZ (孙济中), Zhang XL (张献龙), Nie YC (聂以春), Liu JL (刘金兰), Feng CD (冯纯大) (2000) Constructing a linkage map of upland cotton (Gossypium hirsutum L.) using RFLP, RAPD and SSR markers. Journal of Huazhong Agricutral University (华中农业大学学报), 19,190-193. (in Chinese with English abstract) |
| [1] | 蔡新宇, 毛晓伟, 赵毅强. 家养动物驯化起源的研究方法与进展[J]. 生物多样性, 2022, 30(4): 21457-. |
| [2] | 张亚红, 贾会霞, 王志彬, 孙佩, 曹德美, 胡建军. 滇杨种群遗传多样性与遗传结构[J]. 生物多样性, 2019, 27(4): 355-365. |
| [3] | 莫日根高娃, 商辉, 刘保东, 康明, 严岳鸿. 一个种还是多个种? 简化基因组及其形态学证据揭示中国白桫椤植物的物种多样性分化[J]. 生物多样性, 2019, 27(11): 1196-1204. |
| [4] | 赵颖, 马荣, 尹永香, 张志东, 田呈明. 新疆不同来源金黄壳囊孢的多样性[J]. 生物多样性, 2019, 27(10): 1122-1131. |
| [5] | 李淑英, 朱加保, 路献勇, 程福如, 郑曙峰, 崔金杰, 雒珺瑜, 马艳. 转RRM2基因棉田昆虫群落多样性及其时序动态[J]. 生物多样性, 2018, 26(11): 1190-1203. |
| [6] | 盛芳, 陈淑英, 田嘉, 李鹏, 秦雪, 罗淑萍, 李疆. 新疆准噶尔山楂不同居群的遗传多样性[J]. 生物多样性, 2017, 25(5): 518-530. |
| [7] | 雒珺瑜, 张帅, 朱香镇, 王春义, 吕丽敏, 李春花, 崔金杰. 盐碱旱地转基因抗虫棉田昆虫群落多样性[J]. 生物多样性, 2016, 24(3): 332-340. |
| [8] | 杨洁, 赫佳, 王丹碧, 施恩, 杨文宇, 耿其芳, 王中生. InDel标记的研究和应用进展[J]. 生物多样性, 2016, 24(2): 237-243. |
| [9] | 刘华, 常晓蕾, 蒋玮, 白蓝, 郑曙峰, 王金斌, 王维, 潘爱虎, 王荣谈, 唐雪明. 华东三省转Bt基因棉花种植对边际水体中Cry1Ab/c蛋白残留的影响[J]. 生物多样性, 2016, 24(12): 1373-1380. |
| [10] | 徐刚标, 梁艳, 蒋燚, 刘雄盛, 胡尚力, 肖玉菲, 郝博搏. 伯乐树种群遗传多样性及遗传结构[J]. 生物多样性, 2013, 21(6): 723-731. |
| [11] | 吴雪琴, 徐刚标, 梁艳, 申响保. 南岭地区观光木自然和人工迁地保护种群的遗传多样性[J]. 生物多样性, 2013, 21(1): 71-79. |
| [12] | 向妮, 肖炎农, 段灿星, 王晓鸣, 朱振东. 利用SSR标记分析茄镰孢豌豆专化型的遗传多样性[J]. 生物多样性, 2012, 20(6): 693-702. |
| [13] | 马洪峥, 李珊珊, 葛颂, 戴思兰, 陈文俐. 能源作物芒属双药芒组SSR引物的筛选及其评价[J]. 生物多样性, 2011, 19(5): 535-542. |
| [14] | 王丽侠, 程须珍, 王素华. 基于SSR标记分析小豆及其近缘植物的遗传关系[J]. 生物多样性, 2011, 19(1): 17-23. |
| [15] | 张丽娜, 叶武威, 王俊娟, 樊保香, 王德龙. 棉花耐盐相关种质资源遗传多样性分析[J]. 生物多样性, 2010, 18(2): 137-144. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn