



Biodiv Sci ›› 2016, Vol. 24 ›› Issue (10): 1189-1196. DOI: 10.17520/biods.2016265 cstr: 32101.14.biods.2016265
• Methodologies • Previous Articles Next Articles
Gengping Zhu1,*( ), Huijie Qiao2
), Huijie Qiao2
Received:2016-09-20
Accepted:2016-10-28
Online:2016-10-20
Published:2016-11-10
Contact:
Zhu Gengping
Gengping Zhu, Huijie Qiao. Effect of the Maxent model’s complexity on the prediction of species potential distributions[J]. Biodiv Sci, 2016, 24(10): 1189-1196.
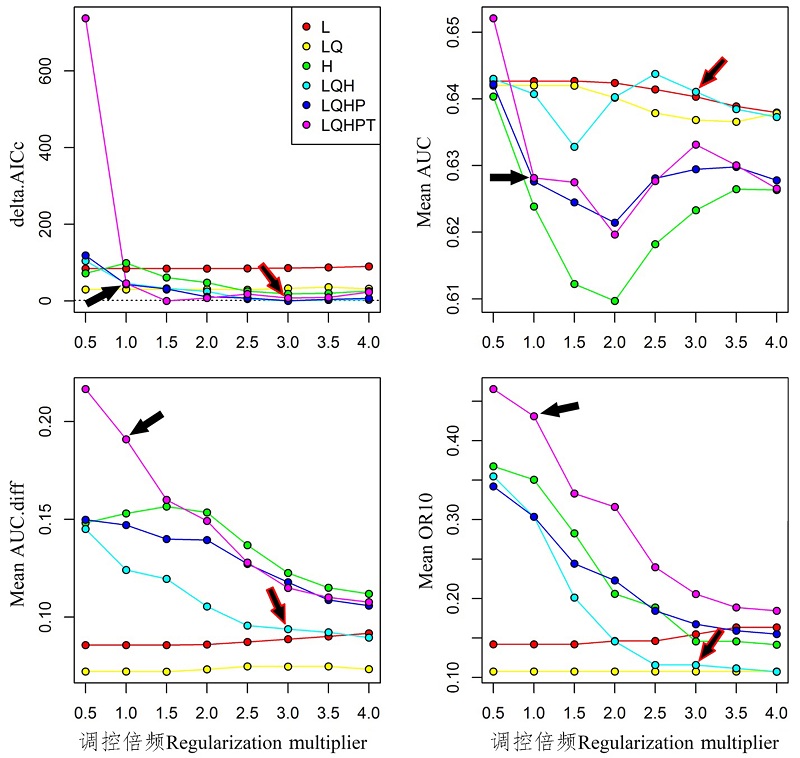
Fig. 1 Performances of native niche model of Halyomorpha halys under different settings. Black arrow indicates default setting, rededge arrow indicates the AICc-chosen setting.
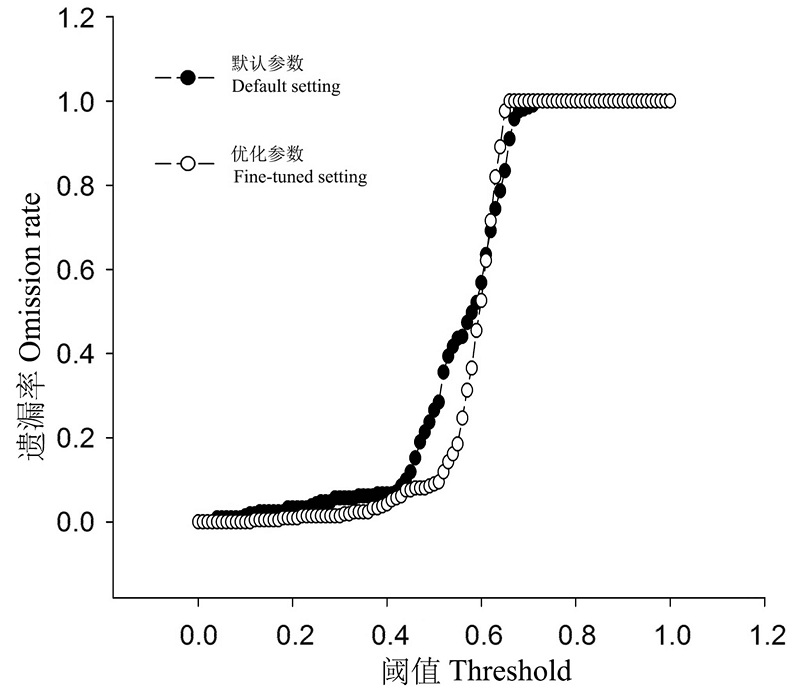
Fig. 4 Omission rates of North American records of Halyomorpha halys based on its native Maxent models predictions using the default and fine-tuned settings.
| 1 | Ahmed SE, McInerny G, O’Hara K, Harper R, Salido L, Emmott S, Joppa LN (2015) Scientists and software - surveying the species distribution modelling community. Diversity and Distributions, 21, 258-267. |
| 2 | Akaike H (1973) Information theory and an extension of the maximum likelihood principle. In: 2nd International Symposium on Information Theory (eds Petrov BN, Csáki F), pp. 267-281. Akadémiai Kiadó, Budapest. |
| 3 | Barbosa FG, Schneck F (2015) Characteristics of the top-cited papers in species distribution predictive models. Ecological Modelling, 313, 77-83. |
| 4 | Elith J, Phillips SJ, Hastie T, Dudík D, Chee YE, Yates CJ (2010) A statistical explanation of MaxEnt for ecologists. Diversity and Distributions, 17, 43-57. |
| 5 | Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965-1978. |
| 6 | Jiménez-Valverde A, Peterson AT, Soberón J, Overton JM, Aragón P, Lobo JM (2011) Use of niche models in invasive species risk assessments. Biological Invasions, 13, 2785-2797. |
| 7 | Kearney MR, Wintle BA, Porter WP (2010) Correlative and mechanistic models of species distribution provide congruent forecasts under climate change. Conservation Letters, 3, 203-213. |
| 8 | Kriticos DJ, Webber BL, Leriche A, Ota N, Macadam I, Bathols J, Scott JK (2011) CliMond: global high resolution historical and future scenario climate surfaces for bioclimatic modeling. Methods in Ecology and Evolution, 3, 53-64. |
| 9 | Muscarella R, Galante PJ, Soley-Guardia M, Boria RA, Kass JM, Uriarte M, Anderson RP (2014) ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for MAXENT ecological niche models. Methods in Ecology and Evolution, 5, 1198-1205. |
| 10 | Peterson AT, Papeş M, Soberón J (2008) Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecological Modelling, 213, 63-72. |
| 11 | Peterson AT, Soberón J (2012) Species distribution modeling and ecological niche modeling: getting the concepts right. Natureza & Conservacao, 10, 102-107. |
| 12 | Peterson AT, Soberón J, Pearson RG, Anderson RP, Nakamura M, Martínez-Meyer E, Araújo MB (2011) Ecological Niches and Geographical Distributions. Princeton University Press, New Jersey. |
| 13 | Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecological Modelling, 190, 231-259. |
| 14 | Phillips SJ, Dudík MM (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography, 31, 161-175. |
| 15 | Qiao HJ, Hu JH, Huang JH (2013) Theoretical basis, future directions, and challenges for ecological niche models. Scientia Sinica Vitae, 43, 915-927. (in Chinese with English abstract) |
| [乔慧捷, 胡军华, 黄继红 (2013) 生态位模型的理论基础、发展方向与挑战. 中国科学: 生命科学, 43, 915-927.] | |
| 16 | Qiao HJ, Soberón J, Peterson AT (2015) No silver bullets in correlative ecological niche modeling: insights from testing among many potential algorithms for niche estimation. Methods in Ecology and Evolution, 6, 1126-1136. |
| 17 | Soberón J, Peterson AT (2005) Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodiversity Informatics, 2, 1-10. |
| 18 | Vaz UL, Cunha HF, Nabout JC (2015) Trends and biases in global scientific literature about ecological niche models. Brazilian Journal of Biology, 75, 17-24. |
| 19 | Warren DL, Seifert SN (2011) Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecology Applications, 21, 335-342. |
| 20 | Warren DL, Wright AN, Seifert SN, Shaffer HB (2014) Incorporating model complexity and spatial sampling bias into ecological niche models of climate change risks faced by 90 California vertebrate species of concern. Diversity and Distributions, 20, 334-343. |
| 21 | Zhu GP, Bu WJ, Gao YB, Liu GQ (2012) Potential geographic distribution of brown marmorated stink bug invasion (Halyomorpha halys). PLoS ONE, 7, e31246. |
| 22 | Zhu GP, Gao YB, Zhu L (2013) Delimiting the coastal geographic background to predict potential distribution of Spartina alterniflora. Hydrobiologia, 717, 177-187. |
| 23 | Zhu GP, Redei D, Kment P, Bu WJ (2014) Effect of geographic background and equilibrium state on niche model transferability: predicting areas of invasion of Leptoglossus occidentalis. Biological Invasions, 16, 1069-1081. |
| 24 | Zhu GP, Gariepy TD, Haye T, Bu WJ (2016) Patterns of niche filling and expansion across the invaded ranges of Halyomorpha halys in North America and Europe. Journal of Pest Science, doi:10.1007/s10340-016-0786-z. |
| 25 | Zhu GP, Liu GQ, Bu WJ, Gao YB (2013) Ecological niche modeling and its applications in biodiversity conservation. Biodiversity Science, 21, 90-98. (in Chinese with English abstract) |
| [朱耿平, 刘国卿, 卜文俊, 高玉葆 (2013) 生态位模型的基本原理及其在生物多样性保护中的应用. 生物多样性, 21, 90-98.] | |
| 26 | Zhu GP, Liu Q, Gao YB (2014) Improving ecological niche model transferability to predict the potential distribution of invasive exotic species. Biodiversity Science, 22, 223-230. (in Chinese with English abstract) |
| [朱耿平, 刘强, 高玉葆 (2014) 提高生态位模型转移能力来模拟入侵物种的潜在分布. 生物多样性, 22, 223-230.] |
| [1] | Fu Mengdi, Zhu Yanpeng, Ren Yueheng, Li Shuang, Qin Le, Xie Zhengjun, Wang Qingchun, Zhang Libo. Research on the optimization of wildlife passage spatial layout in Xinjiang [J]. Biodiv Sci, 2025, 33(3): 24346-. |
| [2] | Xuejiao Yuan, Yuanyuan Zhang, Yanliang Zhang, Luyi Hu, Weiguo Sang, Zheng Yang, Qi Chen. Investigating the prediction ability of the species distribution model fitted with the historical distribution records of Chromolaena odorata [J]. Biodiv Sci, 2024, 32(11): 24288-. |
| [3] | Tingwei Dong, Meiling Huang, Xu Wei, Shuo Ma, Qu Yue, Wenli Liu, Jiaxin Zheng, Gang Wang, Rui Ma, Youzhong Ding, Shunqi Bo, Zhenghuan Wang. Potential spatial distribution pattern and landscape connectivity of Pelophylax plancyi in Shanghai, China [J]. Biodiv Sci, 2023, 31(8): 22692-. |
| [4] | Wei Liu, Ruge Wang, Tianqiao Fan, Nayiman Abudulijiang, Xinhang Song, Shuping Xiao, Ning Guo, Lingying Shuai. Habitat suitability for the Aviceda leuphotes in Mingxi County, Fujian Province [J]. Biodiv Sci, 2023, 31(7): 22660-. |
| [5] | Chang Deng, Jiewei Hao, De Gao, Mingxun Ren, Lina Zhang. Identification and protection of suitable habitat hotspots for threatened bryophytes in Hainan [J]. Biodiv Sci, 2023, 31(4): 22580-. |
| [6] | Qiongyue Zhang, Zhuodi Deng, Xuebin Hu, Zhifeng Ding, Rongbo Xiao, Chen Xiu, Zhenghao Wu, Guang Wang, Donghui Han, Yuke Zhang, Jianchao Liang, Huijian Hu. The impact of urbanization on regional bird distribution and habitat connectivity in the Guangdong-Hong Kong-Macao Greater Bay Area [J]. Biodiv Sci, 2023, 31(3): 22161-. |
| [7] | Bo Wei, Linshan Liu, Changjun Gu, Haibin Yu, Yili Zhang, Binghua Zhang, Bohao Cui, Dianqing Gong, Yanli Tu. The climate niche is stable and the distribution area of Ageratina adenophora is predicted to expand in China [J]. Biodiv Sci, 2022, 30(8): 21443-. |
| [8] | Jirong Teng, Xingming Liu, Liwen He, Junliang Wang, Jian Huang, Jie Feng, Fang Wang, Yue Weng. The spatio-temporal impact of domestic dogs (Canis familiaris) on giant panda (Ailuropoda melanoleuca) in Baishuijiang National Nature Reserve [J]. Biodiv Sci, 2022, 30(1): 21204-. |
| [9] | Chen Zhang, Wei Ma, Chen Chen, Muyang Wang, Wenxuan Xu, Weikang Yang. Changes of habitat pattern for goitered gazelle in the Xinjiang Kalamaili Mountain Ungulate Nature Reserve under the influence of major projects [J]. Biodiv Sci, 2022, 30(1): 21176-. |
| [10] | Xing Ma, Hao Wang, Wei Yu, Yong Du, Jianchao Liang, Huijian Hu, Shengrong Qiu, Lu Liu. Analysis on the hotspot and conservation gaps of bird biodiversity in Guangdong Province based on MaxEnt model [J]. Biodiv Sci, 2021, 29(8): 1097-1107. |
| [11] | Run Zhou, Xiuqin Ci, Jianhua Xiao, Guanlong Cao, Jie Li. Effects and conservation assessment of climate change on the dominant group—The genusCinnamomum of subtropical evergreen broad-leaved forests [J]. Biodiv Sci, 2021, 29(6): 697-711. |
| [12] | Yuhan Shi, Zongxin Ren, Weijia Wang, Xin Xu, Jie Liu, Yanhui Zhao, Hong Wang. Predicting the spatial distribution of three Astragalusspecies and their pollinating bumblebees in the Sino-Himalayas [J]. Biodiv Sci, 2021, 29(6): 759-769. |
| [13] | Ran Wang, Huijie Qiao. Matters needing attention about invoking ecological niche model in epidemiology [J]. Biodiv Sci, 2020, 28(5): 579-586. |
| [14] | Hai-Sheng Yuan, Yulian Wei, Liwei Zhou, Wenmin Qin, Baokai Cui, Shuanghui He. Potential distribution and ecological niches of four butt-rot pathogenic fungi in Northeast China [J]. Biodiv Sci, 2019, 27(8): 873-879. |
| [15] | Yifeng Hu, Wenhua Yu, Yang Yue, Zhenglanyi Huang, Yuchun Li, Yi Wu. Species diversity and potential distribution of Chiroptera on Hainan Island, China [J]. Biodiv Sci, 2019, 27(4): 400-408. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2026 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn