



生物多样性 ›› 2011, Vol. 19 ›› Issue (4): 485-493. DOI: 10.3724/SP.J.1003.2011.09232 cstr: 32101.14.SP.J.1003.2011.09232
所属专题: 土壤生物与土壤健康
收稿日期:2010-09-25
接受日期:2011-02-19
出版日期:2011-07-20
发布日期:2011-07-29
通讯作者:
崔海瑞
作者简介:* E-mail: hrcui@zju.edu.cn基金资助:
Chunnan Li1,2, Hairui Cui1,*( ), Weibo Wang1
), Weibo Wang1
Received:2010-09-25
Accepted:2011-02-19
Online:2011-07-20
Published:2011-07-29
Contact:
Hairui Cui
摘要:
将近年来新建立的分子标记技术——相关序列扩增多态性(sequence-related amplified polymorphism, SRAP)应用于土壤微生物遗传多样性研究。采用22对引物组合对20种植物根际土壤微生物进行了分析, 共获得237个扩增位点, 其中多态性位点221个, 占93.2%。平均每对引物组合的多态位点比例(PPL)、多态信息含量(PIC)、等位基因单体型(Ah)和遗传杂合度(He)分别为93.78%、0.94、18.05和0.92, 说明SRAP对根际土壤微生物具有较强的鉴别能力, 也反映了本研究中20种不同植物的根际土壤微生物具有丰富的遗传多样性。水稻的2个不同种植地和4个不同发育时期间的根际土壤微生物遗传距离差异极显著, 但2个不同水稻品种间的差异不显著。Shannon多样性指数揭示, 水稻根际土壤微生物的遗传多样性最低, 莴苣的最高。按照非加权类平均法(UPGMA)聚类, 在遗传距离0.454的水平上, 可将20种植物根际土壤微生物分为三类: 第一类是水稻根际土壤微生物, 第二类是种植于大棚温室的芹菜根际土壤微生物, 第三类为其余18种旱作植物根际土壤微生物。本研究结果证明SRAP是分析土壤微生物遗传多样性的有效手段。
李春楠, 崔海瑞, 王伟博 (2011) 用SRAP标记研究根际土壤微生物的遗传多样性. 生物多样性, 19, 485-493. DOI: 10.3724/SP.J.1003.2011.09232.
Chunnan Li, Hairui Cui, Weibo Wang (2011) Genetic diversity in rhizosphere soil microbes detected with SRAP markers. Biodiversity Science, 19, 485-493. DOI: 10.3724/SP.J.1003.2011.09232.
| 编号 Code | 植物 Plant species |
|---|---|
| A 区 Zone A | |
| OS | 水稻 Rice (Oryza sativa) |
| B 区 Zone B | |
| AG | 芹菜 Celery (Apium graveolens) |
| AS | 大蒜 Garlic (Allium sativum) |
| AT | 韭菜 Chinese chives (Allium tuberosum) |
| BC | 白菜 Chinese cabbage (Brassica campestris ssp. pekinensis) |
| BN | 油菜 Rapeseed (Brassica napus) |
| CS | 茶树 Tea plant (Camellia sinensis) |
| GM | 大豆 Soybean (Glycine max) |
| LE | 番茄 Tomato (Lycopersicon esculentum) |
| LS | 莴苣 Lettuce (Lactuca sativa) |
| PS | 豌豆 Pea (Pisum sativum) |
| RS | 萝卜 Radish (Raphanus sativus) |
| SM | 茄子 Eggplant (Solanum melongena) |
| ST | 马铃薯 Potato (Solanum tuberosum) |
| TA | 普通小麦 Wheat (Triticum aestivum) |
| ZM | 玉米 Maize (Zea mays) |
| C 区 Zone C | |
| BO | 甘蓝 Cabbage (Brassica oleracea) |
| FA | 高羊茅 Tall fescue (Festuca arundinacea) |
| HV | 大麦 Barley (Hordeum vulgare) |
| VF | 蚕豆 Broad bean (Vicia faba) |
表1 20种植物根际土壤取样编号
Table 1 Codes of rhizosphere soils sampled from 20 plant species
| 编号 Code | 植物 Plant species |
|---|---|
| A 区 Zone A | |
| OS | 水稻 Rice (Oryza sativa) |
| B 区 Zone B | |
| AG | 芹菜 Celery (Apium graveolens) |
| AS | 大蒜 Garlic (Allium sativum) |
| AT | 韭菜 Chinese chives (Allium tuberosum) |
| BC | 白菜 Chinese cabbage (Brassica campestris ssp. pekinensis) |
| BN | 油菜 Rapeseed (Brassica napus) |
| CS | 茶树 Tea plant (Camellia sinensis) |
| GM | 大豆 Soybean (Glycine max) |
| LE | 番茄 Tomato (Lycopersicon esculentum) |
| LS | 莴苣 Lettuce (Lactuca sativa) |
| PS | 豌豆 Pea (Pisum sativum) |
| RS | 萝卜 Radish (Raphanus sativus) |
| SM | 茄子 Eggplant (Solanum melongena) |
| ST | 马铃薯 Potato (Solanum tuberosum) |
| TA | 普通小麦 Wheat (Triticum aestivum) |
| ZM | 玉米 Maize (Zea mays) |
| C 区 Zone C | |
| BO | 甘蓝 Cabbage (Brassica oleracea) |
| FA | 高羊茅 Tall fescue (Festuca arundinacea) |
| HV | 大麦 Barley (Hordeum vulgare) |
| VF | 蚕豆 Broad bean (Vicia faba) |
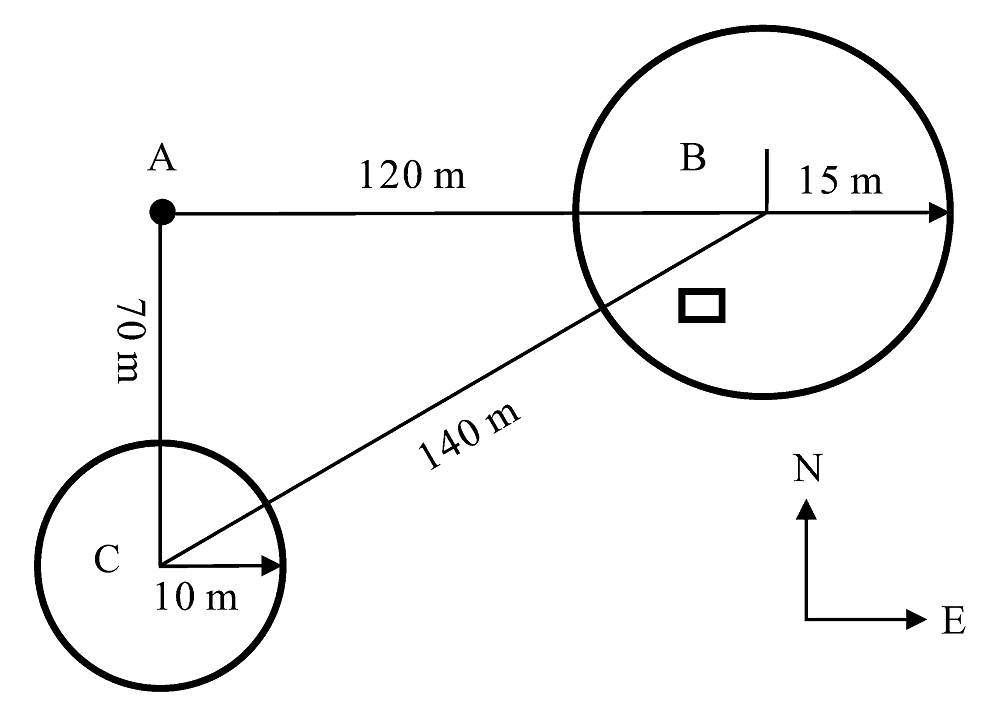
图1 20种植物根际土壤取样分布图(B中?表示种植芹菜的塑料大棚)
Fig. 1 Distribution of rhizosphere soils sampled from 20 plant species (? in B stands for plastic green house for planting celery)
| 取样地点 Sites | pH | 有机质 Organic matter (g/kg) | 全氮 Total N (g/kg) | 速效P Available phosphorus (mg/kg) | 速效K Available potassium (mg/kg) |
|---|---|---|---|---|---|
| A区 | 6.47 | 18.59 | 1.13 | 18.40 | 43.80 |
| B区 | 7.20 | 16.70 | 1.44 | 52.54 | 61.76 |
| C区 | 7.15 | 15.10 | 1.80 | 48.52 | 58.10 |
表2 3个取样区域土壤的理化性质
Table 2 Physical and chemical properties of soil in the three sampling zones
| 取样地点 Sites | pH | 有机质 Organic matter (g/kg) | 全氮 Total N (g/kg) | 速效P Available phosphorus (mg/kg) | 速效K Available potassium (mg/kg) |
|---|---|---|---|---|---|
| A区 | 6.47 | 18.59 | 1.13 | 18.40 | 43.80 |
| B区 | 7.20 | 16.70 | 1.44 | 52.54 | 61.76 |
| C区 | 7.15 | 15.10 | 1.80 | 48.52 | 58.10 |
| 编号 Code | 正向引物序列 Forward primer (5'-3') | 编号 Code | 反向引物序列 Reverse primer (5′-3′) |
|---|---|---|---|
| F1 | TGAGTCCAAACCGGATA | R1 | GACTGCGTACGAATTAAT |
| F2 | TGAGTCCAAACCGGAGC | R2 | GACTGCGTACGAATTTGC |
| F3 | TGAGTCCAAACCGGAAT | R3 | GACTGCGTACGAATTGAC |
| F4 | TGAGTCCAAACCGGACC | R4 | GACTGCGTACGAATTTGA |
| F5 | TGAGTCCAAACCGGAAG | R5 | GACTGCGTACGAATTAAC |
| F6 | TGAGTCCAAACCGGTAA | R6 | GACTGCGTACGAATTGCA |
| F7 | TGAGTCCAAACCGGTCC | R7 | GACTGCGTACGAATTCAA |
| F8 | TGAGTCCAAACCGGTGC | R8 | GACTGCGTACGAATTAGC |
表3 本研究所用SRAP正反引物序列
Table 3 Sequences of SRAP forward and reverse primers used in the present study
| 编号 Code | 正向引物序列 Forward primer (5'-3') | 编号 Code | 反向引物序列 Reverse primer (5′-3′) |
|---|---|---|---|
| F1 | TGAGTCCAAACCGGATA | R1 | GACTGCGTACGAATTAAT |
| F2 | TGAGTCCAAACCGGAGC | R2 | GACTGCGTACGAATTTGC |
| F3 | TGAGTCCAAACCGGAAT | R3 | GACTGCGTACGAATTGAC |
| F4 | TGAGTCCAAACCGGACC | R4 | GACTGCGTACGAATTTGA |
| F5 | TGAGTCCAAACCGGAAG | R5 | GACTGCGTACGAATTAAC |
| F6 | TGAGTCCAAACCGGTAA | R6 | GACTGCGTACGAATTGCA |
| F7 | TGAGTCCAAACCGGTCC | R7 | GACTGCGTACGAATTCAA |
| F8 | TGAGTCCAAACCGGTGC | R8 | GACTGCGTACGAATTAGC |
| 引物组合 Primer combination | 扩增片段大小 Fragment size (bp) | 位点总数 Total fragments | 多态位点比例 PPL (%) | 等位基因单体型 Ah | 多态信息含量 PIC | 遗传杂合度 He |
|---|---|---|---|---|---|---|
| F1/R3* | 110-310 | 14 | 92.86 | 20 | 0.95 | 0.9127 |
| F1/R6* | 180-300 | 9 | 88.89 | 18 | 0.94 | 0.8732 |
| F1/R7* | 120-290 | 7 | 85.71 | 13 | 0.90 | 0.9909 |
| F1/R8 | 70-280 | 17 | 76.47 | 19 | 0.95 | 0.9137 |
| F2/R3* | 90-300 | 13 | 100.00 | 20 | 0.95 | 0.9921 |
| F2/R4* | 50-300 | 16 | 87.50 | 19 | 0.95 | 0.9975 |
| F2/R7* | 100-300 | 10 | 100.00 | 17 | 0.94 | 0.9527 |
| F3/R2 | 120-280 | 9 | 100.00 | 19 | 0.95 | 0.8854 |
| F3/R3* | 100-290 | 11 | 81.82 | 16 | 0.93 | 0.9770 |
| F3/R8* | 110-270 | 8 | 75.00 | 15 | 0.91 | 0.9944 |
| F4/R2 | 90-310 | 7 | 100.00 | 12 | 0.90 | 0.8884 |
| F4/R4 | 130-480 | 9 | 100.00 | 18 | 0.94 | 0.9912 |
| F5/R1 | 140-280 | 11 | 100.00 | 19 | 0.95 | 0.9077 |
| F5/R2 | 130-500 | 10 | 100.00 | 19 | 0.95 | 0.8878 |
| F5/R3 | 60-310 | 12 | 91.67 | 19 | 0.95 | 0.8651 |
| F5/R6 | 130-290 | 10 | 100.00 | 20 | 0.95 | 0.8368 |
| F5/R7 | 80-480 | 15 | 93.33 | 20 | 0.95 | 0.8909 |
| F6/R1 | 130-400 | 9 | 100.00 | 20 | 0.95 | 0.8732 |
| F6/R7* | 110-450 | 10 | 90.00 | 16 | 0.93 | 0.8979 |
| F6/R8 | 100-470 | 8 | 100.00 | 18 | 0.94 | 0.8872 |
| F8/R5* | 80-460 | 12 | 100.00 | 20 | 0.95 | 0.9222 |
| F8/R7* | 120-470 | 10 | 100.00 | 20 | 0.95 | 0.8668 |
表4 22对SRAP引物组合在20种植物根际土壤微生物中的多态性信息
Table 4 Polymorphic information of rhizosphere soil microbes from 20 plant species with 22 SRAP primer combinations
| 引物组合 Primer combination | 扩增片段大小 Fragment size (bp) | 位点总数 Total fragments | 多态位点比例 PPL (%) | 等位基因单体型 Ah | 多态信息含量 PIC | 遗传杂合度 He |
|---|---|---|---|---|---|---|
| F1/R3* | 110-310 | 14 | 92.86 | 20 | 0.95 | 0.9127 |
| F1/R6* | 180-300 | 9 | 88.89 | 18 | 0.94 | 0.8732 |
| F1/R7* | 120-290 | 7 | 85.71 | 13 | 0.90 | 0.9909 |
| F1/R8 | 70-280 | 17 | 76.47 | 19 | 0.95 | 0.9137 |
| F2/R3* | 90-300 | 13 | 100.00 | 20 | 0.95 | 0.9921 |
| F2/R4* | 50-300 | 16 | 87.50 | 19 | 0.95 | 0.9975 |
| F2/R7* | 100-300 | 10 | 100.00 | 17 | 0.94 | 0.9527 |
| F3/R2 | 120-280 | 9 | 100.00 | 19 | 0.95 | 0.8854 |
| F3/R3* | 100-290 | 11 | 81.82 | 16 | 0.93 | 0.9770 |
| F3/R8* | 110-270 | 8 | 75.00 | 15 | 0.91 | 0.9944 |
| F4/R2 | 90-310 | 7 | 100.00 | 12 | 0.90 | 0.8884 |
| F4/R4 | 130-480 | 9 | 100.00 | 18 | 0.94 | 0.9912 |
| F5/R1 | 140-280 | 11 | 100.00 | 19 | 0.95 | 0.9077 |
| F5/R2 | 130-500 | 10 | 100.00 | 19 | 0.95 | 0.8878 |
| F5/R3 | 60-310 | 12 | 91.67 | 19 | 0.95 | 0.8651 |
| F5/R6 | 130-290 | 10 | 100.00 | 20 | 0.95 | 0.8368 |
| F5/R7 | 80-480 | 15 | 93.33 | 20 | 0.95 | 0.8909 |
| F6/R1 | 130-400 | 9 | 100.00 | 20 | 0.95 | 0.8732 |
| F6/R7* | 110-450 | 10 | 90.00 | 16 | 0.93 | 0.8979 |
| F6/R8 | 100-470 | 8 | 100.00 | 18 | 0.94 | 0.8872 |
| F8/R5* | 80-460 | 12 | 100.00 | 20 | 0.95 | 0.9222 |
| F8/R7* | 120-470 | 10 | 100.00 | 20 | 0.95 | 0.8668 |
| 样品编号 Sample codes | 位点总数 Total fragments | 多态位点数 No. of polymorphic loci | 多态位点比例 PPL (%) | Shannon多样性指数 I |
|---|---|---|---|---|
| 水稻 OS | 116 | 99 | 85.34 | 0.2558 |
| 芹菜 AG | 118 | 101 | 85.59 | 0.2579 |
| 大蒜 AS | 149 | 132 | 88.59 | 0.3156 |
| 韭菜 AT | 129 | 112 | 86.82 | 0.2774 |
| 白菜 BC | 152 | 135 | 88.82 | 0.3205 |
| 油菜 BN | 144 | 127 | 88.19 | 0.3019 |
| 茶树 CS | 149 | 132 | 88.59 | 0.3144 |
| 大豆 GM | 145 | 128 | 88.28 | 0.3086 |
| 番茄 LE | 157 | 140 | 89.17 | 0.3281 |
| 莴苣 LS | 166 | 149 | 89.76 | 0.3457 |
| 豌豆 PS | 156 | 139 | 89.10 | 0.3258 |
| 萝卜 RS | 138 | 121 | 87.68 | 0.2945 |
| 茄子 SM | 132 | 115 | 87.12 | 0.2823 |
| 马铃薯 ST | 154 | 137 | 88.96 | 0.3240 |
| 普通小麦 TA | 125 | 108 | 86.40 | 0.2714 |
| 玉米 ZM | 144 | 127 | 88.19 | 0.3066 |
| 甘蓝 BO | 154 | 137 | 88.96 | 0.3243 |
| 高羊茅 FA | 126 | 109 | 86.51 | 0.2728 |
| 大麦 HV | 141 | 124 | 87.94 | 0.2993 |
| 蚕豆 VF | 131 | 114 | 87.02 | 0.2817 |
表6 20种植物根际土壤微生物的SRAP扩增和Shannon多样性指数
Table 6 SRAP amplification and Shannon diversity index of rhizosphere soil microbes from 20 plant species
| 样品编号 Sample codes | 位点总数 Total fragments | 多态位点数 No. of polymorphic loci | 多态位点比例 PPL (%) | Shannon多样性指数 I |
|---|---|---|---|---|
| 水稻 OS | 116 | 99 | 85.34 | 0.2558 |
| 芹菜 AG | 118 | 101 | 85.59 | 0.2579 |
| 大蒜 AS | 149 | 132 | 88.59 | 0.3156 |
| 韭菜 AT | 129 | 112 | 86.82 | 0.2774 |
| 白菜 BC | 152 | 135 | 88.82 | 0.3205 |
| 油菜 BN | 144 | 127 | 88.19 | 0.3019 |
| 茶树 CS | 149 | 132 | 88.59 | 0.3144 |
| 大豆 GM | 145 | 128 | 88.28 | 0.3086 |
| 番茄 LE | 157 | 140 | 89.17 | 0.3281 |
| 莴苣 LS | 166 | 149 | 89.76 | 0.3457 |
| 豌豆 PS | 156 | 139 | 89.10 | 0.3258 |
| 萝卜 RS | 138 | 121 | 87.68 | 0.2945 |
| 茄子 SM | 132 | 115 | 87.12 | 0.2823 |
| 马铃薯 ST | 154 | 137 | 88.96 | 0.3240 |
| 普通小麦 TA | 125 | 108 | 86.40 | 0.2714 |
| 玉米 ZM | 144 | 127 | 88.19 | 0.3066 |
| 甘蓝 BO | 154 | 137 | 88.96 | 0.3243 |
| 高羊茅 FA | 126 | 109 | 86.51 | 0.2728 |
| 大麦 HV | 141 | 124 | 87.94 | 0.2993 |
| 蚕豆 VF | 131 | 114 | 87.02 | 0.2817 |
| 地点 Location | 变化范围 Range | 变异系数 CV (%) | 标准差 SD | 平均 Average |
|---|---|---|---|---|
| 华家池 HJC | 0.0907-0.1771 | 16.66 | 0.0207 | 0.1243A |
| 建德 JD | 0.0256-0.0821 | 29.27 | 0.0160 | 0.0546B |
表7 华家池和建德两个种植地点水稻根际土壤微生物的遗传距离参数
Table 7 Parameters of genetic distance of rice rhizosphere soil microbes at Huajiachi (HJC) and Jiande (JD)
| 地点 Location | 变化范围 Range | 变异系数 CV (%) | 标准差 SD | 平均 Average |
|---|---|---|---|---|
| 华家池 HJC | 0.0907-0.1771 | 16.66 | 0.0207 | 0.1243A |
| 建德 JD | 0.0256-0.0821 | 29.27 | 0.0160 | 0.0546B |
| 品种/地点 Variety/location | 变化范围 Range | 变异系数 CV (%) | 标准差 SD | 平均 Average |
|---|---|---|---|---|
| 明恢63/华家池 Minghui 63/HJC | 0.0958-0.1438 | 15.45 | 0.0189 | 0.1226 A |
| 嘉早935/华家池 Jiazao 935/HJC | 0.1002-0.1339 | 11.04 | 0.0128 | 0.1157A |
| 明恢63/建德 Minghui 63/JD | 0.0256-0.0520 | 25.24 | 0.0104 | 0.0412B |
| 嘉早935/建德 Jiazao 935/JD | 0.0334-0.0455 | 10.82 | 0.0044 | 0.0404 B |
表8 嘉早935和明恢63两水稻品种间根际土壤微生物遗传距离参数比较
Table 8 Parameters of genetic distance between Jiazao 935 and Minghui 63 at Huajiachi (HJC) and Jiande (JD)
| 品种/地点 Variety/location | 变化范围 Range | 变异系数 CV (%) | 标准差 SD | 平均 Average |
|---|---|---|---|---|
| 明恢63/华家池 Minghui 63/HJC | 0.0958-0.1438 | 15.45 | 0.0189 | 0.1226 A |
| 嘉早935/华家池 Jiazao 935/HJC | 0.1002-0.1339 | 11.04 | 0.0128 | 0.1157A |
| 明恢63/建德 Minghui 63/JD | 0.0256-0.0520 | 25.24 | 0.0104 | 0.0412B |
| 嘉早935/建德 Jiazao 935/JD | 0.0334-0.0455 | 10.82 | 0.0044 | 0.0404 B |
| 取样时期 Stage | 变化范围 Range | 变异系数 CV (%) | 标准差 SD | 平均 Average |
|---|---|---|---|---|
| 华家池 Huajiachi | ||||
| 成熟期 Maturation | 0.1050-0.1771 | 15.56 | 0.0215 | 0.1381A |
| 抽穗期 Heading | 0.1002-0.1580 | 15.01 | 0.0185 | 0.1232B |
| 分蘖期 Tillering | 0.0907-0.1620 | 17.20 | 0.0205 | 0.1189B |
| 灌浆期 Filling | 0.0958-0.1548 | 14.94 | 0.0175 | 0.1171B |
| 建德 Jiande | ||||
| 成熟期 Maturation | 0.0256-0.0821 | 34.05 | 0.0195 | 0.0573C |
| 灌浆期 Filling | 0.0404-0.0821 | 25.12 | 0.0142 | 0.0565C |
| 分蘖期 Tillering | 0.0333-0.0800 | 30.99 | 0.0165 | 0.0532C |
| 抽穗期 Heading | 0.0256-0.0701 | 28.03 | 0.0144 | 0.0515C |
表9 两个种植地点水稻不同生育期根际土壤微生物的遗传距离参数
Table 9 Parameters of genetic distance of rice rhizosphere soil microbes at different developmental stages in Huajiachi (HJC) and Jiande (JD)
| 取样时期 Stage | 变化范围 Range | 变异系数 CV (%) | 标准差 SD | 平均 Average |
|---|---|---|---|---|
| 华家池 Huajiachi | ||||
| 成熟期 Maturation | 0.1050-0.1771 | 15.56 | 0.0215 | 0.1381A |
| 抽穗期 Heading | 0.1002-0.1580 | 15.01 | 0.0185 | 0.1232B |
| 分蘖期 Tillering | 0.0907-0.1620 | 17.20 | 0.0205 | 0.1189B |
| 灌浆期 Filling | 0.0958-0.1548 | 14.94 | 0.0175 | 0.1171B |
| 建德 Jiande | ||||
| 成熟期 Maturation | 0.0256-0.0821 | 34.05 | 0.0195 | 0.0573C |
| 灌浆期 Filling | 0.0404-0.0821 | 25.12 | 0.0142 | 0.0565C |
| 分蘖期 Tillering | 0.0333-0.0800 | 30.99 | 0.0165 | 0.0532C |
| 抽穗期 Heading | 0.0256-0.0701 | 28.03 | 0.0144 | 0.0515C |
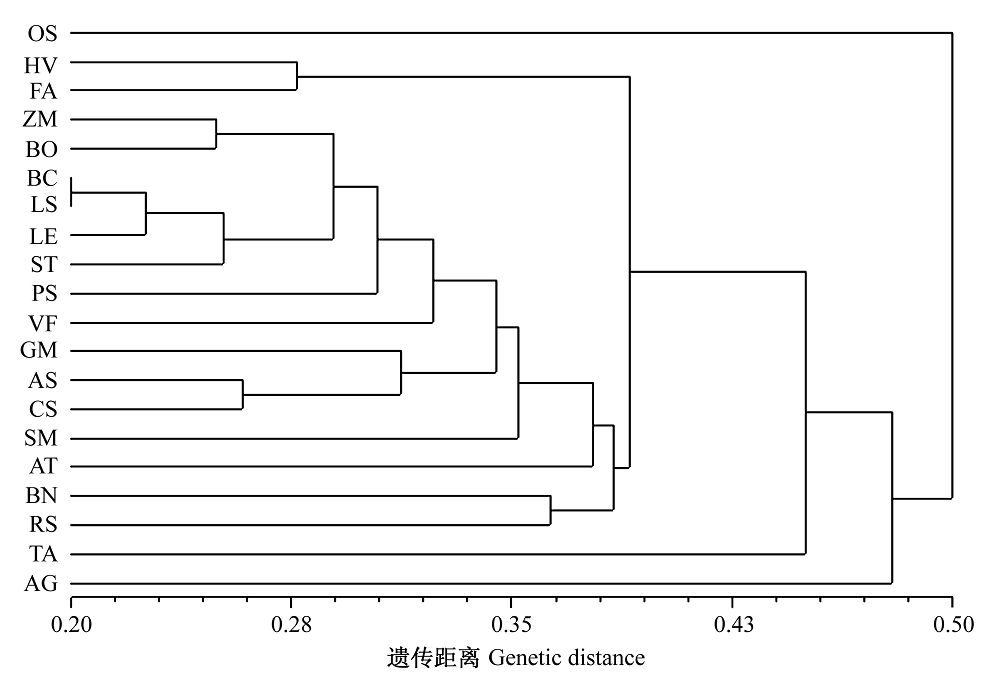
图2 20种植物根际土壤微生物基于SRAP的UPGMA聚类图(土壤取样编号同表1)
Fig. 2 UPGMA dendrogram of rhizosphere soil microbes from 20 plant species generated by SRAP markers. Codes of soil samples see Table 1.
| [1] | Bi JT (毕江涛), He DH (贺达汉) (2009) Research advances in effects of plant on soil microbial diversity. Chinese Agricultural Science Bulletin (中国农学通报), 25, 244-250. (in Chinese with English abstract) |
| [2] |
Borneman J, Triplett EW (1997) Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Applied and Environmental Microbiology, 63, 2647-2653.
DOI URL PMID |
| [3] |
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic-linkage map in man using restriction fragment length polymorphisms. The American Journal of Human Genetics, 32, 314-331.
URL PMID |
| [4] | Carelli M, Gnocchi S, Fancelli S, Mengoni A, Paffetti D, Scotti C, Bazzicalupo M (2000) Genetic diversity and dynamics of Sinorhizobium meliloti populations nodulating different alfalfa cultivars in Italian soils. Applied and Environmental Microbiology, 66, 4785-4789. |
| [5] | Chen J (陈静), Hu XH (胡晓辉), Miao HR (苗华荣), Cui FG (崔凤高), Yu SL (禹山林) (2008) Genome DNA extracted with CTAB method and its use for SSR and SRAP. Journal of Peanut Science (花生学报), 37, 29-31. (in Chinese with English abstract) |
| [6] | Chen XY (陈旭玉), Zhou YK (周亚奎), Yu XM (余贤美), Zheng FC (郑服丛) (2008) An affection method for DNA extraction from soil microorganisms. Chinese Agricultural Science Bulletin (中国农学通报), 24, 33-36. (in Chinese with English abstract) |
| [7] |
Cho JC, Tiedje JM (2001) Bacterial species determination from DNA-DNA hybridization by using genome fragments and DNA microarrays. Applied and Environmental Microbiology, 67, 3677-3682.
URL PMID |
| [8] | Daniel R (2005) The metagenomics of soil. Nature Reviews Microbiology, 3, 470-478. |
| [9] | Fu HT (傅洪拓), Qiao H (乔慧), Yao JH (姚建华), Gong YS (龚永生), Wu Y (吴滟), Jiang SF (蒋速飞), Xiong YW (熊贻伟) (2010) Genetic diversity in five Macrobrachium hainanense populations using SRAP markers. Biodiversity Science (生物多样性), 18, 145-149. (in Chinese with English abstract) |
| [10] |
Head IM, Saunders JR, Pickup RW (1998) Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microbial Ecology, 35, 1-21.
URL PMID |
| [11] | Hugenholtz P, Goebel BM, Pace NR (1998) Impact of culture independent studies on the emerging phylogenetic view of bacterial diversity. Journal of Bacteriology, 180, 4765-4774. |
| [12] |
Hubert C, Shen Y, Voordouw G (1999) Composition of toluene-degrading microbial communities from soil at different concentrations of toluene. Applied and Environmental Microbiology, 65, 3064-3070.
URL PMID |
| [13] | Kirk JL, Beaudette LA, Hart M, Moutoglis P, Klironomos JN, Lee H, Trevors JT (2004) Methods of studying soil microbial diversity. Journal of Microbiological Methods, 58, 169-188. |
| [14] | Lee DH, Zo YG, Kim SJ (1996) Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation polymorphism. Applied and Environmental Microbiology, 62, 3112-3120. |
| [15] | Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theoretical and Applied Genetics, 103, 455-461. |
| [16] | Li Y (李严), Zhang CQ (张春庆) (2005) A molecular marker—SRAP technique optimization and application analysis. Chinese Agricultural Science Bulletin (中国农学通报), 21, 108-112. (in Chinese with English abstract) |
| [17] | Liu LW (柳李旺), Gong YQ (龚义勤), Huang H (黄浩), Zhu XW (朱献文 ) (2004) Novel molecular marker systems—SRAP and TRAP and their application.Hereditas (遗传), 26, 777-781. (in Chinese with English abstract) |
| [18] |
Liu WT, Marsh TL, Cheng H, Forney LJ (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Applied and Environmental Microbiology, 63, 4516-4522.
URL PMID |
| [19] | Muyzer G (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Current Opinion in Microbiology, 2, 317-322. |
| [20] | Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Applied and Environmental Microbiology, 59, 695-700. |
| [21] | Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences, USA, 76, 569-573. |
| [22] | Osborn AM, Moore ERB, Timmis KN (2000) An evaluation of terminal restriction fragment length polymorphisms (T-RFLP) analysis for the study of microbial community structure and dynamics. Environmental Microbiology, 2, 39-50. |
| [23] | Ren M (任民), Jia XH (贾兴华), Jiang CH (蒋彩虹), Yang AG (杨爱国), Wang RJ (王日新) (2008) Comparison study of Bassam and Sanguinetti silver staining in the detecting of SRAP and TRAP. Biotechnology Bulletin (生物技术通报), 1, 113-116. (in Chinese with English abstract) |
| [24] | Rheims H, Spröer C, Rainey FA, Stackebrandt E (1996) Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology, 142, 2863-2870. |
| [25] | Rohlf FJ (2000) NTSYS-PC: Numerical Taxonomy and Multivariate Analysis System, Version 2.1. Exeter Software. Setauket, NewYork. |
| [26] | Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Applied and Environmental Microbiology, 66, 2541-2547. |
| [27] | Terefework Z, Kaijalainen S, Lindström K (2001) AFLP fingerprinting as a tool to study the genetic diversity of Rhizobium galegae isolated from Galega orientalis and Galega officianlis. Journal of Biotechnology, 91, 169-180. |
| [28] | Willems A, Doignon-Bourcier F, Coopman R, Hoste B, de Lajudie P, Gillis M (2000) AFLP fingerprint analysis of Bradyrhizobium strains isolated from Faidherbia albida and Aeschynommene species. Systematic and Applied Microbiology, 23, 137-147. |
| [29] | Xia X, Bollinger J, Ogram A (1995) Molecular genetic analysis of the response of three soil microbial communities to the application of 2,4-D. Molecular Ecology, 4, 17-28. |
| [30] | Zhang JE (章家恩), Liu WG (刘文高), Hu G (胡刚) (2002) The relationship between quantity index of soil microorganisms and soil fertility of different land use systems. Soil and Environmental Sciences (土壤与环境), 11, 140-143. (in Chinese with English abstract) |
| [31] | Zhao X (赵雪), Xie H (谢华), Ma RC (马荣才) (2007) New functional molecular markers for plants in the functional genomics era. China Biotechnology (中国生物工程杂志), 27(8), 104-110. (in Chinese with English abstract) |
| [32] | Zhou J (周桔), Lei T (雷霆) (2007) Review and prospects on methodology and affecting factors of soil microbial diversity. Biodiversity Science (生物多样性), 15, 306-311. (in Chinese with English abstract) |
| [1] | 尚华丹, 张楚晴, 王梅, 裴文娅, 李国宏, 王鸿斌. 中国杨树害虫物种多样性及其地理分布[J]. 生物多样性, 2025, 33(2): 24370-. |
| [2] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [3] | 吴相獐, 雷富民, 单壹壹, 于晶. 上海城市公园苔藓植物多样性分布格局及其环境影响因子[J]. 生物多样性, 2024, 32(2): 23364-. |
| [4] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [5] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [6] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [7] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [8] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [9] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [10] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [11] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [12] | 崔静, 徐明芳, 章群, 李瑶, 曾晓舒, 李莎. 基于3种线粒体标记探讨中日沿海角木叶鲽遗传多样性差异[J]. 生物多样性, 2022, 30(5): 21485-. |
| [13] | 孙军, 宋煜尧, 施义锋, 翟键, 燕文卓. 近十年中国海洋生物多样性研究进展[J]. 生物多样性, 2022, 30(10): 22526-. |
| [14] | 栗冬梅, 杨卫红, 李庆多, 韩茜, 宋秀平, 潘虹, 冯云. 巴尔通体在滇西南蝙蝠中高度流行并具有丰富的遗传变异特征[J]. 生物多样性, 2021, 29(9): 1245-1255. |
| [15] | 姚志, 郭军, 金晨钟, 刘勇波. 中国纳入一级保护的极小种群野生植物濒危机制[J]. 生物多样性, 2021, 29(3): 394-408. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn