



生物多样性 ›› 2025, Vol. 33 ›› Issue (9): 25190. DOI: 10.17520/biods.2025190
洪欣艺1, 蔡易朗1, 方嘉乐1, 姚可侃2, 李佳乐3, 王懿祥4, 白尚斌1, 王楠1, 周秀梅1,*( )
)
收稿日期:2025-05-22
接受日期:2025-07-26
出版日期:2025-09-20
发布日期:2025-10-31
通讯作者:
*E-mail: zhouxm0324@163.com
基金资助:
Xinyi Hong1, Yilang Cai1, Jiale Fang1, Kekan Yao2, Jiale Li3, Yixiang Wang4, Shangbin Bai1, Nan Wang1, Xiumei Zhou1,*( )
)
Received:2025-05-22
Accepted:2025-07-26
Online:2025-09-20
Published:2025-10-31
Contact:
*E-mail: zhouxm0324@163.com
Supported by:摘要:
湿地土壤病毒在有机碳循环中发挥关键作用, 其携带的碳水化合物活性酶(CAZymes)基因可辅助碳代谢过程。为揭示城市湿地土壤病毒多样性特征及其碳代谢功能, 本研究以杭州西溪国家湿地公园为研究对象, 选取乔灌草地、灌草地、芦苇(Phragmites australis)地、池塘和浅滩5种典型生境, 通过病毒宏基因组测序与生物信息学分析, 系统探究了不同湿地生境类型土壤中病毒的多样性特征及其碳代谢基因组成。研究结果显示, 5种生境的病毒丰富度、多样性存在显著差异(P < 0.05), 由高到低依次为灌草地 > 乔灌草地 > 芦苇地 > 浅滩 > 池塘; 结构方程模型分析表明, 该格局主要受土壤理化性质影响, 对病毒多样性效应值排序依次为pH > 土壤湿度 > 土壤有机碳(SOC) ≈ 全氮(TN) > 土壤温度, 对病毒宿主与病毒多样性的综合效应值贡献度依次为TN > SOC > 土壤温度, 病毒宿主对病毒多样性强显著(效应值 = 0.87); 研究共鉴定出158个独特的碳水化合物运输与代谢(G)基因, 包含13种碳水化合物活性酶基因。其中糖基转移酶占比最高(65.8%), 表明土壤病毒在湿地碳循环中可能发挥重要作用。病毒功能基因的表达与分布受SOC和TN含量的显著影响, 说明土壤养分状况与病毒生态功能有密切的联系。研究结果为深入理解碳循环的微生物调控机制提供了重要科学依据, 对湿地保护与管理具有重要实践意义。
洪欣艺, 蔡易朗, 方嘉乐, 姚可侃, 李佳乐, 王懿祥, 白尚斌, 王楠, 周秀梅 (2025) 西溪湿地土壤病毒多样性及其碳代谢基因解析. 生物多样性, 33, 25190. DOI: 10.17520/biods.2025190.
Xinyi Hong, Yilang Cai, Jiale Fang, Kekan Yao, Jiale Li, Yixiang Wang, Shangbin Bai, Nan Wang, Xiumei Zhou (2025) Soil viral diversity and carbon metabolism genes profiling in Xixi Wetland. Biodiversity Science, 33, 25190. DOI: 10.17520/biods.2025190.
| 土壤温度 Soil temperature (℃) | 土壤湿度 Soil moisture (%) | pH | 土壤有机碳 Soil organic carbon (g/kg) | 土壤全氮 Total nitrogen (g/kg) | |
|---|---|---|---|---|---|
| 乔灌草地 Trees, shrubs and grasslands | 9.1 ± 0.2ab | 32.60 ± 14.06bc | 6.84 ± 0.44a | 41.3 ± 24.5a | 3.4 ± 1.8a |
| 灌草地 Shrub grasslands | 8.9 ± 1.4b | 25.29 ± 3.67c | 6.36 ± 0.50a | 30.8 ± 9.6ab | 2.8 ± 0.3ab |
| 芦苇地 Reed marshes | 7.1 ± 0.3c | 39.24 ± 11.89b | 5.57 ± 0.41b | 26.0 ± 10.4b | 2.2 ± 0.8b |
| 池塘 Ponds | 7.4 ± 0.1c | 51.70 ± 10.56ab | 6.56 ± 0.04a | 13.6 ± 0.8c | 1.1 ± 0.1c |
| 浅滩 Shoals | 11.1 ± 0.5a | 74.13 ± 8.35a | 5.47 ± 0.12b | 16.2 ± 1.1bc | 1.4 ± 0.1c |
表1 西溪湿地不同斑块类型的土壤理化性质。数据为平均值 ± 标准差。
Table 1 Soil physicochemical properties across different habitat types in Xixi Wetland. The data is the mean ± SD.
| 土壤温度 Soil temperature (℃) | 土壤湿度 Soil moisture (%) | pH | 土壤有机碳 Soil organic carbon (g/kg) | 土壤全氮 Total nitrogen (g/kg) | |
|---|---|---|---|---|---|
| 乔灌草地 Trees, shrubs and grasslands | 9.1 ± 0.2ab | 32.60 ± 14.06bc | 6.84 ± 0.44a | 41.3 ± 24.5a | 3.4 ± 1.8a |
| 灌草地 Shrub grasslands | 8.9 ± 1.4b | 25.29 ± 3.67c | 6.36 ± 0.50a | 30.8 ± 9.6ab | 2.8 ± 0.3ab |
| 芦苇地 Reed marshes | 7.1 ± 0.3c | 39.24 ± 11.89b | 5.57 ± 0.41b | 26.0 ± 10.4b | 2.2 ± 0.8b |
| 池塘 Ponds | 7.4 ± 0.1c | 51.70 ± 10.56ab | 6.56 ± 0.04a | 13.6 ± 0.8c | 1.1 ± 0.1c |
| 浅滩 Shoals | 11.1 ± 0.5a | 74.13 ± 8.35a | 5.47 ± 0.12b | 16.2 ± 1.1bc | 1.4 ± 0.1c |
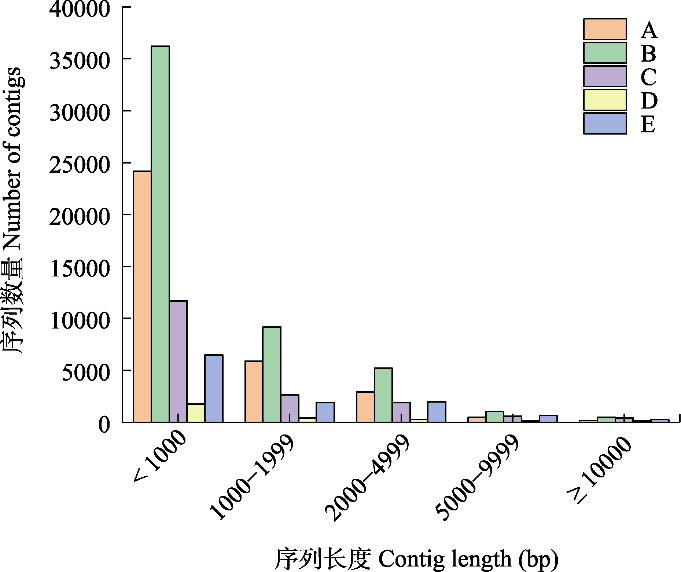
图1 西溪湿地不同斑块类型的contig长度分布图。A: 乔灌草地; B: 灌草地; C: 芦苇地; D: 池塘; E: 浅滩。
Fig. 1 The distribution of contig lengths for different habitat types in Xixi Wetland. A, Trees, shrubs and grasslands; B, Shrub grasslandes; C, Reed marshes; D, Ponds; E, Shoals.
| 宿主属 Host genus | 病毒序列数量 Number of viral sequences | 占总预测病毒宿主序 列数的比值 Proportion of predicted viral host sequences (%) | 乔灌草地 Trees, shrubs and grasslands | 灌草地 Shrub grasslands | 芦苇地 Reed marshes | 池塘 Ponds | 浅滩 Shoals |
|---|---|---|---|---|---|---|---|
| 不动杆菌属 Acinetobacter | 76 | 1.23 | 57 | 69 | 12 | 2 | 2 |
| 伯克霍尔德氏菌属 Burkholderia | 105 | 1.70 | 58 | 87 | 45 | 2 | 2 |
| 汉密尔顿菌属(暂定类群) Candidatus_Hamiltonella | 76 | 1.23 | 33 | 64 | 6 | 0 | 0 |
| 蛭弧菌属 Bdellovibrio | 1,939 | 31.38 | 1,372 | 1,563 | 99 | 4 | 2 |
| 柄杆菌属 Caulobacter | 155 | 2.51 | 105 | 136 | 55 | 2 | 2 |
| 埃希氏菌属 Escherichia | 375 | 6.07 | 257 | 320 | 62 | 6 | 1 |
| 地芽孢杆菌属 Geobacillus | 63 | 1.02 | 39 | 52 | 4 | 0 | 0 |
| 螺杆菌属 Helicobacter | 61 | 0.99 | 39 | 46 | 6 | 2 | 0 |
| 黏液乳杆菌属 Limosilactobacillus | 104 | 1.68 | 55 | 82 | 14 | 1 | 0 |
| 李斯特菌属 Listeria | 120 | 1.94 | 50 | 86 | 27 | 10 | 0 |
| 中慢生根瘤菌属 Mesorhizobium | 76 | 1.23 | 52 | 60 | 21 | 0 | 0 |
| 分枝杆菌属 Mycolicibacterium | 114 | 1.84 | 71 | 89 | 39 | 0 | 0 |
| 副拟杆菌属 Parabacteroides | 68 | 1.10 | 51 | 54 | 8 | 0 | 0 |
| 土球腔菌属 Pedosphaera | 50 | 0.81 | 31 | 47 | 12 | 0 | 0 |
| 普雷斯科特氏菌属 Prescottella | 77 | 1.25 | 53 | 70 | 7 | 0 | 0 |
| 原绿球藻属 Prochlorococcus | 50 | 0.81 | 39 | 32 | 3 | 0 | 0 |
| 假单胞菌属 Pseudomonas | 203 | 3.28 | 109 | 173 | 34 | 2 | 2 |
| 红杆菌属 Rhodobacter | 69 | 1.12 | 57 | 61 | 26 | 2 | 3 |
| 红育菌属 Rhodovulum | 53 | 0.86 | 34 | 45 | 14 | 2 | 2 |
| 罗格氏菌属 Ruegeria | 54 | 0.87 | 38 | 41 | 10 | 1 | 1 |
| 中华根瘤菌属 Sinorhizobium | 57 | 0.92 | 40 | 42 | 32 | 1 | 1 |
| 弧菌属 Vibrio | 129 | 2.09 | 84 | 102 | 26 | 4 | 2 |
| 未知 Unknown | 266 | 4.30 | 161 | 213 | 105 | 3 | 2 |
表2 所预测的病毒宿主大于50条的数量及其在不同湿地斑块类型的分布
Table 2 Summary of the predicted viral hosts with over 50 sequence counts and their distribution across different habitat types
| 宿主属 Host genus | 病毒序列数量 Number of viral sequences | 占总预测病毒宿主序 列数的比值 Proportion of predicted viral host sequences (%) | 乔灌草地 Trees, shrubs and grasslands | 灌草地 Shrub grasslands | 芦苇地 Reed marshes | 池塘 Ponds | 浅滩 Shoals |
|---|---|---|---|---|---|---|---|
| 不动杆菌属 Acinetobacter | 76 | 1.23 | 57 | 69 | 12 | 2 | 2 |
| 伯克霍尔德氏菌属 Burkholderia | 105 | 1.70 | 58 | 87 | 45 | 2 | 2 |
| 汉密尔顿菌属(暂定类群) Candidatus_Hamiltonella | 76 | 1.23 | 33 | 64 | 6 | 0 | 0 |
| 蛭弧菌属 Bdellovibrio | 1,939 | 31.38 | 1,372 | 1,563 | 99 | 4 | 2 |
| 柄杆菌属 Caulobacter | 155 | 2.51 | 105 | 136 | 55 | 2 | 2 |
| 埃希氏菌属 Escherichia | 375 | 6.07 | 257 | 320 | 62 | 6 | 1 |
| 地芽孢杆菌属 Geobacillus | 63 | 1.02 | 39 | 52 | 4 | 0 | 0 |
| 螺杆菌属 Helicobacter | 61 | 0.99 | 39 | 46 | 6 | 2 | 0 |
| 黏液乳杆菌属 Limosilactobacillus | 104 | 1.68 | 55 | 82 | 14 | 1 | 0 |
| 李斯特菌属 Listeria | 120 | 1.94 | 50 | 86 | 27 | 10 | 0 |
| 中慢生根瘤菌属 Mesorhizobium | 76 | 1.23 | 52 | 60 | 21 | 0 | 0 |
| 分枝杆菌属 Mycolicibacterium | 114 | 1.84 | 71 | 89 | 39 | 0 | 0 |
| 副拟杆菌属 Parabacteroides | 68 | 1.10 | 51 | 54 | 8 | 0 | 0 |
| 土球腔菌属 Pedosphaera | 50 | 0.81 | 31 | 47 | 12 | 0 | 0 |
| 普雷斯科特氏菌属 Prescottella | 77 | 1.25 | 53 | 70 | 7 | 0 | 0 |
| 原绿球藻属 Prochlorococcus | 50 | 0.81 | 39 | 32 | 3 | 0 | 0 |
| 假单胞菌属 Pseudomonas | 203 | 3.28 | 109 | 173 | 34 | 2 | 2 |
| 红杆菌属 Rhodobacter | 69 | 1.12 | 57 | 61 | 26 | 2 | 3 |
| 红育菌属 Rhodovulum | 53 | 0.86 | 34 | 45 | 14 | 2 | 2 |
| 罗格氏菌属 Ruegeria | 54 | 0.87 | 38 | 41 | 10 | 1 | 1 |
| 中华根瘤菌属 Sinorhizobium | 57 | 0.92 | 40 | 42 | 32 | 1 | 1 |
| 弧菌属 Vibrio | 129 | 2.09 | 84 | 102 | 26 | 4 | 2 |
| 未知 Unknown | 266 | 4.30 | 161 | 213 | 105 | 3 | 2 |
| 多样性指数 Diversity index | 乔灌草地 Trees, shrubs and grasslands | 灌草地 Shrub grasslands | 芦苇地 Reed marshes | 池塘 Ponds | 浅滩 Shoals |
|---|---|---|---|---|---|
| Shannon指数 Shannon index | 6.30b | 8.26a | 7.50ab | 3.88c | 5.83bc |
| Simpson指数 Simpson index | 0.95a | 0.96a | 0.88b | 0.84b | 0.86b |
| Chao1指数 Chao1 index | 15,357.66ab | 20,083.64a | 4,650.26b | 257.23c | 543.23bc |
表3 西溪湿地不同斑块类型的病毒Shannon指数、Simpson指数和Chao1指数
Table 3 Shannon index, Simpson index and Chao1 index of virus across different habitat types in Xixi Wetland
| 多样性指数 Diversity index | 乔灌草地 Trees, shrubs and grasslands | 灌草地 Shrub grasslands | 芦苇地 Reed marshes | 池塘 Ponds | 浅滩 Shoals |
|---|---|---|---|---|---|
| Shannon指数 Shannon index | 6.30b | 8.26a | 7.50ab | 3.88c | 5.83bc |
| Simpson指数 Simpson index | 0.95a | 0.96a | 0.88b | 0.84b | 0.86b |
| Chao1指数 Chao1 index | 15,357.66ab | 20,083.64a | 4,650.26b | 257.23c | 543.23bc |
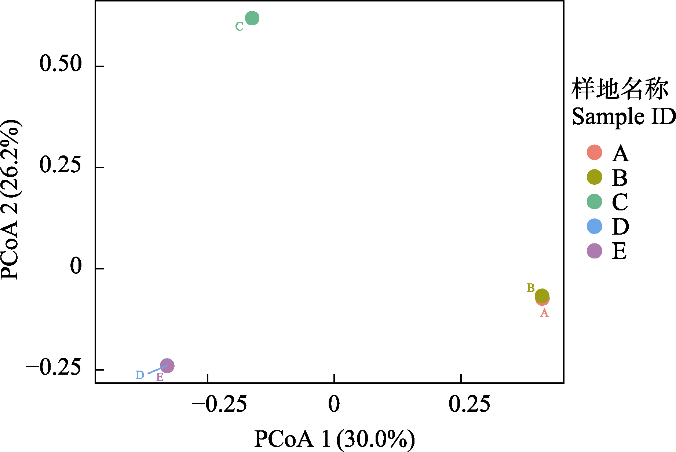
图2 西溪湿地不同斑块类型的病毒群落结构。A: 乔灌草地; B: 灌草地; C: 芦苇地; D: 池塘; E: 浅滩。
Fig. 2 Viral community structure of different habitat types in Xixi Wetland. A, Trees, shrubs and grasslands; B, Shrub grasslands; C, Reed marshes; D, Ponds; E, Shoals.
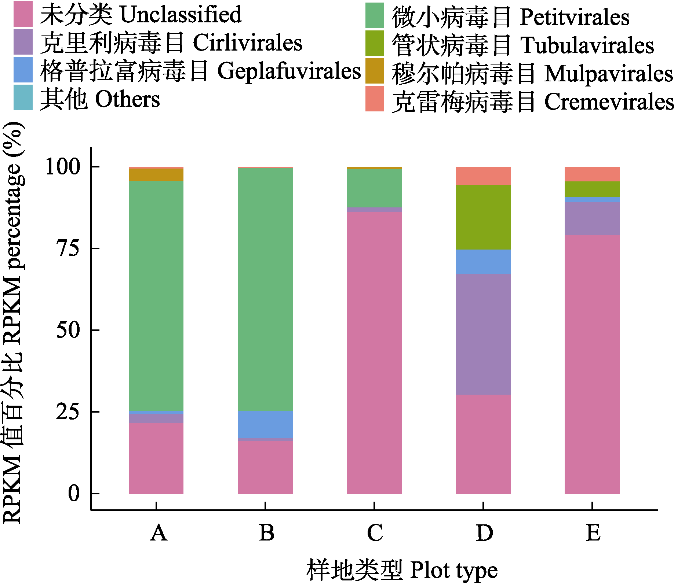
图3 西溪湿地不同斑块病毒目水平群落结构组成。RPKM: 每百万条映射读数中每千碱基的读数。
Fig. 3 Composition of horizontal community structure of viral order level in different habitat types of Xixi Wetland. RPKM, Reads per Kilobase per Million mapped reads.
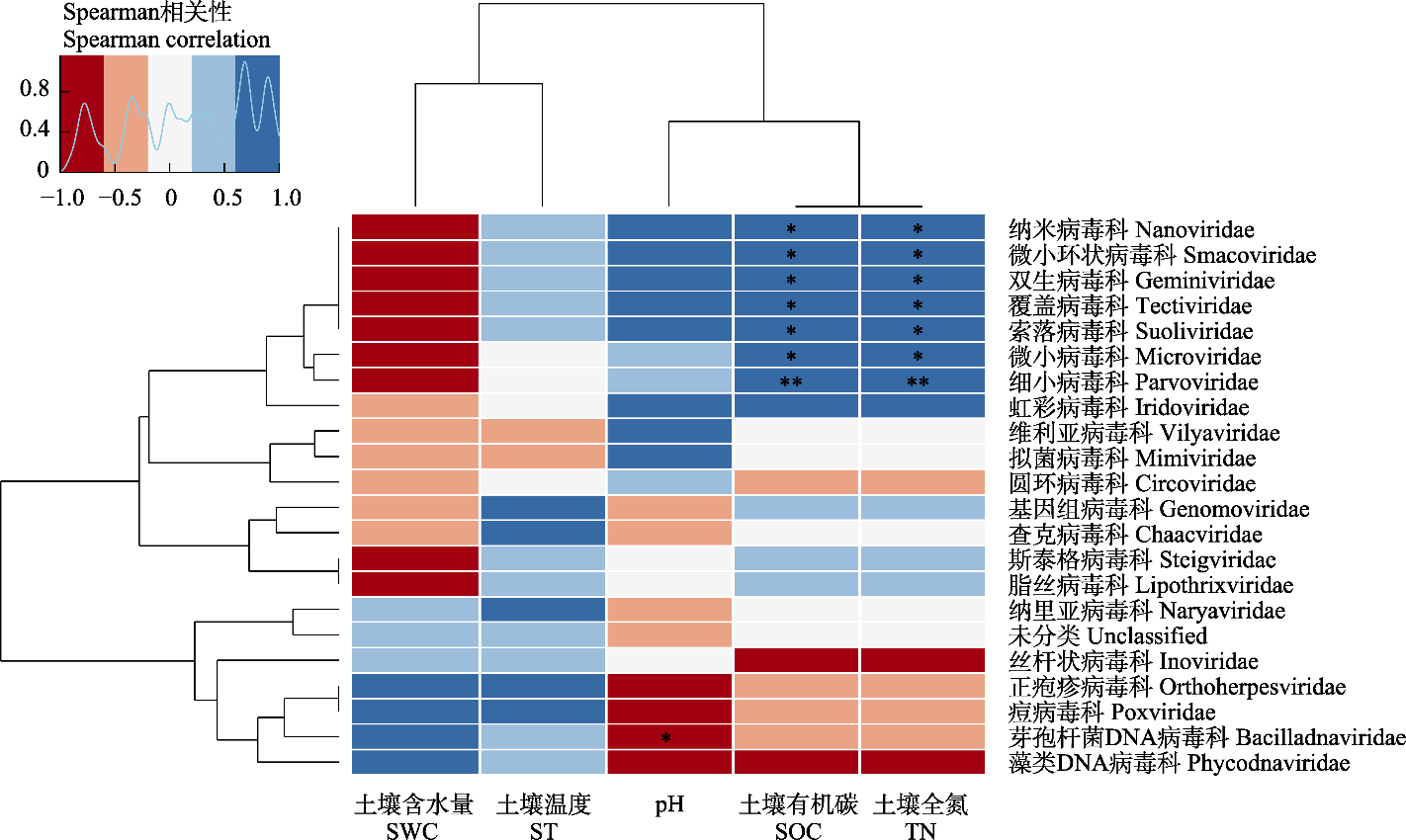
图4 西溪湿地病毒科水平与土壤理化性质的相关关系。* P < 0.05, ** P < 0.01。
Fig. 4 Correlation between viral families composition and soil physicochemical properties in Xixi Wetland. SWC, Soil water content; ST, Soil temperature; SOC, Soil organic carbon; TN, Total nitrogen. * P < 0.05, ** P < 0.01.
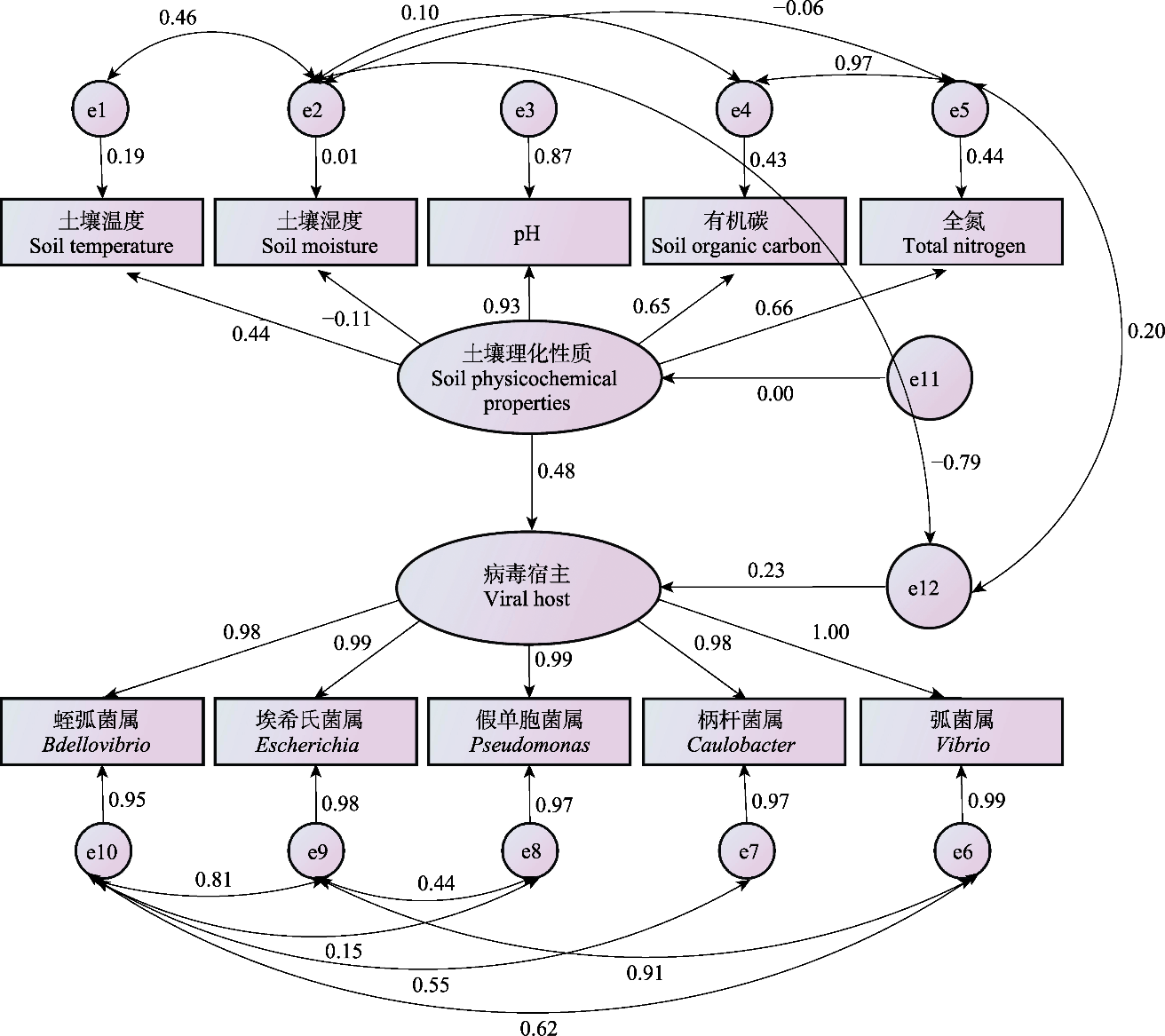
图5 土壤理化性质与病毒宿主结构方程模型。e为残差项; 单箭头为变量间的直接影响关系, 其正值代表正相关(越高相关性越大), 负值代表负相关(其绝对值越高相关性越大); 双箭头为拟合线, 其正值表示观察值大于模型预测值, 负值代表观察值小于模型预测值。
Fig. 5 The structural equation modeling of soil physicochemical properties and viral host. The e represents the residual term; The one-way arrow indicates the direct effect relationship between variables, where positive values indicate a positive correlation (the higher the value, the stronger the correlation) and negative values indicate a negative correlation (the larger the absolute value, the stronger the correlation); The double arrow indicates fitted lines, with positive values meaning observed values are greater than model-predicted values and negative values meaning observed values are less than model-predicted values.
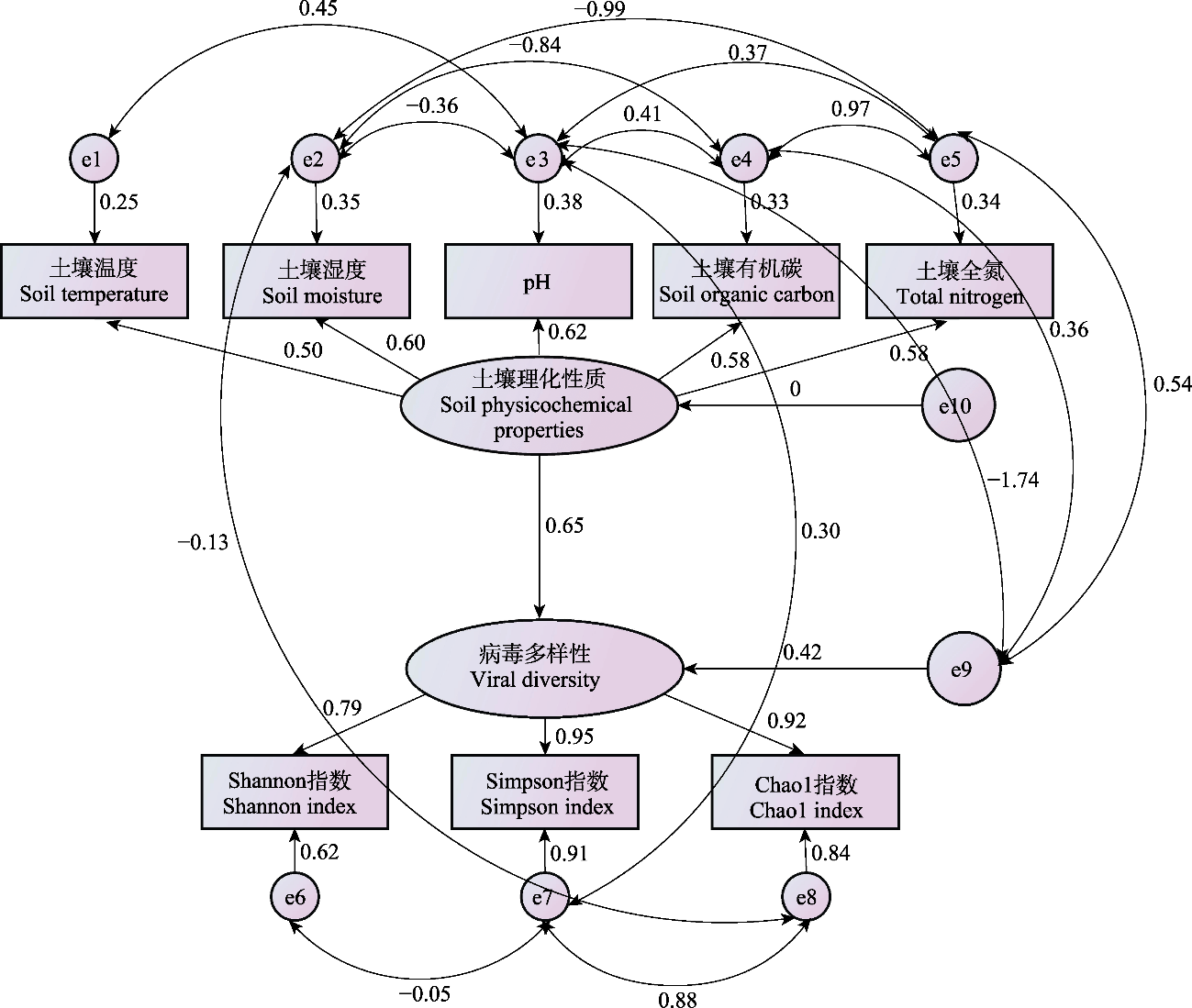
图6 土壤理化性质与病毒多样性的结构方程模型。e为残差项; 单箭头为变量间的直接影响关系, 其正值代表正相关(越高相关性越大), 负值代表负相关(其绝对值越高相关性越大); 双箭头为拟合线, 其正值表示观察值大于模型预测值, 负值代表观察值小于模型预测值。
Fig. 6 The structural equation modeling of soil physicochemical properties and viral diversity. The e represents the residual term; The one-way arrow indicates the direct effect relationship between variables, where positive values indicate a positive correlation (the higher the value, the stronger the correlation) and negative values indicate a negative correlation (the larger the absolute value, the stronger the correlation); The double arrow indicates fitted lines, with positive values meaning observed values are greater than model-predicted values and negative values meaning observed values are less than model-predicted values.
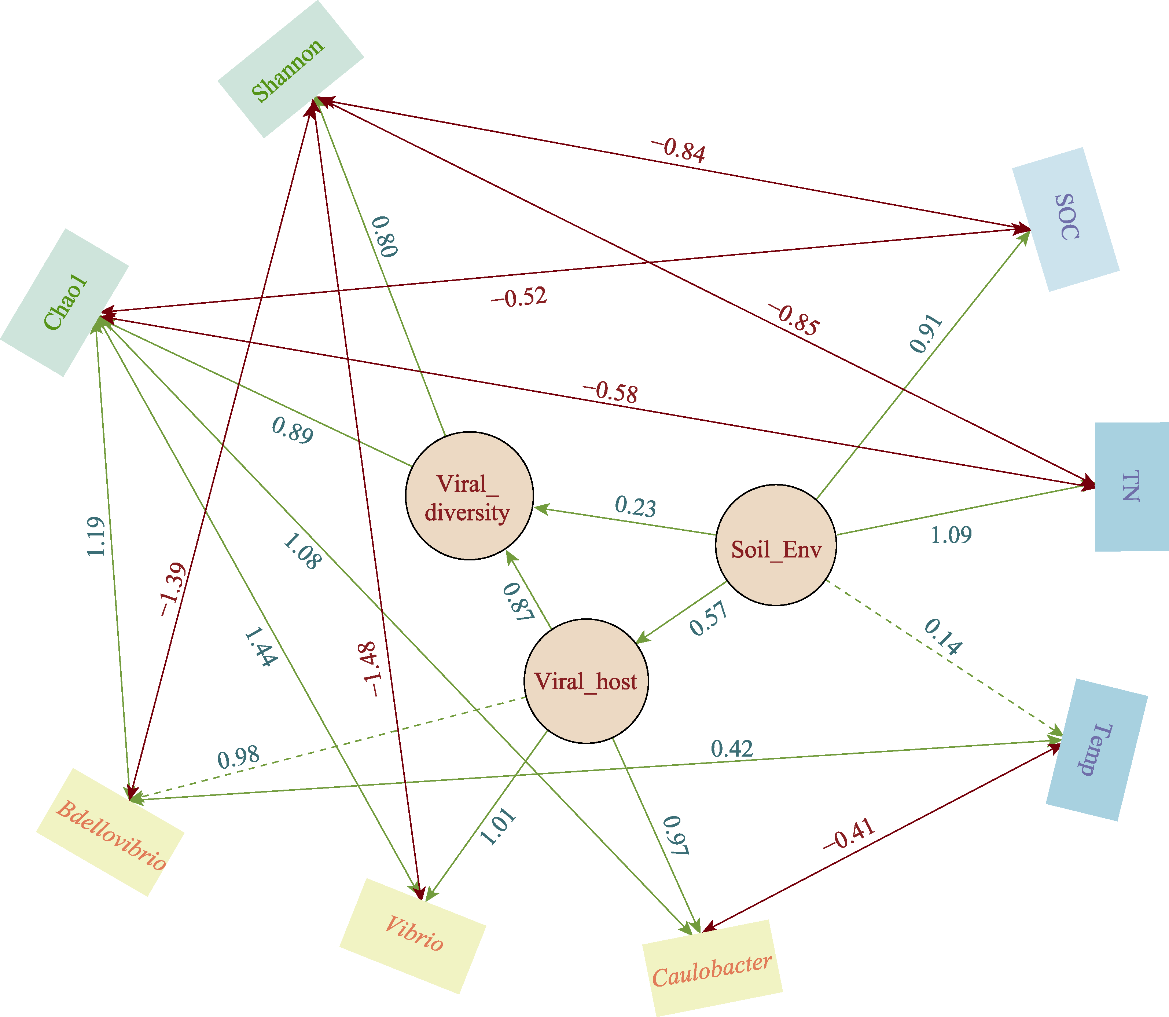
图7 土壤理化性质-病毒宿主-病毒多样性的结构方程模型。绿实线单向箭头(指向观测变量)表示直接正相关, 绿虚线单向箭头表示间接相关性, 双箭头为拟合线, 实心红线表示直接负相关; 负值代表负面(抑制性效应), 正值则代表正面效应。Bdellovibrio: 蛭弧菌属; Vibrio: 弧菌属; Caulobacter: 柄杆菌属; Chao1: Chao1指数; Shannon: Shannon指数; SOC: 土壤有机碳; TN: 土壤全氮; Temp: 温度; Viral_diversity: 病毒多样性; Viral_host: 病毒宿主; Soil_Env: 土壤环境。
Fig. 7 The structural equation modeling of soil physicochemical properties-viral host-viral diversity. A solid green one-way arrow (pointing to the observed variable) indicates a direct positive correlation, while a dashed green one-way arrow indicates an indirect correlation, and the double arrow indicates the fitting line, the solid red line indicates a direct negative correlation. Negative values represent a negative (inhibitory) effect, while positive values represent a positive effect. Chao1, Chao1 index; Shannon, Shannon index; SOC, Soil organic carbon; TN, Total nitrogen; Temp, Temperature; Viral_diversity, Viral diversity; Viral_host, Viral host; Soil_Env, Soil environment.
| [1] | Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F (2006) The marine viromes of four oceanic regions. PLoS Biology, 4, e368. |
| [2] |
Aziz RK, Breitbart M, Edwards RA (2010) Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Research, 38, 4207-4217.
DOI PMID |
| [3] |
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology, 19, 455-477.
DOI PMID |
| [4] | Bi L, Du S, Yu DT, Zhang LM, He JZ, Han LL (2021) Compositional and functional characteristics of viruses in soil under two types of land-use in Xinjiang, China. Acta Ecologica Sinica, 41, 2728-2737. (in Chinese with English abstract) |
| [毕丽, 杜帅, 于丹婷, 张丽梅, 贺纪正, 韩丽丽 (2021) 新疆两种土地利用方式下土壤病毒的群落组成与功能特征. 生态学报, 41, 2728-2737.] | |
| [5] |
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114-2120.
DOI PMID |
| [6] |
Brussaard CPD, Wilhelm SW, Thingstad F, Weinbauer MG, Bratbak G, Heldal M, Kimmance SA, Middelboe M, Nagasaki K, Paul JH, Schroeder DC, Suttle CA, Vaqué D, Wommack KE (2008) Global-scale processes with a nanoscale drive: The role of marine viruses. The ISME Journal, 2, 575-578.
DOI URL |
| [7] |
Córdova-Kreylos AL, Cao YP, Green PG, Hwang HM, Kuivila KM, Lamontagne MG, Van De Werfhorst LC, Holden PA, Scow KM (2006) Diversity, composition, and geographical distribution of microbial communities in California salt marsh sediments. Applied and Environmental Microbiology, 72, 3357-3366.
DOI PMID |
| [8] | Dell’Anno A, Corinaldesi C, Danovaro R (2015) Virus decomposition provides an important contribution to benthic deep-sea ecosystem functioning. Proceedings of the National Academy of Sciences, USA, 112, E2014-E2019. |
| [9] |
Ding J, Wang Y, Yu S (2025) Ecotone-driven vegetation transitions reshape soil nitrogen cycling functional genes in black soils of Northeast China. Biology, 14, 1474.
DOI URL |
| [10] | Gao RC, Hu M, Li FB, Chen GH, Fang LP (2022) Research progress and ecological function of phages in soil. Journal of South China Agricultural University, 43, 1-11. (in Chinese with English abstract) |
| [高瑞川, 胡敏, 李芳柏, 陈冠虹, 方利平 (2022) 土壤噬菌体的研究进展及生态功能解析. 华南农业大学学报, 43, 1-11.] | |
| [11] |
Guo JR, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, Akbar Pratama, Mart Krupovic, Dean V, Sullivan BM (2021) VirSorter2: A multi-classifier, expert-guided approach to detect diverse DNA and RNAviruses. Microbiome, 9, 37.
DOI URL |
| [12] | He L, Yan YT, Yuan CY, Lin QS, Yu DT (2024) Characteristics of soil viral communities in Cunninghamia lanceolata plantations with different stand ages. Chinese Journal of Applied Ecology, 35, 2543-2551. (in Chinese with English abstract) |
|
[和莉, 严雨亭, 袁程昱, 林秋沙, 于丹婷 (2024) 不同林龄杉木人工林土壤病毒群落特征. 应用生态学报, 35, 2543-2551.]
DOI |
|
| [13] | Hulo C, de Castro E, Masson P, Bougueleret L, Bairoch A, Xenarios I, Le Mercier P (2011) ViralZone: A knowledge resource to understand virus diversity. Nucleic Acids Research, 39, D576-D582. |
| [14] | Jiang JZ, Yuan WG, Shang J, Ying HS, Yang LL, Li M, Zhang P, Jiang T, Sun YN, Li HY (2023) Virus classification for viral genomic fragments using PhaGCN2. Brief Bioinformatics, 24, bbac505. |
| [15] |
Kuzyakov Y, Mason-Jones K (2018) Viruses in soil: Nano-scale undead drivers of microbial life, biogeochemical turnover and ecosystem functions. Soil Biology and Biochemistry, 127, 305-317.
DOI URL |
| [16] |
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754-1760.
DOI PMID |
| [17] |
Li RQ, Li YR, Kristiansen K, Wang J (2008) SOAP: Short oligonucleotide alignment program. Bioinformatics, 24, 713-714.
DOI PMID |
| [18] |
Liang C, Amelung W, Lehmann J, Kästner M (2019) Quantitative assessment of microbial necromass contribution to soil organic matter. Global Change Biology, 25, 3578-3590.
DOI PMID |
| [19] |
Liang C, Schimel JP, Jastrow JD (2017) The importance of anabolism in microbial control over soil carbon storage. Nature Microbiology, 2, 17105.
DOI PMID |
| [20] |
Lu CY, Zhang Z, Cai ZN, Zhu ZZ, Qiu Y, Wu AP, Jiang TJ, Zheng HP, Peng YS (2021) Prokaryotic virus host predictor: A Gaussian model for host prediction of prokaryotic viruses in metagenomics. BMC Biology, 19, 5.
DOI PMID |
| [21] | Lü MZ, Sheng LX, Zhang L (2013) A review on carbon fluxes for typical wetlands in different climates of China. Wetland Science, 11, 114-120. (in Chinese with English abstract) |
| [吕铭志, 盛连喜, 张立 (2013) 中国典型湿地生态系统碳汇功能比较. 湿地科学, 11, 114-120.] | |
| [22] |
Ma B, Lu CY, Wang YL, Yu JW, Zhao KK, Xue R, Ren H, Lv XF, Pan RH, Zhang JB, Zhu YG, Xu JM (2023) A genomic catalogue of soil microbiomes boosts mining of biodiversity and genetic resources. Nature Communications, 14, 7318.
DOI PMID |
| [23] | Man BY, Xiang X, Luo Y, Mao XT, Zhang C, Sun BH, Wang X (2021) Characteristics and influencing factors of soil fungal community of typical vegetation types in Mount Huangshan, East China. Mycosystema, 40, 2735-2751. (in Chinese with English abstract) |
|
[满百膺, 向兴, 罗洋, 毛小涛, 张超, 孙丙华, 王希 (2021) 黄山典型植被类型土壤真菌群落特征及其影响因素. 菌物学报, 40, 2735-2751.]
DOI |
|
| [24] |
Mao YE, Zhou XM, Wang N, Li XX, You YK, Bai SB (2023) Impact of Phyllostachys edulis expansion to Chinese fir forest on the soil bacterial community. Biodiversity Science, 31, 22659. (in Chinese with English abstract)
DOI URL |
|
[毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌 (2023) 毛竹扩张对杉木林土壤细菌群落的影响. 生物多样性, 31, 22659.]
DOI |
|
| [25] |
Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC (2021) CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nature Biotechnology, 39, 578-585.
DOI PMID |
| [26] |
Peng Y, Leung HCM, Yiu SM, Chin FYL (2012) IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics, 28, 1420-1428.
DOI PMID |
| [27] | Qin JQ, Xiao ZR, Ming AG, Zhu H, Teng JQ, Liang ZL, Tao Y, Qin L (2023) Effect of monoculture and mixed plantation with coniferous and broadleaved tree species on soil microbial carbon cycle functional gene abundance. Ecology and Environmental Sciences, 32, 1719-1731. (in Chinese with English abstract) |
|
[秦佳琪, 肖指柔, 明安刚, 朱豪, 滕金倩, 梁泽丽, 陶怡, 覃林 (2023) 针阔人工混交林及其纯林对土壤微生物碳循环功能基因丰度的影响. 生态环境学报, 32, 1719-1731.]
DOI |
|
| [28] |
Sakschewski B, von Bloh W, Boit A, Poorter L, Peña-Claros M, Heinke J, Joshi J, Thonicke K (2016) Resilience of Amazon forests emerges from plant trait diversity. Nature Climate Change, 6, 1032-1036.
DOI |
| [29] |
Seemann T (2014) Prokka: Rapid prokaryotic genome annotation. Bioinformatics, 30, 2068-2069.
DOI PMID |
| [30] | Shang JY, Sun YN (2022) CHERRY: A computational metHod for accuratE pRediction of virus-pRokarYotic interactions using a graph encoder-decoder model. Briefings in Bioinformatics, 23, bbac182. |
| [31] |
Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE (2008) Phages across the biosphere: Contrasts of viruses in soil and aquatic environments. Research in Microbiology, 159, 349-357.
DOI PMID |
| [32] |
Suttle CA (2005) Viruses in the sea. Nature, 437, 356-361.
DOI |
| [33] |
Trubl G, Hyman P, Roux S, Abedon ST (2020) Coming-of-age characterization of soil viruses: A user’s guide to virus isolation, detection within metagenomes, and viromics. Soil Systems, 4, 23.
DOI URL |
| [34] | Wang GH, Liu JJ, Zhu D, Ye M, Zhu YG (2020) A review of researches on viruses in soil, advancement and challenges. Acta Pedologica Sinica, 57, 1319-1332. (in Chinese with English abstract) |
| [王光华, 刘俊杰, 朱冬, 叶茂, 朱永官 (2020) 土壤病毒的研究进展与挑战. 土壤学报, 57, 1319-1332.] | |
| [35] | Wu HQ, Ruan CJ, Han M, Wang G (2024) Mystery of soil viruses: Advances, challenges, and perspectives. Acta Microbiologica Sinica, 64, 1824-1847. (in Chinese with English abstract) |
| [吴汉卿, 阮楚晋, 韩苗, 王钢 (2024) 土壤病毒之奥秘: 研究进展、挑战及未来展望. 微生物学报, 64, 1824-1847.] | |
| [36] | Yu HT, Huang SH, Liu YJ, Kou WB, Liu YZ, Wu L (2017) Profile distribution characteristics of soil enzymes and microbial biomass in the Poyang Lake Wetland. Research of Environmental Sciences, 30, 1715-1722. (in Chinese with English abstract) |
| [于昊天, 黄时豪, 刘亚军, 寇文伯, 刘以珍, 吴兰 (2017) 鄱阳湖湿地土壤酶及微生物生物量的剖面分布特征. 环境科学研究, 30, 1715-1722.] | |
| [37] | Zhai XJ, Cui LJ, Li W, Zhao XS, Zhang MY, Kang XM (2024) Carbon sequestration by typical wetland ecosystems in China. Journal of Hydroecology, 45, 76-85. (in Chinese with English abstract) |
| [翟夏杰, 崔丽娟, 李伟, 赵欣胜, 张曼胤, 康晓明 (2024) 中国典型湿地生态系统的固碳价值研究. 水生态学杂志, 45, 76-85.] | |
| [38] |
Zhou GX, Chen L, Zhang CZ, Ma DH, Zhang JB (2023) Bacteria-virus interactions are more crucial in soil organic carbon storage than iron protection in biochar-amended paddy soils. Environmental Science & Technology, 57, 19713-19722.
DOI URL |
| [1] | 孙济伦, 谢将剑, 张长春, 张军国. 基于YOLO-DAS模型的湿地水鸟检测方法: 以内蒙古南海子湿地为例[J]. 生物多样性, 2025, 33(9): 25237-. |
| [2] | 王欣, 鲍风宇. 基于鸟类多样性提升的南滇池国家湿地公园生态修复效果[J]. 生物多样性, 2025, 33(5): 24531-. |
| [3] | 张明燡, 王晓梅, 郑言鑫, 吴楠, 李东浩, 樊恩源, 李娜, 单秀娟, 于涛, 赵春暖, 李波, 徐帅, 吴玉萍, 任利群. 黄河口典型牡蛎礁分布区资源状况和栖息地功能[J]. 生物多样性, 2025, 33(4): 24208-. |
| [4] | 陈丁松, 刘子恺, 贺子洋, 陈伟东. 缓步动物多样性、分布特征和生态功能研究进展[J]. 生物多样性, 2025, 33(2): 24406-. |
| [5] | 宋远昊, 龚吕, 李贲, 胡阳, 李秀珍. 辽河口不同退塘还湿方式对大型底栖动物的影响[J]. 生物多样性, 2025, 33(2): 24316-. |
| [6] | 刘源, 杜剑卿, 马丽媛, 杨刚, 田建卿. 纳木措流域岸边带湿地产甲烷古菌群落多样性与分布特征[J]. 生物多样性, 2025, 33(1): 24247-. |
| [7] | 张乃鹏, 梁洪儒, 张焱, 孙超, 陈勇, 王路路, 夏江宝, 高芳磊. 土壤类型和地下水埋深对黄河三角洲典型盐沼植物群落空间分异的影响[J]. 生物多样性, 2024, 32(2): 23370-. |
| [8] | 胡婉君, 郝泽周, 夏灿玮, 谢将剑. 湿地声景录音策略及面向声景分类的特征选择[J]. 生物多样性, 2024, 32(10): 24121-. |
| [9] | 吴浩, 余玉蓉, 王佳钰, 赵媛博, 高娅菲, 李小玲, 卜贵军, 薛丹, 吴林. 低水位增加灌木多样性和生物量但降低土壤有机碳含量: 以鄂西南贫营养泥炭地为例[J]. 生物多样性, 2023, 31(3): 22600-. |
| [10] | 黄雨菲, 路春燕, 贾明明, 王自立, 苏越, 苏艳琳. 基于无人机影像与面向对象-深度学习的滨海湿地植物物种分类[J]. 生物多样性, 2023, 31(3): 22411-. |
| [11] | 王士政, 孙翊斐, 李珍珍, 舒越, 冯佳伟, 王天明. 鸟类迁徙对图们江下游湿地声景时间格局的影响[J]. 生物多样性, 2023, 31(1): 22337-. |
| [12] | 汪婷, 周立志. 合肥市小微湿地鸟类多样性的时空格局及其影响因素[J]. 生物多样性, 2022, 30(7): 21445-. |
| [13] | 付裕, 黄康祥, 蔡锦枫, 陈慧敏, 任久生, 万松泽, 张扬, 任珩, 毛瑢, 石福习. 三江平原沼泽湿地4种优势植物空间格局对不同水位环境的响应[J]. 生物多样性, 2022, 30(3): 21392-. |
| [14] | 姚保民, 曾青, 张丽梅. 土壤原生生物多样性及其生态功能研究进展[J]. 生物多样性, 2022, 30(12): 22353-. |
| [15] | 高洁, 李德浩, 姜海波, 邓光怡, 张超凡, 何春光, 孙鹏. 白鹤利用农田作为中转取食生境的原因[J]. 生物多样性, 2022, 30(11): 22093-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2026 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()