



生物多样性 ›› 2025, Vol. 33 ›› Issue (8): 25072. DOI: 10.17520/biods.2025072 cstr: 32101.14.biods.2025072
范平1,2( ), 王欢2, 温知新2(
), 王欢2, 温知新2( ), 宋刚2(
), 宋刚2( ), 雷富民2,*(
), 雷富民2,*( )
)
收稿日期:2025-02-27
接受日期:2025-06-25
出版日期:2025-08-20
发布日期:2025-09-17
通讯作者:
*E-mail: leifm@ioz.ac.cn
基金资助:
Ping Fan1,2( ), Huan Wang2, Zhixin Wen2(
), Huan Wang2, Zhixin Wen2( ), Gang Song2(
), Gang Song2( ), Fuming Lei2,*(
), Fuming Lei2,*( )
)
Received:2025-02-27
Accepted:2025-06-25
Online:2025-08-20
Published:2025-09-17
Contact:
*E-mail: leifm@ioz.ac.cn
Supported by:摘要:
探讨遗传多样性与物种分布面积之间的关系及其成因, 对于生物多样性保护策略的制定具有举足轻重的意义。遗传多样性蕴含着物种对气候变化的潜在适应能力, 而物种分布面积则是气候变化与物种适应能力共同作用的结果。因此, 气候因子可能是连接遗传多样性和物种分布面积的关键纽带, 对两者之间的关系起到重要的调节作用。为了验证这一假设, 本研究以全球鸟类为研究对象, 选取细胞色素c氧化酶亚基I (COI)基因片段为分子遗传标记, 分析了单倍型多样性和核苷酸多样性这两个遗传多样性关键指标, 探讨8个气候因子(年均温、年均温范围、年降水、年降水范围、降水季节性、降水季节性范围、温度季节性、温度季节性范围)在单倍型多样性-物种分布面积关系和核苷酸多样性-物种分布面积关系中的作用。研究结果表明, 鸟类总体的核苷酸多样性为0.008 ± 0.001 (平均值 ± 标准误差), 单倍型多样性为0.699 ± 0.011。气候因子对核苷酸多样性与单倍型多样性表征的遗传多样性-物种分布面积关系的影响不同: 对于单倍型多样性而言, 气候因子主要通过影响物种的分布面积来调节单倍型多样性-分布面积关系(df = 6, χ2 = 10.77, AIC = 2,231.8, BIC = 2,270.5); 而对于核苷酸多样性, 气候因子可以同时通过影响物种分布面积和核苷酸多样性来调节多样性-分布面积关系(df = 0, χ2 = 0, AIC = 2,155.0, BIC = 2,219.6)。基于上述结果, 我们建议在探讨气候变化对遗传多样性的影响时, 应综合考虑不同的遗传多样性指标, 以便制定更为有效的生物多样性保护策略。
范平, 王欢, 温知新, 宋刚, 雷富民 (2025) 气候因子对鸟类遗传多样性与物种分布面积关系的影响. 生物多样性, 33, 25072. DOI: 10.17520/biods.2025072.
Ping Fan, Huan Wang, Zhixin Wen, Gang Song, Fuming Lei (2025) Impact of climatic factors on the genetic diversity-species area relationship of birds. Biodiversity Science, 33, 25072. DOI: 10.17520/biods.2025072.
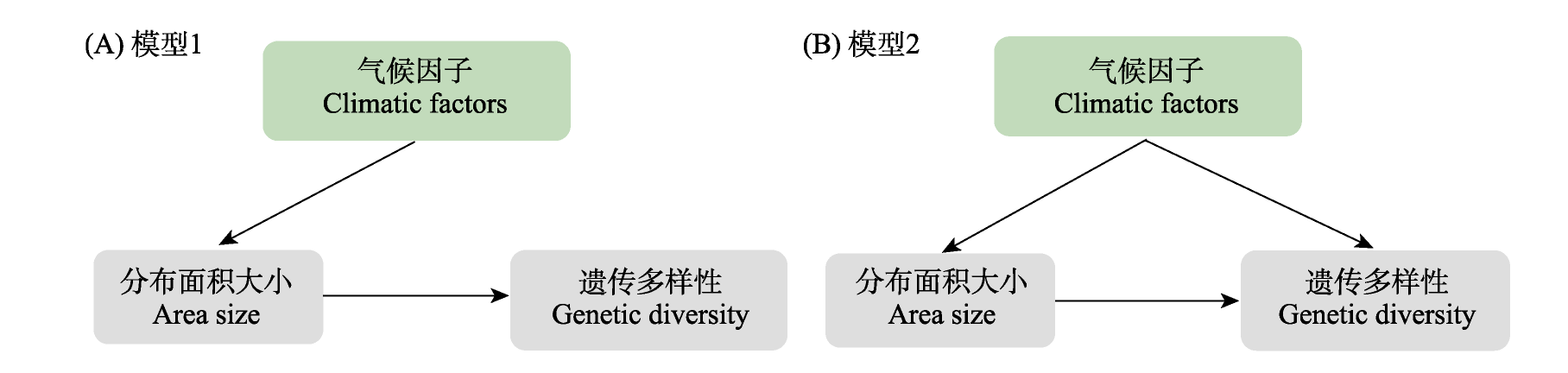
图1 结构方程模型检测的气候因子、分布面积大小和遗传多样性之间的关系。黑色箭头展示了两个因子之间的相互关系。(A)模型1: 遗传多样性与物种分布面积大小直接关联, 其中物种分布面积大小受气候因子的直接影响; 遗传多样性不受气候因子的直接影响; (B)模型2: 遗传多样性与物种分布面积大小直接关联, 物种分布面积大小与遗传多样性均直接受到气候因子的影响。
Fig. 1 Models tested by structural equation models of the relationships among genetic diversity, area size, and climatic factors. Black arrows indicate the relationships between each two factors. (A) Model 1: Climatic factors are directly associated with area size of species, while area size is directly associated with genetic diversity. (B) Model 2: Climatic factors are directly associated with both area size and genetic diversity, and area size is directly associated with genetic diversity.
| 模型 Models | df | χ2 | P | AIC | BIC | |
|---|---|---|---|---|---|---|
| 单倍型多样性 Haplotype diversity | 模型1 Model 1 | 6 | 10.77 | > 0.05 | 2,231.8 | 2,270.5 |
| 模型2 Model 2 | 0 | 0 | 1 | 2,233.0 | 2,297.5 | |
| 核苷酸多样性 Nucleotide diversity | 模型1 Model 1 | 6 | 87.809 | < 0.05 | 2,230.8 | 2,269.6 |
| 模型2 Model 2 | 0 | 0 | 1 | 2,155.0 | 2,219.6 |
表1 鸟类整体结构方程模型结果
Table 1 Structural equation model results of birds
| 模型 Models | df | χ2 | P | AIC | BIC | |
|---|---|---|---|---|---|---|
| 单倍型多样性 Haplotype diversity | 模型1 Model 1 | 6 | 10.77 | > 0.05 | 2,231.8 | 2,270.5 |
| 模型2 Model 2 | 0 | 0 | 1 | 2,233.0 | 2,297.5 | |
| 核苷酸多样性 Nucleotide diversity | 模型1 Model 1 | 6 | 87.809 | < 0.05 | 2,230.8 | 2,269.6 |
| 模型2 Model 2 | 0 | 0 | 1 | 2,155.0 | 2,219.6 |
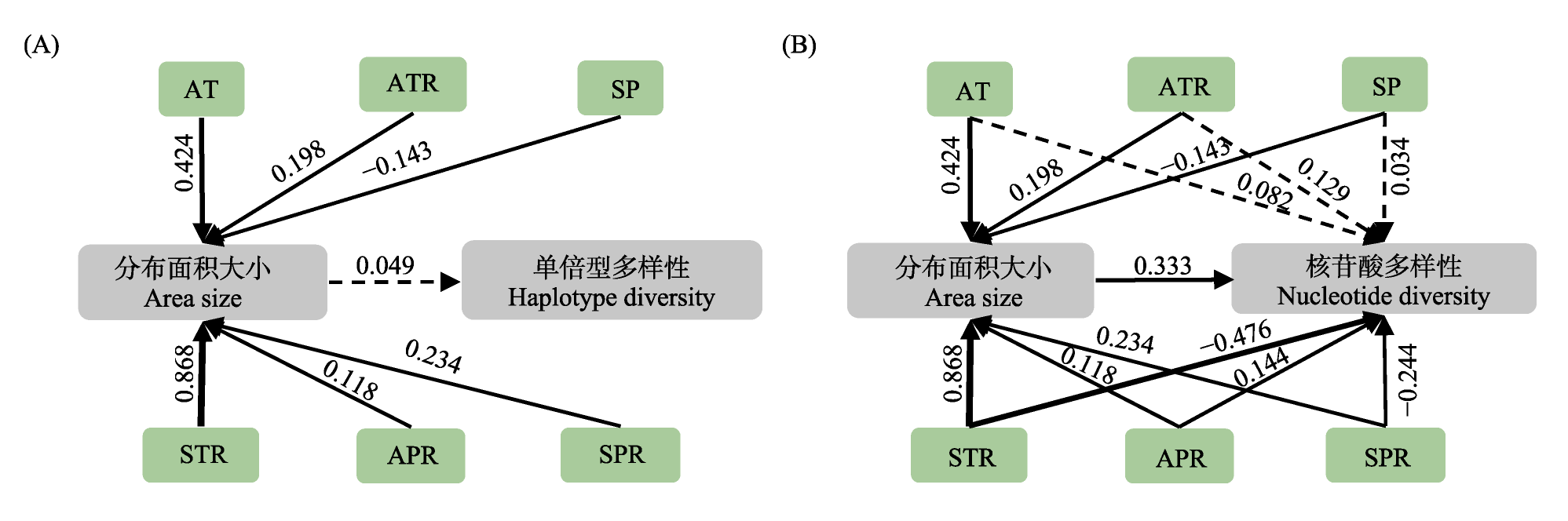
图3 气候因子、分布面积和遗传多样性(A: 单倍型多样性; B: 核苷酸多样性)的最优结构方程模型。黑色实线/虚线箭头分别表示类群特异的显著/不显著的相关方向。图中线段的粗细对应于其所标注的标准化路径系数的大小。图中涉及的气候因子为年均温(AT)、年均温范围(ATR)、温度季节性范围(STR)、年降水范围(APR)、降水季节性(SP)和降水季节性范围(SPR)。
Fig. 3 The optimal structural equation model illustrating the relationships among climatic factors, area size, and genetic diversity (A, Haplotype diversity; B, Nucleotide diversity). Black solid/dashed arrows indicate correlations with significant/non-significant directions. The thickness of each line segment in the diagram is proportionally representative of the magnitude of the standardized path coefficient indicated adjacent to it. The climatic factors used in this study are annual temperature (AT), annual temperature range (ATR), temperature seasonality range (STR), annual precipitation range (APR), precipitation seasonality (SP), and precipitation seasonality range (SPR).
| 生态类群 Ecological groups | 模型1 Model 1 | 模型2 Model 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| df | χ2 | P | AIC | BIC | df | χ2 | P | AIC | BIC | ||
| 单倍型多样性 Haplotype diversity | 游禽 Natatores (n = 34) | 6 | 10.075 | > 0.05 | 148.732 | 162.469 | 0 | 0 | 1 | 150.657 | 173.552 |
| 涉禽 Wading birds (n = 40) | 6 | 10.986 | > 0.05 | 172.220 | 187.420 | 0 | 0 | 1 | 173.234 | 198.568 | |
| 陆禽 Landfowl (n = 30) | 6 | 4.882 | > 0.05 | 143.666 | 156.277 | 0 | 0 | 1 | 150.784 | 171.802 | |
| 攀禽 Climbing birds ( n = 73) | 6 | 10.601 | > 0.05 | 327.464 | 348.078 | 0 | 0 | 1 | 328.863 | 363.220 | |
| 猛禽 Raptors (n = 32) | 6 | 6.707 | > 0.05 | 130.392 | 143.584 | 0 | 0 | 1 | 135.685 | 157.671 | |
| 鸣禽 Songbirds (n = 377) | 6 | 5.264 | > 0.05 | 1,392.849 | 1,427.849 | 0 | 0 | 1 | 399.585 | 1,456.752 | |
| 核苷酸多样性 Nucleotide diversity | 游禽 Natatores (n = 34) | 6 | 6.020 | > 0.05 | 151.392 | 165.129 | 0 | 0 | 1 | 157.372 | 180.268 |
| 涉禽 Wading birds (n = 40) | 6 | 11.259 | > 0.05 | 170.348 | 185.548 | 0 | 0 | 1 | 171.089 | 196.422 | |
| 陆禽 Landfowl (n = 30) | 6 | 8.130 | > 0.05 | 138.462 | 151.073 | 0 | 0 | 1 | 142.332 | 163.350 | |
| 攀禽 Climbing birds (n = 73) | 6 | 21.202 | > 0.05 | 327.575 | 348.189 | 0 | 0 | 1 | 318.373 | 352.730 | |
| 猛禽 Raptors (n = 32) | 6 | 10.006 | > 0.05 | 129.977 | 143.169 | 0 | 0 | 1 | 131.972 | 153.958 | |
| 鸣禽 Songbirds (n = 377) | 6 | 71.636 | > 0.05 | 1,395.794 | 1,430.094 | 0 | 0 | 1 | 1,336.158 | 1,393.325 | |
表2 鸟类6种生态类群结构方程模型结果
Table 2 Results of the structural equation model for the six major ecological groups of birds
| 生态类群 Ecological groups | 模型1 Model 1 | 模型2 Model 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| df | χ2 | P | AIC | BIC | df | χ2 | P | AIC | BIC | ||
| 单倍型多样性 Haplotype diversity | 游禽 Natatores (n = 34) | 6 | 10.075 | > 0.05 | 148.732 | 162.469 | 0 | 0 | 1 | 150.657 | 173.552 |
| 涉禽 Wading birds (n = 40) | 6 | 10.986 | > 0.05 | 172.220 | 187.420 | 0 | 0 | 1 | 173.234 | 198.568 | |
| 陆禽 Landfowl (n = 30) | 6 | 4.882 | > 0.05 | 143.666 | 156.277 | 0 | 0 | 1 | 150.784 | 171.802 | |
| 攀禽 Climbing birds ( n = 73) | 6 | 10.601 | > 0.05 | 327.464 | 348.078 | 0 | 0 | 1 | 328.863 | 363.220 | |
| 猛禽 Raptors (n = 32) | 6 | 6.707 | > 0.05 | 130.392 | 143.584 | 0 | 0 | 1 | 135.685 | 157.671 | |
| 鸣禽 Songbirds (n = 377) | 6 | 5.264 | > 0.05 | 1,392.849 | 1,427.849 | 0 | 0 | 1 | 399.585 | 1,456.752 | |
| 核苷酸多样性 Nucleotide diversity | 游禽 Natatores (n = 34) | 6 | 6.020 | > 0.05 | 151.392 | 165.129 | 0 | 0 | 1 | 157.372 | 180.268 |
| 涉禽 Wading birds (n = 40) | 6 | 11.259 | > 0.05 | 170.348 | 185.548 | 0 | 0 | 1 | 171.089 | 196.422 | |
| 陆禽 Landfowl (n = 30) | 6 | 8.130 | > 0.05 | 138.462 | 151.073 | 0 | 0 | 1 | 142.332 | 163.350 | |
| 攀禽 Climbing birds (n = 73) | 6 | 21.202 | > 0.05 | 327.575 | 348.189 | 0 | 0 | 1 | 318.373 | 352.730 | |
| 猛禽 Raptors (n = 32) | 6 | 10.006 | > 0.05 | 129.977 | 143.169 | 0 | 0 | 1 | 131.972 | 153.958 | |
| 鸣禽 Songbirds (n = 377) | 6 | 71.636 | > 0.05 | 1,395.794 | 1,430.094 | 0 | 0 | 1 | 1,336.158 | 1,393.325 | |
| [1] |
Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F (2012) Impacts of climate change on the future of biodiversity. Ecology Letters, 15, 365-377.
DOI PMID |
| [2] |
Blomberg SP, Garland TJ, Ives AR (2003) Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution, 57, 717-745.
DOI PMID |
| [3] |
Brambilla M, Scridel D, Bazzi G, Ilahiane L, Iemma A, Pedrini P, Bassi E, Bionda R, Marchesi L, Genero F, Teufelbauer N, Probst R, Vrezec A, Kmecl P, Mihelič T, Bogliani G, Schmid H, Assandri G, Pontarini R, Braunisch V, Arlettaz R, Chamberlain D (2020) Species interactions and climate change: How the disruption of species co-occurrence will impact on an avian forest guild. Global Change Biology, 26, 1212-1224.
DOI PMID |
| [4] |
Chapin III FS, Zavaleta ES, Eviner VT, Naylor RL, Vitousek PM, Reynolds HL, Hooper DU, Lavorel S, Sala OE, Hobbie SE, Mack MC, Díaz SM (2000) Consequences of changing biodiversity. Nature, 405, 234-242.
DOI |
| [5] | Chen IC, Hill JK, Ohlemüller R, Roy DB, Thomas CD (2011) Rapid range shifts of species associated with high levels of climate warming. Science, 333, 1024-1026. |
| [6] |
Cramer MJ, Willig MR (2005) Habitat heterogeneity, species diversity and null models. Oikos, 108, 209-218.
DOI URL |
| [7] |
De Kort H, Prunier JG, Ducatez S, Honnay O, Baguette M, Stevens VM, Blanchet S (2021) Life history, climate and biogeography interactively affect worldwide genetic diversity of plant and animal populations. Nature Communications, 12, 516.
DOI PMID |
| [8] | DeAngelis DL (1995) Relationships Between the Energetics of Species and Large-scale Species Richness. Springer, New York. |
| [9] |
Doyle JM, Hacking CC, Willoughby JR, Sundaram M, DeWoody JA (2015) Mammalian genetic diversity as a function of habitat, body size, trophic class, and conservation status. Journal of Mammalogy, 96, 564-572.
DOI URL |
| [10] |
Dziak JJ, Coffman DL, Lanza ST, Li RZ, Jermiin LS (2020) Sensitivity and specificity of information criteria. Briefings in Bioinformatics, 21, 553-565.
DOI PMID |
| [11] |
Evans MC, Jarman PJ (1999) Diets and feeding selectivities of bridled nailtail wallabies and black-striped wallabies. Wildlife Research, 26, 1-19
DOI URL |
| [12] |
Fan H, Zhang Q, Rao J, Cao J, Lu X (2019) Genetic diversity-area relationships across bird species. The American Naturalist, 194, 736-740.
DOI PMID |
| [13] |
Fan P, Fjeldså J, Liu X, Dong YF, Chang YB, Qu YH, Song G, Lei FM (2021) An approach for estimating haplotype diversity from sequences with unequal lengths. Methods in Ecology and Evolution, 12, 1658-1667.
DOI URL |
| [14] | Fan P, Song G, Qiao HJ, Zhang DZ, Ji YZ, Qu YH, Fjeldså J, Lei FM (2025) Revaluation of the genetic diversity-area relationship by integrating nucleotide and haplotype diversity. Current Zoology, doi: 10.1093/cz/zoae078. |
| [15] |
Fan P, Wen ZX, Song G (2025) The effect of climatic factors and anthropogenic activities on different genetic diversity indicators of amphibians and mammals. Biodiversity Science, 33, 25022. (in Chinese with English abstract)
DOI |
|
[范平, 温知新, 宋刚 (2025) 气候因子和人类活动对两栖及哺乳动物不同遗传多样性指标的影响. 生物多样性, 33, 25022.]
DOI |
|
| [16] |
Gillman LN, Wright SD (2014) Species richness and evolutionary speed: The influence of temperature, water and area. Journal of Biogeography, 41, 39-51.
DOI URL |
| [17] |
Graham RW, Lundelius EL, Graham MA, Schroeder EK, Toomey RS, Anderson E, Barnosky AD, Burns JA, Churcher CS, Grayson DK, Guthrie RD, Harington CR, Jefferson GT, Martin LD, McDonald HG, Morlan RE, Semken HA, Webb SD, Werdelin L, Wilson MC (1996) Spatial response of mammals to late quaternary environmental fluctuations. Science, 272, 1601-1606.
PMID |
| [18] |
Grant W, Bowen B (1998) Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89, 415-426.
DOI URL |
| [19] |
Gratton P, Marta S, Bocksberger G, Winter M, Trucchi E, Kühl H (2017) A world of sequences: Can we use georeferenced nucleotide databases for a robust automated phylogeography? Journal of Biogeography, 44, 475-486.
DOI URL |
| [20] | Haberl H, Erb KH, Krausmann F, Gaube V, Bondeau A, Plutzar C, Gingrich S, Lucht W, Fischer-Kowalski M (2007) Quantifying and mapping the human appropriation of net primary production in earth’s terrestrial ecosystems. Proceedings of the National Academy of Sciences, USA, 104, 12942-12947. |
| [21] |
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society, 58, 247-276.
DOI URL |
| [22] |
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965-1978.
DOI URL |
| [23] | Hof C (2021) Towards more integration of physiology, dispersal and land-use change to understand the responses of species to climate change. The Journal of Experimental Biology, 224, jeb238352. |
| [24] | Hofreiter M, Rödder D, Lawing AM, Flecks M, Ahmadzadeh F, Dambach J, Engler JO, Habel JC, Hartmann T, Hörnes D, Ihlow F, Schidelko K, Stiels D, Polly PD (2013) Evaluating the significance of paleophylogeographic species distribution models in reconstructing quaternary range-shifts of Nearctic chelonians. PLoS ONE, 8, e72855. |
| [25] |
Huston MA (1979) A general hypothesis of species diversity. The American Naturalist, 113, 81-101.
DOI URL |
| [26] |
Janzen DH (1967) Why mountain passes are higher in the tropics. The American Naturalist, 101, 233-249.
DOI URL |
| [27] |
Lefcheck JS (2016) piecewiseSEM: Piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods in Ecology and Evolution, 7, 573-579.
DOI URL |
| [28] | Lv L, van de Pol M, Osmond HL, Liu Y, Cockburn A, Kruuk LEB (2023) Winter mortality of a passerine bird increases following hotter summers and during winters with higher maximum temperatures. Science Advances, 9, eabm0197. |
| [29] |
McDonald KW, McClure CJW, Rolek BW, Hill GE (2012) Diversity of birds in eastern North America shifts north with global warming. Ecology and Evolution, 2, 3052-3060.
DOI PMID |
| [30] |
McGlaughlin ME, Wallace LE, Wheeler GL, Bresowar G, Riley L, Britten NR, Helenurm K (2014) Do the island biogeography predictions of MacArthur and Wilson hold when examining genetic diversity on the near mainland California Channel Islands? Examples from endemic Acmispon (Fabaceae). Botanical Journal of the Linnean Society, 174, 289-304.
DOI URL |
| [31] |
Meng LH, Chen G, Li ZH, Yang YP, Wang ZK, Wang LY (2015) Refugial isolation and range expansions drive the genetic structure of Oxyria sinensis (Polygonaceae) in the Himalaya-Hengduan Mountains. Scientific Reports, 5, 10396.
DOI |
| [32] |
Mimura M, Yahara T, Faith DP, Vázquez-Domínguez E, Colautti RI, Araki H, Javadi F, Núñez-Farfán J, Mori AS, Zhou SL, Hollingsworth PM, Neaves LE, Fukano Y, Smith GF, Sato YI, Tachida H, Hendry AP (2017) Understanding and monitoring the consequences of human impacts on intraspecific variation. Evolutionary Applications, 10, 121-139.
DOI PMID |
| [33] |
Miraldo A, Li S, Borregaard MK, Flórez-Rodríguez A, Gopalakrishnan S, Rizvanovic M, Wang ZH, Rahbek C, Marske KA, Nogués-Bravo D (2016) An Anthropocene map of genetic diversity. Science, 353, 1532-1535.
PMID |
| [34] |
Neto EDC, de Oliveira VM, Rosas A, Campos PRA (2011) The effect of spatially correlated environments on genetic diversity-area relationships. Journal of Theoretical Biology, 288, 57-65.
DOI PMID |
| [35] | Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO (2014) The biodiversity of species and their rates of extinction, distribution, and protection. Science, 344, 1246752. |
| [36] |
Qu YH, Ericson PGP, Quan Q, Song G, Zhang RY, Gao B, Lei FM (2014) Long-term isolation and stability explain high genetic diversity in the Eastern Himalaya. Molecular Ecology, 23, 705-720.
PMID |
| [37] |
Romiguier J, Gayral P, Ballenghien M, Bernard A, Cahais V, Chenuil A, Chiari Y, Dernat R, Duret L, Faivre N, Loire E, Lourenco JM, Nabholz B, Roux C, Tsagkogeorga G, Weber AAT, Weinert LA, Belkhir K, Bierne N, Glémin S, Galtier N (2014) Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature, 515, 261-263.
DOI |
| [38] | Sillero N, Huey RB, Gilchrist G, Rissler L, Pascual M (2020) Distribution modelling of an introduced species:Do adaptive genetic markers affect potential range? Proceedings of the Royal Society B: Biological Sciences, 287, 20201791. |
| [39] |
Slatyer RA, Hirst M, Sexton JP (2013) Niche breadth predicts geographical range size: A general ecological pattern. Ecology Letters, 16, 1104-1114.
DOI PMID |
| [40] |
Stein CM, Morris NJ, Nock NL (2012). Structural equation modeling. Methods in Molecular Biology, 850, 495-512.
DOI PMID |
| [41] |
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics, 105, 437-460.
DOI PMID |
| [42] |
Tajima F (1993) Statistical analysis of DNA polymorphism. The Japanese Journal of Genetics, 68, 567-595.
DOI URL |
| [43] |
Tang ZY, Qiao XJ, Fang JY (2009) Species-area relationship in biological communities. Biodiversity Science, 17, 549-559. (in Chinese with English abstract)
DOI URL |
|
[唐志尧, 乔秀娟, 方精云 (2009) 生物群落的种-面积关系. 生物多样性, 17, 549-559.]
DOI |
|
| [44] | Teixeira JC, Huber CD (2021) The inflated significance of neutral genetic diversity in conservation genetics. Proceedings of the National Academy of Sciences, USA, 118, e2015096118. |
| [45] |
Theodoridis S, Fordham DA, Brown SC, Li S, Rahbek C, Nogues-Bravo D (2020) Evolutionary history and past climate change shape the distribution of genetic diversity in terrestrial mammals. Nature Communications, 11, 2557.
DOI PMID |
| [46] |
Virkkala R, Lehikoinen A (2017) Birds on the move in the face of climate change: High species turnover in northern Europe. Ecology and Evolution, 7, 8201-8209.
DOI PMID |
| [47] |
Voelker G, Wogan GOU, Huntley JW, Kaliba PM, DE Swardt DH, Bowie RCK (2025) Climate cycling did not affect haplotype distribution in an abundant Southern African avian habitat generalist species, the familiar chat (Oenanthe familiaris). Integrative Zoology, 20, 595-607.
DOI URL |
| [48] |
Wang SP, Zhu W, Gao X, Li XP, Yan SF, Liu X, Yang J, Gao ZX, Li YM (2014) Population size and time since island isolation determine genetic diversity loss in insular frog populations. Molecular Ecology, 23, 637-648.
DOI PMID |
| [49] | Zillig MW, Brooks W, Fleishman E (2024) Shifts in elevational distributions of montane birds in an arid ecosystem. Ecography, 2024, e06780. |
| [1] | 柴恩平, 葛苏婷, 王锡茂, 杨州, 向昭宜, 张美惠, 李漫淑, 沈瑶, 斯幸峰. 基于高频监测研究人为干扰对城市夜鹭巢址分布动态及繁殖成功的影响[J]. 生物多样性, 2025, 33(9): 25025-. |
| [2] | 徐欢, 辛凤飞, 施宏亮, 袁琳, 薄顺奇, 赵欣怡, 邓帅涛, 潘婷婷, 余婧, 孙赛赛, 薛程. 生态修复技术集成应用对长江口北支生境与鸟类多样性提升效果评估[J]. 生物多样性, 2025, 33(5): 24478-. |
| [3] | 李菡, 董伟, 陆江涛, 吴永杰, 何兴成, 刘林, 木留里哈, 张学林. 四川省甘洛县马鞍山自然保护区及周边地区红外相机鸟兽监测数据集[J]. 生物多样性, 2025, 33(10): 25165-. |
| [4] | 胡志清, 董路. 城市化对鸟类参与的种间互作的影响[J]. 生物多样性, 2024, 32(8): 24048-. |
| [5] | 段菲, 刘鸣章, 卜红亮, 俞乐, 李晟. 城市化对鸟类群落组成及功能特征的影响——以京津冀地区为例[J]. 生物多样性, 2024, 32(8): 23473-. |
| [6] | 王秦韵, 张玉泉, 刘浩, 李明, 刘菲, 赵宁, 陈鹏, 齐敦武, 阙品甲. 成都大熊猫繁育研究基地鸟类多样性[J]. 生物多样性, 2024, 32(8): 24066-. |
| [7] | 白皓天, 余上, 潘新园, 凌嘉乐, 吴娟, 谢恺琪, 刘阳, 陈学业. AI辅助识别的鸟类被动声学监测在城市湿地公园中的应用[J]. 生物多样性, 2024, 32(8): 24188-. |
| [8] | 吴琼, 赵梓羲, 孙桃柱, 赵雨梦, 于丛, 祝芹, 李忠秋. 城市道路特征及自然景观对动物路杀的影响: 以南京为例[J]. 生物多样性, 2024, 32(8): 24141-. |
| [9] | 顾燚芸, 薛嘉祈, 高金会, 谢心仪, 韦铭, 雷进宇, 闻丞. 一种基于公众科学数据的区域性鸟类多样性评价方法[J]. 生物多样性, 2024, 32(7): 24080-. |
| [10] | 李柏灿, 张军国, 张长春, 王丽凤, 徐基良, 刘利. 基于TC-YOLO模型的北京珍稀鸟类识别方法[J]. 生物多样性, 2024, 32(5): 24056-. |
| [11] | 李斌, 宋鹏飞, 顾海峰, 徐波, 刘道鑫, 江峰, 梁程博, 张萌, 高红梅, 蔡振媛, 张同作. 昆仑山青海片区鸟类群落多样性格局及其驱动因素[J]. 生物多样性, 2024, 32(4): 23406-. |
| [12] | 王鹏, 隋佳容, 丁欣瑶, 王伟中, 曹雪倩, 赵海鹏, 王彦平. 郑州城市公园鸟类群落嵌套分布格局及其影响因素[J]. 生物多样性, 2024, 32(3): 23359-. |
| [13] | 郝泽周, 张承云, 李乐, 高丙涛, 曾伟, 王淳, 王梓炫, 黄万涛, 张悦, 裴男才, 肖治术. 城市鸟类多样性被动声学监测与评价技术应用[J]. 生物多样性, 2024, 32(10): 24123-. |
| [14] | 李乐, 张承云, 裴男才, 高丙涛, 王娜, 李嘉睿, 武瑞琛, 郝泽周. 基于被动声学监测技术的城市绿地景观格局与鸟类多样性关联分析[J]. 生物多样性, 2024, 32(10): 24296-. |
| [15] | 谭娟, 朱丹丹, 王卿, 王敏. 被动声学技术在城市公园绿地鸟类多样性监测中的应用: 以上海闵行区春申公园为例[J]. 生物多样性, 2024, 32(10): 24262-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2026 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()