



生物多样性 ›› 2018, Vol. 26 ›› Issue (3): 238-247. DOI: 10.17520/biods.2017259 cstr: 32101.14.biods.2017259
黄勋和1,2, 余哲琪1, 翁茁先1,2,3, 何丹林4, 易振华4, 李威娜1,2, 陈洁波1,2, 张细权4, 杜炳旺5, 钟福生1,2,3,*( )
)
收稿日期:2017-10-08
接受日期:2017-12-27
出版日期:2018-03-20
发布日期:2018-05-05
通讯作者:
钟福生
作者简介:# 共同第一作者
基金资助:
Xunhe Huang1,2, Zheqi Yu1, Zhuoxian Weng1,2,3, Danlin He4, Zhenhua Yi4, Weina Li1,2, Jiebo Chen1,2, Xiquan Zhang4, Bingwang Du5, Fusheng Zhong1,2,3,*( )
)
Received:2017-10-08
Accepted:2017-12-27
Online:2018-03-20
Published:2018-05-05
Contact:
Zhong Fusheng
About author:# Co-first authors
摘要:
系统评估地方鸡的遗传变异水平并追溯其母系起源, 可为保护利用优质家禽种质资源库提供科学依据。本研究测定了广东省和邻省共12个地方鸡品种的线粒体DNA D-loop序列, 分析品种间的遗传距离与系统关系, 并构建单倍型系统发生树和中介网络图。360份样品共检测到60个突变位点, 均为转换。定义了85种单倍型, 归属于单倍型类群A、B、C和E, 在12个鸡品种中均有分布, 其中B是优势单倍型类群(187个, 51.94%), E次之(76个, 21.11%)。B02 和C01是优势单倍型(85个, 23.61%; 48个, 13.33%), 为12个鸡品种共有; E03位居第三(35个, 9.72%), 杏花鸡、黄郎鸡和宁都三黄鸡未见此单倍型。杏花鸡集中分布在单倍型类群B, 惠阳胡须鸡和中山沙栏鸡则主要分布在单倍型类群E; 怀乡鸡的单倍型数量最多, 中山沙栏鸡的最少。广东地方鸡品种间遗传距离为0.012-0.015, 单倍型多样性0.805 ± 0.047至0.949 ± 0.026, 核苷酸多样性0.0102 ± 0.0017至0.0138 ± 0.0009。邻接树和中介网络图将85种单倍型划分为进化枝A、B、C和E, 广东省与邻省地方鸡单倍型的地理分布模式相似。中性检验显示广东地方鸡未经历明显的群体历史扩张。结果表明广东地方鸡处于较好的保护状态, 遗传多样性水平较高, 品种的形成受到邻省和北方家鸡的影响, 东南亚红原鸡对广东地方鸡也有重要的遗传贡献。
黄勋和, 余哲琪, 翁茁先, 何丹林, 易振华, 李威娜, 陈洁波, 张细权, 杜炳旺, 钟福生 (2018) 广东省地方鸡线粒体遗传多样性与母系起源. 生物多样性, 26, 238-247. DOI: 10.17520/biods.2017259.
Xunhe Huang, Zheqi Yu, Zhuoxian Weng, Danlin He, Zhenhua Yi, Weina Li, Jiebo Chen, Xiquan Zhang, Bingwang Du, Fusheng Zhong (2018) Mitochondrial genetic diversity and maternal origin of Guangdong indigenous chickens. Biodiversity Science, 26, 238-247. DOI: 10.17520/biods.2017259.
| 品种 Breed | 代号 Code | 产地 Origin |
|---|---|---|
| 怀乡鸡 Huaixiang chicken | HX | 广东省信宜市 Xinyi, Guangdong |
| 杏花鸡 Xinghua chicken | XH | 广东省封开县 Fengkai, Guangdong |
| 阳山鸡 Yangshan chicken | YS | 广东省阳山县 Yangshan, Guangdong |
| 清远麻鸡 Qingyuan spotted chicken | QY | 广东省清远市 Qingyuan, Guangdong |
| 惠阳胡须鸡 Huiyang bearded chicken | HY | 广东省惠州市 Huizhou, Guangdong |
| 五华三黄鸡 Wuhua yellow chicken | WH | 广东省五华县 Wuhua, Guangdong |
| 中山沙栏鸡 Zhongshan shalan chicken | SL | 广东省中山市 Zhongshan, Guangdong |
| 广西三黄鸡 Guangxi yellow chicken | GX | 广西省桂平市 Guiping, Guangxi |
| 文昌鸡 Wenchang chicken | WC | 海南省文昌市 Wenchang, Hainan |
| 河田鸡 Hetian chicken | HT | 福建省长汀县 Changting, Fujian |
| 黄郎鸡 Huanglang chicken | HL | 湖南省衡南县 Hengnan, Hunan |
| 宁都三黄鸡 Ningdu yellow chicken | ND | 江西省宁都县 Ningdu, Jiangxi |
表1 本研究的样品信息(每个品种均取30个样本)
Table 1 Sample information of the present study (30 samples for every chicken breed)
| 品种 Breed | 代号 Code | 产地 Origin |
|---|---|---|
| 怀乡鸡 Huaixiang chicken | HX | 广东省信宜市 Xinyi, Guangdong |
| 杏花鸡 Xinghua chicken | XH | 广东省封开县 Fengkai, Guangdong |
| 阳山鸡 Yangshan chicken | YS | 广东省阳山县 Yangshan, Guangdong |
| 清远麻鸡 Qingyuan spotted chicken | QY | 广东省清远市 Qingyuan, Guangdong |
| 惠阳胡须鸡 Huiyang bearded chicken | HY | 广东省惠州市 Huizhou, Guangdong |
| 五华三黄鸡 Wuhua yellow chicken | WH | 广东省五华县 Wuhua, Guangdong |
| 中山沙栏鸡 Zhongshan shalan chicken | SL | 广东省中山市 Zhongshan, Guangdong |
| 广西三黄鸡 Guangxi yellow chicken | GX | 广西省桂平市 Guiping, Guangxi |
| 文昌鸡 Wenchang chicken | WC | 海南省文昌市 Wenchang, Hainan |
| 河田鸡 Hetian chicken | HT | 福建省长汀县 Changting, Fujian |
| 黄郎鸡 Huanglang chicken | HL | 湖南省衡南县 Hengnan, Hunan |
| 宁都三黄鸡 Ningdu yellow chicken | ND | 江西省宁都县 Ningdu, Jiangxi |
| 单倍型 Haplotype | 变异位点* Variable sites | 单倍型的品种分布(频率) Haplotype distribution in breeds (frequency) | 合计 Total |
|---|---|---|---|
NC_007235 | 111111222222222222222222222222223333333333333333333334444444 356899011112223334445556788989990011123333445566689991334467 387279102790250792362691413162694603590135274523741690456960 TGTTTTTCATCTACTAAGTTATATAAAAAACTTTCCTACTTAAATTCCTACTGTAGCTAT | ||
| A01 | ..C.....G....T.....C...C............C....................... | HX2, HY1, QY3, WH4, XH2, YS3, SL2, GX1, WC1, HL1 | 20 |
| A02 | ..C.....G....T.....C...C............C.......C............... | XH1 | 1 |
| A03 | ..C.....G....T.....C...C........G...C....................... | XH1 | 1 |
| A04 | ..C.....G....T..G..C...C............C....................... | HY1, YS2 | 3 |
| A05 | ..C....TG....T.....C...C............C....................... | HX1 | 1 |
| A06 | ..C.....G....T.....C...C............C.........T.....A....... | HY1 | 1 |
| A07 | ..C.....G....T.....C...C............C...............A....... | HX1, HT1, HL2 | 4 |
| A08 | ..C.....G....T.....C...C............C...C................... | HY1 | 1 |
| A09 | ..C.....G....T.....C...C..G.........C..............C......G. | HL1 | 1 |
| A10 | ..C.....G....T.....C...C............C........C......A....... | HL1 | 1 |
| A11 | ..C.....G....T..G..C...C............C...............A....... | HT1 | 1 |
| A12 | ..C.....G..C.T.....C...C............C...............A....... | HT2 | 2 |
| B01 | ............................................................ | XH2, ND1 | 3 |
| B02 | .......................C.................................... | HX6, HY6, QY7, WH5, XH8, YS9, SL5, HT1, GX20, WC13, HL2, ND3 | 85 |
| B03 | .......................C......T............................. | XH2, GX1, WC1, HL1 | 5 |
| B04 | .......................C.....................C.............. | XH3 | 3 |
| B05 | ......................GC.........C.......................... | XH3 | 3 |
| B06 | .....................A.C.................................... | XH2 | 2 |
| B07 | .......................C.............................C...... | XH1 | 1 |
| B08 | .....................C.C.................................... | WH2, YS2, GX1, WC1, HL1 | 7 |
| B09 | .......................C..........................T......... | HX1, WH1, YS1, HT1, ND1 | 5 |
| B10 | ...................C...C.................................... | HX1 | 1 |
| B11 | ..............C........C.................................... | HX1 | 1 |
| B12 | .......................C............................A....... | HX3, WH3, HT6, HL2, ND9 | 23 |
| B13 | ..........T............C............................A....... | HX1, ND2 | 3 |
| B14 | .......................C......T.....................A....... | HX1 | 1 |
| B15 | .......................C..........................T.A....... | HX1 | 1 |
| B16 | ............................................C............... | WH1 | 1 |
| B17 | ............................................C.......A....... | WH1 | 1 |
| B18 | .......................C.......C............................ | QY1 | 1 |
| B19 | .......................C.........C.......................... | QY1, WC1 | 2 |
| B20 | .......................C.G.................................. | QY1 | 1 |
| B21 | ...C...................C.................................... | QY1 | 1 |
| B22 | .......................C.............G...................... | QY1 | 1 |
| B23 | ..................C....C.................................... | SL2 | 2 |
| B24 | .......................CG................................... | SL1 | 1 |
| B25 | .......................C...............C.................... | HY1 | 1 |
| 单倍型 Haplotype | 变异位点* Variable sites | 单倍型的品种分布(频率) Haplotype distribution in breeds (frequency) | 合计 Total |
| B26 | .......................C.........................G.......... | GX1 | 1 |
| B27 | .......................C.................G.................. | GX1 | 1 |
| B28 | ......C................C.....G.............................. | GX1 | 1 |
| B29 | ......................GC.........C....T..................... | GX1 | 1 |
| B30 | .......................C...........T........................ | WC1 | 1 |
| B31 | .......................C.................................C.. | WC1 | 1 |
| B32 | .......................C..........................A......... | WC1 | 1 |
| B33 | .......................C....................C............... | HL2 | 2 |
| B34 | .......................C.....................C......A....... | HL1 | 1 |
| B35 | .....................C.C...............................C.... | HL1 | 1 |
| B36 | .............T.........C............................A....... | HL1 | 1 |
| B37 | .......................C..............................G.T..C | HL1 | 1 |
| B38 | .......................C...G........................A....... | HL1 | 1 |
| B39 | ..........T............C.................................... | ND1 | 1 |
| B40 | ................C......C.................................... | ND1 | 1 |
| B41 | .......................C.........C..................A....... | ND2 | 2 |
| B42 | .................................C..................A....... | ND3 | 3 |
| B43 | .......................C.....G.............................. | HT1 | 1 |
| B44 | .......................C.....G......................A....... | HT3 | 3 |
| B45 | .......................C...........................C........ | HT1 | 1 |
| B46 | C.........T............C.................................... | HT1 | 1 |
| B47 | .......................C...................G........A....... | HT1 | 1 |
| B48 | .....................C.C............................A....... | HX2, ND1 | 3 |
| C01 | ........G........ACC.C.C.G........T.C.....G....TC........... | HX2, HY7, QY2, WH4, XH3, YS6, SL6, HT2, GX2, WC4, HL5, ND5 | 48 |
| C02 | ........G........ACC.C.C.G........T.C.....G....TC.T......... | XH1, HL2 | 3 |
| C03 | ........G........ACC.C.C.G.......CT.C..........TC........... | HX2 | 2 |
| C04 | .......TG........ACC.C.C.G........T.C.....G....TC........... | WH1 | 1 |
| C05 | ........G........ACC.C.C.G........T.C.T...G....TC.T......... | HL1 | 1 |
| C06 | ....C...G........ACC.C.C.G........T.C..........TC........... | HL1 | 1 |
| C07 | ........G........ACC.C.C..........T.C.....G....TC........... | HT2 | 2 |
| C08 | ........G........ACC.C...G........T.C.....G....TC........... | HT2 | 2 |
| E01 | .....C..GC........CC.C............T.C...................T... | HY1, QY1, WH1, XH1, YS1, SL3, WC1 | 9 |
| E02 | ........GC........CC.C............T.C.....G.............T... | YS1 | 1 |
| E03 | ........GC........CC.C............T.C...................T... | HX1, HY8, QY5, WH2, YS2, SL11, HT1, GX1, WC4 | 35 |
| E04 | ........GC..G.....CC.C............T.C.T.................T... | QY3, YS2 | 5 |
| E05 | .A......GC........CC.C............T.C...................T... | YS1 | 1 |
| E06 | ........GC........CC.C............T.C.......C...........T... | HX1, HY1 | 2 |
| E07 | ........GC........CC.C............T.C.T.................T... | HX1, HY1, WC1 | 3 |
| E08 | ........GC........CC.C............T.C...............A...T... | HX1, WH2, HT2, HL2 | 7 |
| E09 | .....C..GC........CC.C............T.C...............A...T... | WH3 | 3 |
| E10 | ........GC........CC.C......G.....T.C......T............T... | QY1 | 1 |
| E11 | ........GC........C..C............T.C...................T... | QY1 | 1 |
| E12 | ........GC........CCTC............T.C...................T... | QY1 | 1 |
| E13 | ........GC.....G..CC.C............T.C...................T... | QY1 | 1 |
| E14 | ........GC........CCTC............T.C...............A...T... | HY1 | 1 |
| E15 | ........GC........CC.C............T.C.............A.....T... | HY1 | 1 |
| E16 | ........GC........CC.C............T.C.....G.........A...T... | HT2, HL1 | 3 |
| E17 | ........G..C......CC.C...........CT.C.............A......... | ND1 | 1 |
表2 12种地方鸡mtDNA D-loop的单倍型及其在品种中的分布。*表示参考序列NC_007235对应的变异位点位置, 圆点表示与参考序列具有相同的碱基。
Table 2 mtDNA D-loop haplotypes and their distribution in the 12 indigenous chicken breeds. Star (*) indicate the corresponding position with the reference sequence NC_007235, Dots (.) within the nucleotide position indicate the same nucleotides as given in reference sequences.
| 单倍型 Haplotype | 变异位点* Variable sites | 单倍型的品种分布(频率) Haplotype distribution in breeds (frequency) | 合计 Total |
|---|---|---|---|
NC_007235 | 111111222222222222222222222222223333333333333333333334444444 356899011112223334445556788989990011123333445566689991334467 387279102790250792362691413162694603590135274523741690456960 TGTTTTTCATCTACTAAGTTATATAAAAAACTTTCCTACTTAAATTCCTACTGTAGCTAT | ||
| A01 | ..C.....G....T.....C...C............C....................... | HX2, HY1, QY3, WH4, XH2, YS3, SL2, GX1, WC1, HL1 | 20 |
| A02 | ..C.....G....T.....C...C............C.......C............... | XH1 | 1 |
| A03 | ..C.....G....T.....C...C........G...C....................... | XH1 | 1 |
| A04 | ..C.....G....T..G..C...C............C....................... | HY1, YS2 | 3 |
| A05 | ..C....TG....T.....C...C............C....................... | HX1 | 1 |
| A06 | ..C.....G....T.....C...C............C.........T.....A....... | HY1 | 1 |
| A07 | ..C.....G....T.....C...C............C...............A....... | HX1, HT1, HL2 | 4 |
| A08 | ..C.....G....T.....C...C............C...C................... | HY1 | 1 |
| A09 | ..C.....G....T.....C...C..G.........C..............C......G. | HL1 | 1 |
| A10 | ..C.....G....T.....C...C............C........C......A....... | HL1 | 1 |
| A11 | ..C.....G....T..G..C...C............C...............A....... | HT1 | 1 |
| A12 | ..C.....G..C.T.....C...C............C...............A....... | HT2 | 2 |
| B01 | ............................................................ | XH2, ND1 | 3 |
| B02 | .......................C.................................... | HX6, HY6, QY7, WH5, XH8, YS9, SL5, HT1, GX20, WC13, HL2, ND3 | 85 |
| B03 | .......................C......T............................. | XH2, GX1, WC1, HL1 | 5 |
| B04 | .......................C.....................C.............. | XH3 | 3 |
| B05 | ......................GC.........C.......................... | XH3 | 3 |
| B06 | .....................A.C.................................... | XH2 | 2 |
| B07 | .......................C.............................C...... | XH1 | 1 |
| B08 | .....................C.C.................................... | WH2, YS2, GX1, WC1, HL1 | 7 |
| B09 | .......................C..........................T......... | HX1, WH1, YS1, HT1, ND1 | 5 |
| B10 | ...................C...C.................................... | HX1 | 1 |
| B11 | ..............C........C.................................... | HX1 | 1 |
| B12 | .......................C............................A....... | HX3, WH3, HT6, HL2, ND9 | 23 |
| B13 | ..........T............C............................A....... | HX1, ND2 | 3 |
| B14 | .......................C......T.....................A....... | HX1 | 1 |
| B15 | .......................C..........................T.A....... | HX1 | 1 |
| B16 | ............................................C............... | WH1 | 1 |
| B17 | ............................................C.......A....... | WH1 | 1 |
| B18 | .......................C.......C............................ | QY1 | 1 |
| B19 | .......................C.........C.......................... | QY1, WC1 | 2 |
| B20 | .......................C.G.................................. | QY1 | 1 |
| B21 | ...C...................C.................................... | QY1 | 1 |
| B22 | .......................C.............G...................... | QY1 | 1 |
| B23 | ..................C....C.................................... | SL2 | 2 |
| B24 | .......................CG................................... | SL1 | 1 |
| B25 | .......................C...............C.................... | HY1 | 1 |
| 单倍型 Haplotype | 变异位点* Variable sites | 单倍型的品种分布(频率) Haplotype distribution in breeds (frequency) | 合计 Total |
| B26 | .......................C.........................G.......... | GX1 | 1 |
| B27 | .......................C.................G.................. | GX1 | 1 |
| B28 | ......C................C.....G.............................. | GX1 | 1 |
| B29 | ......................GC.........C....T..................... | GX1 | 1 |
| B30 | .......................C...........T........................ | WC1 | 1 |
| B31 | .......................C.................................C.. | WC1 | 1 |
| B32 | .......................C..........................A......... | WC1 | 1 |
| B33 | .......................C....................C............... | HL2 | 2 |
| B34 | .......................C.....................C......A....... | HL1 | 1 |
| B35 | .....................C.C...............................C.... | HL1 | 1 |
| B36 | .............T.........C............................A....... | HL1 | 1 |
| B37 | .......................C..............................G.T..C | HL1 | 1 |
| B38 | .......................C...G........................A....... | HL1 | 1 |
| B39 | ..........T............C.................................... | ND1 | 1 |
| B40 | ................C......C.................................... | ND1 | 1 |
| B41 | .......................C.........C..................A....... | ND2 | 2 |
| B42 | .................................C..................A....... | ND3 | 3 |
| B43 | .......................C.....G.............................. | HT1 | 1 |
| B44 | .......................C.....G......................A....... | HT3 | 3 |
| B45 | .......................C...........................C........ | HT1 | 1 |
| B46 | C.........T............C.................................... | HT1 | 1 |
| B47 | .......................C...................G........A....... | HT1 | 1 |
| B48 | .....................C.C............................A....... | HX2, ND1 | 3 |
| C01 | ........G........ACC.C.C.G........T.C.....G....TC........... | HX2, HY7, QY2, WH4, XH3, YS6, SL6, HT2, GX2, WC4, HL5, ND5 | 48 |
| C02 | ........G........ACC.C.C.G........T.C.....G....TC.T......... | XH1, HL2 | 3 |
| C03 | ........G........ACC.C.C.G.......CT.C..........TC........... | HX2 | 2 |
| C04 | .......TG........ACC.C.C.G........T.C.....G....TC........... | WH1 | 1 |
| C05 | ........G........ACC.C.C.G........T.C.T...G....TC.T......... | HL1 | 1 |
| C06 | ....C...G........ACC.C.C.G........T.C..........TC........... | HL1 | 1 |
| C07 | ........G........ACC.C.C..........T.C.....G....TC........... | HT2 | 2 |
| C08 | ........G........ACC.C...G........T.C.....G....TC........... | HT2 | 2 |
| E01 | .....C..GC........CC.C............T.C...................T... | HY1, QY1, WH1, XH1, YS1, SL3, WC1 | 9 |
| E02 | ........GC........CC.C............T.C.....G.............T... | YS1 | 1 |
| E03 | ........GC........CC.C............T.C...................T... | HX1, HY8, QY5, WH2, YS2, SL11, HT1, GX1, WC4 | 35 |
| E04 | ........GC..G.....CC.C............T.C.T.................T... | QY3, YS2 | 5 |
| E05 | .A......GC........CC.C............T.C...................T... | YS1 | 1 |
| E06 | ........GC........CC.C............T.C.......C...........T... | HX1, HY1 | 2 |
| E07 | ........GC........CC.C............T.C.T.................T... | HX1, HY1, WC1 | 3 |
| E08 | ........GC........CC.C............T.C...............A...T... | HX1, WH2, HT2, HL2 | 7 |
| E09 | .....C..GC........CC.C............T.C...............A...T... | WH3 | 3 |
| E10 | ........GC........CC.C......G.....T.C......T............T... | QY1 | 1 |
| E11 | ........GC........C..C............T.C...................T... | QY1 | 1 |
| E12 | ........GC........CCTC............T.C...................T... | QY1 | 1 |
| E13 | ........GC.....G..CC.C............T.C...................T... | QY1 | 1 |
| E14 | ........GC........CCTC............T.C...............A...T... | HY1 | 1 |
| E15 | ........GC........CC.C............T.C.............A.....T... | HY1 | 1 |
| E16 | ........GC........CC.C............T.C.....G.........A...T... | HT2, HL1 | 3 |
| E17 | ........G..C......CC.C...........CT.C.............A......... | ND1 | 1 |
| 品种 Breed | 突变位点数 Variable sites | 单倍型数量 No. of haplotypes | 单倍型类群(个体数) Haplogroups (individuals) | 单倍型多样性 Haplotype diversity (SD) | 核苷酸多样性 Nucleotide diversity (SD) | 平均核苷酸差异 Average number of nucleotide differences | Tajima’s D检验 Tajima’s D test |
|---|---|---|---|---|---|---|---|
| HX | 26 | 19 (A=4, B=9, C=2, E=4) | A(5), B(17), C(4), E(4) | 0.949 (0.026) | 0.0123 (0.0014) | 6.382 | -0.0987 |
| HY | 25 | 12(A=3, B=2, C=1, E=6) | A(3), B(7), C(7), E(13) | 0.853 (0.039) | 0.0132 (0.0008) | 6.837 | 0.2969 |
| QY | 27 | 15 (A=1, B=6, C=1, E=7) | A(3), B(12), C(2), E(13) | 0.913 (0.032) | 0.0127 (0.0008) | 6.623 | -0.1011 |
| WH | 21 | 13 (A=1, B=6, C=2, E=4) | A(4), B(13), C(5), E(8) | 0.929 (0.022) | 0.0138 (0.0009) | 7.177 | 1.2395 |
| XH | 25 | 13 (A=3, B=7, C=2, E=1) | A(4), B(21), C(4), E(1) | 0.906 (0.036) | 0.0102 (0.0017) | 5.285 | -0.6955 |
| YS | 22 | 11 (A=2, B=3, C=1, E=5) | A(5), B(12), C(6), E(7) | 0.867 (0.042) | 0.0134 (0.0009) | 6.989 | 0.9093 |
| SL | 18 | 7 (A=1, B=3, C=1, E=2) | A(2), B(8), C(6), E(14) | 0.805 (0.047) | 0.0120 (0.0009) | 6.248 | 1.2923 |
| GX | 24 | 10 (A=1, B=7, C=1, E=1) | A(1), B (26), C(2), E(1) | 0.561 (0.109) | 0.0055 (0.0018) | 2.839 | -1.8847* |
| HL | 30 | 20 (A=4, B=10, C=4, E=2) | A(5), B(13), C(9), E(3) | 0.963 (0.021) | 0.0155 (0.0009) | 8.025 | 0.2160 |
| HT | 25 | 17 (A=3, B=8, C=3, E=3) | A(4), B(15), C(6), E(5) | 0.945 (0.025) | 0.0141 (0.0011) | 7.317 | 0.5680 |
| ND | 18 | 12 (B=10, C=1, E=1) | B(24), C(5), E(1) | 0.876 (0.041) | 0.0097 (0.0019) | 5.039 | 0.1757 |
| WC | 23 | 12 (A=1, B=7, C=1, E=3) | A(1), B(19), C(4), E(6) | 0.793 (0.067) | 0.0108 (0.0015) | 5.630 | -0.1070 |
| 合计Total | 60 | 85 (A=12, B=48, C=8, E=17) | A(37), B(187), C(60), E(76) | 0.909 (0.010) | 0.0130 (0.0003) | 6.739 | -0.9237 |
表3 12个地方鸡品种的线粒体DNA D-loop遗传多样性统计。品种代号同表1。
Table 3 Genetic diversity of mtDNA D-loop in the 12 indigenous chicken breeds. Breed codes are the same with Table 1.
| 品种 Breed | 突变位点数 Variable sites | 单倍型数量 No. of haplotypes | 单倍型类群(个体数) Haplogroups (individuals) | 单倍型多样性 Haplotype diversity (SD) | 核苷酸多样性 Nucleotide diversity (SD) | 平均核苷酸差异 Average number of nucleotide differences | Tajima’s D检验 Tajima’s D test |
|---|---|---|---|---|---|---|---|
| HX | 26 | 19 (A=4, B=9, C=2, E=4) | A(5), B(17), C(4), E(4) | 0.949 (0.026) | 0.0123 (0.0014) | 6.382 | -0.0987 |
| HY | 25 | 12(A=3, B=2, C=1, E=6) | A(3), B(7), C(7), E(13) | 0.853 (0.039) | 0.0132 (0.0008) | 6.837 | 0.2969 |
| QY | 27 | 15 (A=1, B=6, C=1, E=7) | A(3), B(12), C(2), E(13) | 0.913 (0.032) | 0.0127 (0.0008) | 6.623 | -0.1011 |
| WH | 21 | 13 (A=1, B=6, C=2, E=4) | A(4), B(13), C(5), E(8) | 0.929 (0.022) | 0.0138 (0.0009) | 7.177 | 1.2395 |
| XH | 25 | 13 (A=3, B=7, C=2, E=1) | A(4), B(21), C(4), E(1) | 0.906 (0.036) | 0.0102 (0.0017) | 5.285 | -0.6955 |
| YS | 22 | 11 (A=2, B=3, C=1, E=5) | A(5), B(12), C(6), E(7) | 0.867 (0.042) | 0.0134 (0.0009) | 6.989 | 0.9093 |
| SL | 18 | 7 (A=1, B=3, C=1, E=2) | A(2), B(8), C(6), E(14) | 0.805 (0.047) | 0.0120 (0.0009) | 6.248 | 1.2923 |
| GX | 24 | 10 (A=1, B=7, C=1, E=1) | A(1), B (26), C(2), E(1) | 0.561 (0.109) | 0.0055 (0.0018) | 2.839 | -1.8847* |
| HL | 30 | 20 (A=4, B=10, C=4, E=2) | A(5), B(13), C(9), E(3) | 0.963 (0.021) | 0.0155 (0.0009) | 8.025 | 0.2160 |
| HT | 25 | 17 (A=3, B=8, C=3, E=3) | A(4), B(15), C(6), E(5) | 0.945 (0.025) | 0.0141 (0.0011) | 7.317 | 0.5680 |
| ND | 18 | 12 (B=10, C=1, E=1) | B(24), C(5), E(1) | 0.876 (0.041) | 0.0097 (0.0019) | 5.039 | 0.1757 |
| WC | 23 | 12 (A=1, B=7, C=1, E=3) | A(1), B(19), C(4), E(6) | 0.793 (0.067) | 0.0108 (0.0015) | 5.630 | -0.1070 |
| 合计Total | 60 | 85 (A=12, B=48, C=8, E=17) | A(37), B(187), C(60), E(76) | 0.909 (0.010) | 0.0130 (0.0003) | 6.739 | -0.9237 |
| 品种 breed | XH | YS | HX | WH | QY | SL | HY | GX | WC | HL | ND | HT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XH | 0.001 | 0.000 | 0.001 | 0.002 | 0.003 | 0.003 | 0.000 | 0.000 | 0.001 | 0.001 | 0.001 | |
| YS | 0.013 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.002 | 0.000 | |
| HX | 0.012 | 0.013 | 0.000 | 0.001 | 0.002 | 0.002 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | |
| WH | 0.013 | 0.014 | 0.013 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.001 | 0.000 | |
| QY | 0.013 | 0.013 | 0.014 | 0.014 | 0.000 | 0.000 | 0.003 | 0.000 | 0.001 | 0.003 | 0.001 | |
| SL | 0.015 | 0.013 | 0.014 | 0.014 | 0.013 | 0.000 | 0.005 | 0.002 | 0.001 | 0.005 | 0.002 | |
| HY | 0.015 | 0.014 | 0.015 | 0.014 | 0.013 | 0.012 | 0.005 | 0.002 | 0.001 | 0.004 | 0.002 | |
| GX | 0.008 | 0.012 | 0.010 | 0.012 | 0.012 | 0.014 | 0.015 | 0.001 | 0.002 | 0.001 | 0.002 | |
| WC | 0.011 | 0.013 | 0.012 | 0.013 | 0.012 | 0.013 | 0.014 | 0.009 | 0.001 | 0.001 | 0.001 | |
| HL | 0.014 | 0.015 | 0.014 | 0.015 | 0.015 | 0.015 | 0.016 | 0.013 | 0.014 | 0.001 | 0.000 | |
| ND | 0.011 | 0.014 | 0.012 | 0.013 | 0.014 | 0.016 | 0.016 | 0.008 | 0.011 | 0.014 | 0.001 | |
| HT | 0.013 | 0.015 | 0.013 | 0.014 | 0.015 | 0.015 | 0.015 | 0.012 | 0.013 | 0.015 | 0.013 |
表4 12个鸡品种间的净遗传距离(上三角)和Kimura双参数距离(下三角)。品种代号同表1。
Table 4 Net distances (above diagonal) and K2P distance (below diagonal) among 12 chicken breeds. Breed codes are the same with Table 1.
| 品种 breed | XH | YS | HX | WH | QY | SL | HY | GX | WC | HL | ND | HT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XH | 0.001 | 0.000 | 0.001 | 0.002 | 0.003 | 0.003 | 0.000 | 0.000 | 0.001 | 0.001 | 0.001 | |
| YS | 0.013 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.002 | 0.000 | |
| HX | 0.012 | 0.013 | 0.000 | 0.001 | 0.002 | 0.002 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | |
| WH | 0.013 | 0.014 | 0.013 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.001 | 0.000 | |
| QY | 0.013 | 0.013 | 0.014 | 0.014 | 0.000 | 0.000 | 0.003 | 0.000 | 0.001 | 0.003 | 0.001 | |
| SL | 0.015 | 0.013 | 0.014 | 0.014 | 0.013 | 0.000 | 0.005 | 0.002 | 0.001 | 0.005 | 0.002 | |
| HY | 0.015 | 0.014 | 0.015 | 0.014 | 0.013 | 0.012 | 0.005 | 0.002 | 0.001 | 0.004 | 0.002 | |
| GX | 0.008 | 0.012 | 0.010 | 0.012 | 0.012 | 0.014 | 0.015 | 0.001 | 0.002 | 0.001 | 0.002 | |
| WC | 0.011 | 0.013 | 0.012 | 0.013 | 0.012 | 0.013 | 0.014 | 0.009 | 0.001 | 0.001 | 0.001 | |
| HL | 0.014 | 0.015 | 0.014 | 0.015 | 0.015 | 0.015 | 0.016 | 0.013 | 0.014 | 0.001 | 0.000 | |
| ND | 0.011 | 0.014 | 0.012 | 0.013 | 0.014 | 0.016 | 0.016 | 0.008 | 0.011 | 0.014 | 0.001 | |
| HT | 0.013 | 0.015 | 0.013 | 0.014 | 0.015 | 0.015 | 0.015 | 0.012 | 0.013 | 0.015 | 0.013 |
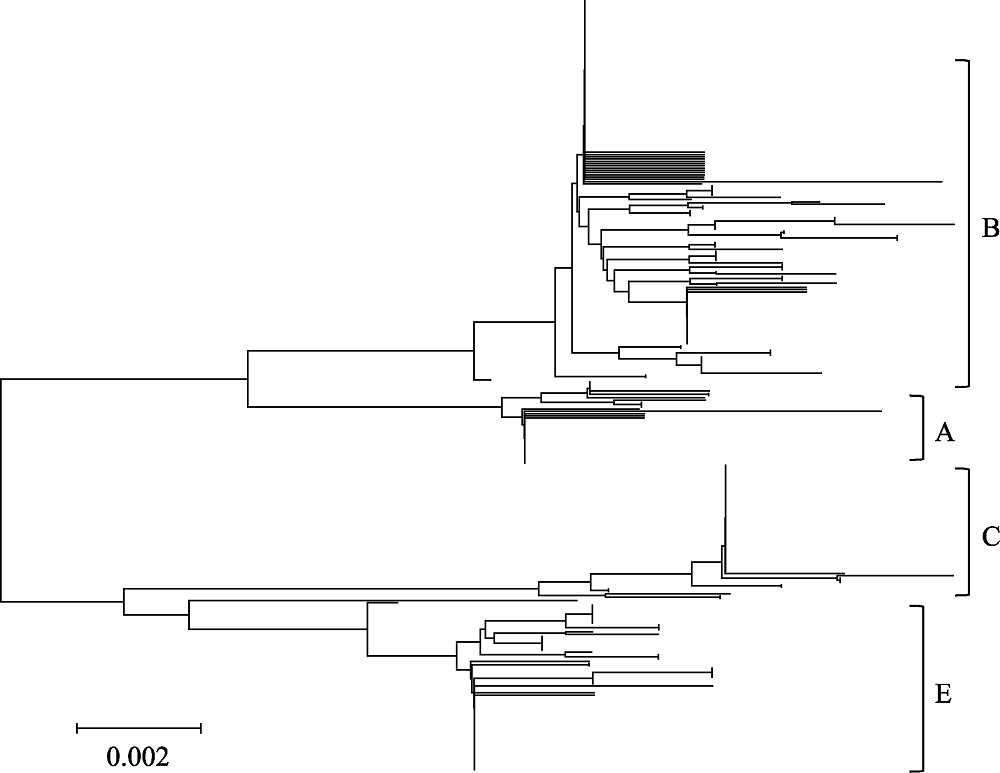
图1 基于85种广东省和邻省家鸡线粒体DNA D-loop单倍型构建的邻接树
Fig. 1 Neighbor-joining tree of 85 mtDNA D-loop haplotypes from Guangdong and its adjacent provinces indigenous chicken breeds
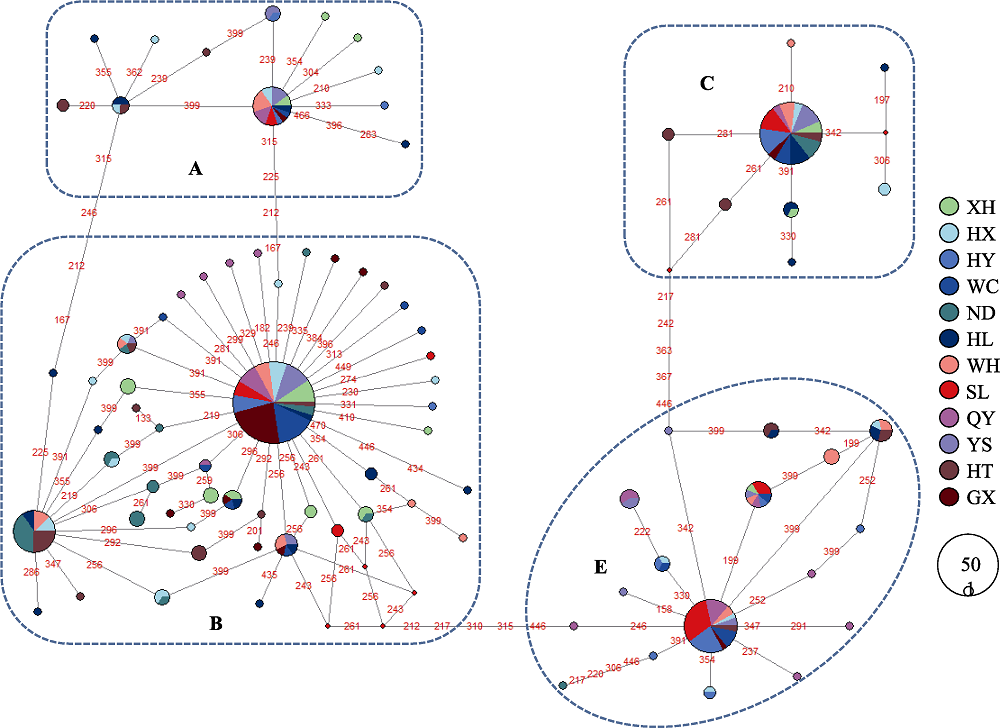
图2 广东省和邻省12种地方鸡线粒体DNA D-loop单倍型的中介网络图。连接点数字表示核苷酸转换的位置; 圆的大小对应单倍型频率; 不同鸡品种用不同颜色标注。品种代号同表1。
Fig. 2 Median-joining network of mtDNA D-loop haplotypes of 12 indigenous chicken breeds from Guangdong and its adjacent provinces. The links are labeled by the nucleotide positions to designate transitions. Cycle size is roughly proportional to the haplotype frequency. The breeds are indicated by different colors. Breed codes are the same with Table 1.
| 地区 Region | 个体数Individuals | 单倍型a/个体数 Haplotypesa/individuals | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | Z | ||
| 广东省 Guangdong | 210 | 8(4)/26 | 26(13)/90 | 5(2)/34 | 15(5)/60 | ||||||
| 中国北方 northern China | 273 | 25(12)/111 | 11(6)/66 | 11(2)/59 | 2/2 | 8/35 | |||||
| 中国南方b southern China | 863 | 28(17)/182 | 67(45)/394 | 22(12)/131 | 5 (3)/13 | 17(3)/137 | 1/1 | 2/4 | 1(1)/1 | ||
| 中国西南 southwest China | 1,153 | 36(19)/366 | 22(12)/260 | 6(2)/29 | 9/98 | 22(16)/152 | 40(31)/237 | 3(3)/11 | |||
| 东南亚 Southeast Asia | 621 | 38(29)/146 | 53(41)/289 | 5(2)/19 | 45(41)/67 | 15(9)/42 | 12(9)/38 | 4(1)/11 | 2 (2)/2 | 3(3)/7 | |
| 南亚 South Asia | 603 | 3(1)/14 | 4(1)/16 | 19(16)/31 | 17(10)/56 | 102(79)/450 | 3(1)/12 | 11(9)/21 | 1(1)/3 | ||
| 合计 Total | 3,723 | ||||||||||
表5 家鸡主要进化枝的地理分布。a单倍型数量及该地区的独享型单倍型(括号数值); b不包括广东省家鸡的数量。
Table 5 Geographical distribution of the major clades in domestic chickens. a, Number of haplotypes and unique haplotypes (in parentheses); b, Excluding those from Guangdong Province.
| 地区 Region | 个体数Individuals | 单倍型a/个体数 Haplotypesa/individuals | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | Z | ||
| 广东省 Guangdong | 210 | 8(4)/26 | 26(13)/90 | 5(2)/34 | 15(5)/60 | ||||||
| 中国北方 northern China | 273 | 25(12)/111 | 11(6)/66 | 11(2)/59 | 2/2 | 8/35 | |||||
| 中国南方b southern China | 863 | 28(17)/182 | 67(45)/394 | 22(12)/131 | 5 (3)/13 | 17(3)/137 | 1/1 | 2/4 | 1(1)/1 | ||
| 中国西南 southwest China | 1,153 | 36(19)/366 | 22(12)/260 | 6(2)/29 | 9/98 | 22(16)/152 | 40(31)/237 | 3(3)/11 | |||
| 东南亚 Southeast Asia | 621 | 38(29)/146 | 53(41)/289 | 5(2)/19 | 45(41)/67 | 15(9)/42 | 12(9)/38 | 4(1)/11 | 2 (2)/2 | 3(3)/7 | |
| 南亚 South Asia | 603 | 3(1)/14 | 4(1)/16 | 19(16)/31 | 17(10)/56 | 102(79)/450 | 3(1)/12 | 11(9)/21 | 1(1)/3 | ||
| 合计 Total | 3,723 | ||||||||||
| [1] |
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48.
DOI URL PMID |
| [2] | Chen GH, Wang KH, Wang JY, Ding C, Yang N (2004) oultry Genetic Resources in China. Shanghai Scientific and Technical Publishers, Shanghai. (in Chinese) |
| [陈国宏, 王克华, 王金玉, 丁铲, 杨宁 (2004) 中国禽类遗传资源. 上海科学技术出版社, 上海.] | |
| [3] | China National Commission of Animal Genetic Resources(2011) Animal Genetic Resources in China: Poultry. China Agriculture Press, Beijing. (in Chinese) |
| [国家畜禽遗传资源委员会(2011) 中国畜禽遗传资源志: 家禽志. 中国农业出版社, 北京.] | |
| [4] | Editorial Committee of Local Historiography of Guangdong(2004) Guangdong Provincial Local-Records Compilation: Executive Summary. Guangdong People Press, Guangzhou. (in Chinese) |
| [广东省地方史志编纂委员会(2004) 广东省志?总述. 广东人民出版社, 广州.] | |
| [5] | Fu Y, Niu D, Luo J, Ruan H, He GQ, Zhang YP (2001) Studies of the origin of Chinese domestic fowls. Acta Genetica Sinica, 28, 411-417. (in Chinese) |
| [傅衍, 牛冬, 罗静, 阮晖, 何国庆, 张亚平 (2001) 中国家鸡的起源探讨. 遗传学报, 28, 411-417.] | |
| [6] |
Fumihito A, Miyake T, Sumi S, Takada M, Ohno S, Kondo N (1994) One subspecies of the red jungle fowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proceedings of the National Academy of Sciences, USA, 91, 12505-12509.
DOI URL PMID |
| [7] |
Gao YS, Jia XX, Tang XJ, Fan YF, Lu JX, Huang SH, Tang MJ (2017) The genetic diversity of chicken breeds from Jiangxi, assessed with BCDO2 and the complete mitochondrial DNA D-loop region. PLoS ONE, 12, e0173192.
DOI URL PMID |
| [8] |
Hall TA (1999) Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98.
DOI URL |
| [9] |
Huang XH, Chen JB, He DL, Zhang XQ, Zhong FS (2016c) DNA barcoding of indigenous chickens in China: A reevaluation. Scientia Agricultura Sinica, 49, 2622-2633. (in Chinese with English abstract)
DOI URL |
|
[黄勋和, 陈洁波, 何丹林, 张细权, 钟福生 (2016c) DNA条形码技术鉴定地方鸡品种的重新评估. 中国农业科学, 49, 2622-2633.]
DOI URL |
|
| [10] | Huang XH, Li LZ, Zhang JF, He DL, Zhang XQ, Chen JB, Li WN, Du BW, Zhong FS (2016a) Evaluation of diversity and evolution of the microsatellite LEI0258 in chicken MHC-B region from South China. Acta Veterinaria et Zootechnica Sinica, 47, 2175-2183. (in Chinese with English abstract) |
| [黄勋和, 李丽芝, 张金枫, 何丹林, 张细权, 陈洁波, 李威娜, 杜炳旺, 钟福生 (2016a) 华南家鸡MHC-B区域复合微卫星位点LEI0258的遗传多样性与进化研究. 畜牧兽医学报, 47, 2175-2183.] | |
| [11] |
Huang XH, Wu YJ, Miao YW, Peng MS, Chen X, He DL, Suwannapoom C, Du BW, Li XY, Weng ZX, Jin SH, Song JJ, Wang MS, Chen JB, Li WN, Otecko NO, Geng ZY, Qu XY, Wu YP, Yang XR, Jin JQ, Han JL, Zhong FS, Zhang XQ, Zhang YP (2018) Was chicken domesticated in northern China? New evidence from mitochondrial genomes. Science Bulletin, DOI: 10.1016/j.scib.2017.12.004.
DOI URL |
| [12] |
Huang XH, Zhang JF, He DL, Zhang XQ, Zhong FS, Li WN, Zheng QM, Chen JB, Du BW (2016) Genetic diversity and population structure of indigenous chicken breeds in South China. Frontiers of Agricultural Science and Engineering, 3, 97-101.
DOI URL |
| [13] |
Huang XH, Zhang JF, Tan KF, Li LZ, Li WN, Zhong FS (2016b) Diversity and evolution of the highly tandem repeat LEI0258 with in MHC-B region in Wuhua three-yellow chicken. Guangdong Agricultural Sciences, 43, 163-168. (in Chinese with English abstract)
DOI URL |
|
[黄勋和, 张金枫, 谭坤凤, 李丽芝, 李威娜, 钟福生 (2016b) 五华三黄鸡MHC-B区域复合微卫星位点LEI0258遗传多样性与进化研究. 广东农业科学, 43, 163-168.]
DOI URL |
|
| [14] |
Jia XX, Tang XJ, Fan YF, Lu JX, Huang SH, Ge QL, Gao YS, Han W (2017) Genetic diversity of local chicken breeds in East China based on mitochondrial DNA D-loop region. Biodiversity Science, 25, 540-548. (in Chinese with English abstract)
DOI URL |
|
[贾晓旭, 唐修君, 樊艳凤, 陆俊贤, 黄胜海, 葛庆联, 高玉时, 韩威 (2017) 华东地区地方鸡品种mtDNA控制区遗传多样性. 生物多样性, 25, 540-548.]
DOI URL |
|
| [15] |
Lan D, Hu YD, Zhu Q, Liu Y (2017) Mitochondrial DNA study in domestic chicken. Mitochondrial DNA Part A: DNA Mapping, Sequencing, and Analysis, 28, 25-29.
DOI URL PMID |
| [16] |
Langford SM, Kraitsek S, Baskerville B, Ho SYW, Gongora J (2013) Australian and Pacific contributions to the genetic diversity of Norfolk Island feral chickens. BMC Genetics, 14, 91.
DOI URL PMID |
| [17] | Lawler A (2015) Why Did the Chicken Cross the World? G Duckworth, New York. |
| [18] |
Liao YY, Mo GD, Sun JL, Wei FY, Liao DJ (2016) Genetic diversity of Guangxi chicken breeds assessed with microsatellites and the mitochondrial DNA D-loop region. Molecular Biology Reports, 43, 415-425.
DOI URL PMID |
| [19] |
Liu YP, Wu GS, Yao YG, Miao YW, Luikart G, Baig M, Beja-Pereira A, Ding ZL, Palanichamy MG, Zhang YP (2006) Multiple maternal origins of chickens: Out of the Asian jungles. Molecular Phylogenetics and Evolution, 38, 12-19.
DOI URL PMID |
| [20] |
Miao YW, Peng MS, Wu GS, Ouyang YN, Yang ZY, Yu N, Liang JP, Pianchou G, Beja-Pereira A, Mitra B, Palanichamy MG, Baig M, Chaudhuri TK, Shen YY, Kong QP, Murphy RW, Yao YG, Zhang YP (2013) Chicken domestication: An updated perspective based on mitochondrial genomes. Heredity, 110, 277-282.
DOI URL |
| [21] |
Peng MS, Shi NN, Yao YG, Zhang YP (2015) Caveats about interpretation of ancient chicken mtDNAs from northern China. Proceedings of the National Academy of Sciences, USA, 112, E1970-E1971.
DOI URL PMID |
| [22] |
Peters J, Lebrasseur O, Best J, Miller H, Fothergill T, Dobney K, Thomas RM, Maltby M, Sykes N, Hanotte O, O’Connor T, Collins MJ, Larson G (2015) Questioning new answers regarding Holocene chicken domestication in China. Proceedings of the National Academy of Sciences, USA, 112, E2416.
DOI URL PMID |
| [23] |
Qu H, Shu DM, Yang CF (2004) The actuality and development proposal in Guangdong quality chicken germplasm resources. Guangdong Agricultural Sciences, 31(6), 4-7. (in Chinese)
DOI URL |
|
[瞿浩, 舒鼎铭, 杨纯芬 (2004) 广东优质鸡种质资源的现状及发展建议. 广东农业科学, 31(6), 4-7.]
DOI URL |
|
| [24] |
Qu LJ, Li XY, Xu GF, Chen KW, Yang HJ, Zhang LC, Wu GQ, Hou ZC, Xu GY, Yang N (2006) Evaluation of genetic diversity in Chinese indigenous chicken breeds using microsatellite markers. Science in China Series C: Life Science, 49, 332-341.
DOI URL PMID |
| [25] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large datasets. Molecular Biology Evolution, 34, 3299-3302.
DOI URL PMID |
| [26] |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution, 30, 2725-2729.
DOI URL PMID |
| [27] |
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882.
DOI URL PMID |
| [28] |
Wu YP, Huo JH, Xie JF, Liu LX, Wei QP, Xie MG, Kang ZF, Ji HY, Ma YH (2014) Phylogeography and origin of Chinese domestic chicken. Mitochondrial DNA, 25, 126-130.
DOI URL PMID |
| [29] | Xiang H, Gao JQ, Yu BQ, Zhou H, Cai DW, Zhang YW, Chen XY, Wang X, Michael H, Zhao XB (2014) Early Holocene chicken domestication in northern China. Proceedings of the National Academy of Sciences, USA, 111, 11564-11569. |
| [30] |
Zhang L, Zhang P, Li QQ, Gaur U, Liu YP, Zhu Q, Zhao XL, Wang Y, Yin HD, Hu YD, Liu AP, Li DY (2017) Genetic evidence from mitochondrial DNA corroborates the origin of Tibetan chickens. PLoS ONE, 12, e0172945.
DOI URL PMID |
| [31] |
Zhang XQ, Liang SD, Zhang DX (2006) Review on new breed selection of quality chicken in Guangdong Province. China Poultry, 28(1), 9-11. (in Chinese)
DOI URL |
|
[张细权, 梁少东, 张德祥 (2006) 广东优质鸡新品种选育回顾. 中国家禽, 28(1), 9-11.]
DOI URL |
| [1] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [2] | 姚祥坦, 张心怡, 陈阳, 袁晔, 程旺大, 王天瑞, 邱英雄. 基于基因组重测序揭示栽培欧菱遗传多样性及‘南湖菱’的起源驯化历史[J]. 生物多样性, 2024, 32(9): 24212-. |
| [3] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [4] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [5] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [6] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [7] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [8] | 朱华. 地质事件和季风气候影响了云南植物区系和植被的演化[J]. 生物多样性, 2023, 31(12): 23262-. |
| [9] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [10] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [11] | 黄宏文, 廖景平. 论我国国家植物园体系建设: 以任务带学科构建国家植物园迁地保护综合体系[J]. 生物多样性, 2022, 30(6): 22220-. |
| [12] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [13] | 崔静, 徐明芳, 章群, 李瑶, 曾晓舒, 李莎. 基于3种线粒体标记探讨中日沿海角木叶鲽遗传多样性差异[J]. 生物多样性, 2022, 30(5): 21485-. |
| [14] | 蔡新宇, 毛晓伟, 赵毅强. 家养动物驯化起源的研究方法与进展[J]. 生物多样性, 2022, 30(4): 21457-. |
| [15] | 孙军, 宋煜尧, 施义锋, 翟键, 燕文卓. 近十年中国海洋生物多样性研究进展[J]. 生物多样性, 2022, 30(10): 22526-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2026 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()