



生物多样性 ›› 2019, Vol. 27 ›› Issue (4): 366-372. DOI: 10.17520/biods.2018332 cstr: 32101.14.biods.2018332
陈志祥1, 姚雪莹1, Stephen R.Downie2, 王奇志1,*( )
)
收稿日期:2018-12-18
接受日期:2019-01-09
出版日期:2019-04-20
发布日期:2019-06-05
通讯作者:
王奇志
基金资助:
Zhixiang Chen1, Xueying Yao1, R. Downie Stephen2, Qizhi Wang1,*( )
)
Received:2018-12-18
Accepted:2019-01-09
Online:2019-04-20
Published:2019-06-05
Contact:
Qizhi Wang
摘要:
直刺变豆菜(Sanicula orthacantha)是中国广泛分布的多年生草本植物, 也是一味著名的民族药。本文通过二代高通量测序平台Illumina HiSeq PE150对直刺变豆菜叶绿体全基因组进行测序, 并通过生物信息学方法对其结构特征进行分析。结果表明: 直刺变豆菜叶绿体全基因组大小为157,163 bp, 包括大单拷贝区(large single copy, LSC)、小单拷贝区(small single copy, SSC)和2个反向重复序列(inverted repeat sequence, IRa和IRb), 长度分别为87,547 bp、17,122 bp和26,247 bp, 具有典型被子植物叶绿体基因组环状四分体结构; 共注释得到129个基因, 包括8个核糖体RNA (rRNA)基因、37个转运RNA (tRNA)基因和84个蛋白质编码基因。直刺变豆菜在叶绿体基因组结构、基因种类、排列顺序上与其他伞形科植物基本一致。直刺变豆菜叶绿体全基因组测序的成功为变豆菜属植物完整叶绿体基因组组装及其特征分析提供了新的方法。
陈志祥, 姚雪莹, Stephen R.Downie, 王奇志 (2019) 直刺变豆菜叶绿体全基因组及其特征. 生物多样性, 27, 366-372. DOI: 10.17520/biods.2018332.
Zhixiang Chen, Xueying Yao, R. Downie Stephen, Qizhi Wang (2019) Assembling and analysis of Sanicula orthacantha chloroplast genome. Biodiversity Science, 27, 366-372. DOI: 10.17520/biods.2018332.
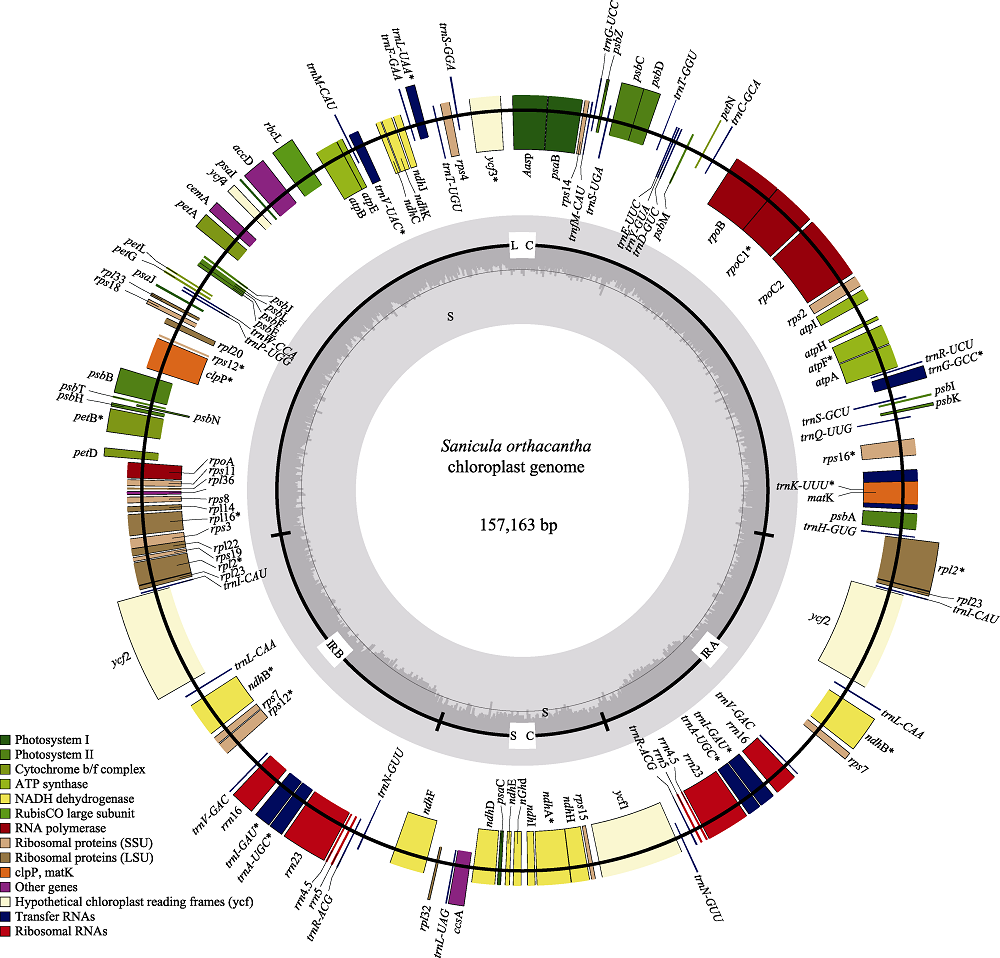
图1 直刺变豆菜叶绿体全基因组。外圈基因逆时针方向转录, 内圈基因顺时针方向转录, 不同的颜色表明基因不同的功能, 小圈中深灰色表示GC含量, 浅灰色表示AT含量。
Fig. 1 Sequence map of the Sanicula orthacantha chloroplast genome. Genes drawn outside of the circle are transcribed counter-clockwise, while genes shown on the inside of the circle are transcribed clockwise. Genes belonging to different functional groups are color-coded. The dark gray in the inner circle indicates GC content, while the light gray corresponds to AT content.
| 基因分类 Category for genes | 基因分组 Group of genes | 基因名称 Name of genes |
|---|---|---|
| 表达相关基因 Self replication | 核糖体RNA基因 Ribosomal RNAs | rrn4.5(×2), rrn5(×2), rrn16(×2), rrn 23(×2) |
| 转运RNA基因 Transfer RNAs | trnA-UGC(×2), trnC-GCA, trnD-GUG, trnE-UCC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU(×2), trnI-GAU(×2), trnK- UUU, trnL-CAA(×2), trnL-UAA, trnL-UAG, trnM-CAU, trnN-GUU (×2) trnP-UGG, trnQ-UUC, trnR-ACG(×2) trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnV-UAC, trnW-CCA, trnY-GUA | |
| 核糖体小亚基基因 Ribosomal small subunit (SSU) | rps16, rps2, rps14, rps4, rps18, rps11, rps8, rps3, rps19, rps7(×2), rps12, rps15 | |
| 核糖体大亚基基因 Ribosomal large subuni (LSU) | rpl33, rpl20, rpl36, rpl14, rpl16, rpl22, rpl2(×2), rpl23(×2), rpl32 | |
| RNA聚合酶亚基基因 RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| 光合作用相关基因 Genes for photosynthesis | 光合系统I基因 Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| 光合系统II基因 Photosystem II | psbA, psbB, psbK, psbI, psbM, psbD, psbC, psbE, psbJ, psbL, psbT, psbH, psbN, psbF, psbZ, psbJ | |
| 细胞色素复合物基因 Cytochrome b/f complex | petA, petD, petG, petL, petN | |
| ATP合酶基因 ATP synthase | atpA, atpF, atpH, atpI, atpE, atpB | |
| 依赖ATP的蛋白酶单元p基因 ATP-dependent protease subunit p gene | clpP | |
| 二磷酸核酮糖羧化酶大亚基基因 RubiscoCO large subunit | rbcL | |
| NADH脱氢酶基因 NADH dehydrogenase | ndhJ, ndhK, ndhC, ndhB(×2), ndhF, ndhD, ndhE, ndhG, ndhI, ndhA, ndhH | |
| 其他基因 Other genes | 成熟酶基因 Maturase | matK |
| 包裹膜蛋白基因 Envelop membrane protein | cemA | |
| 乙酰辅酶A羧化酶亚基基因 Subunit of acetyl-CoA-carboxylase | accD | |
| c型细胞色素合成基因 c-type cytochrome synthesis ccsA gene | ccsA | |
| 转录起始因子基因 Transcription initiation factor IF-1 | InfA | |
| 未知功能基因 Genes of unknown function | 保守开放阅读框 Conserved open reading frames | ycf1, ycf2(×2), ycf3, ycf4 |
表1 直刺变豆菜叶绿体基因组所编码的基因
Table 1 List of genes found in Sanicula orthacantha chloroplast genome
| 基因分类 Category for genes | 基因分组 Group of genes | 基因名称 Name of genes |
|---|---|---|
| 表达相关基因 Self replication | 核糖体RNA基因 Ribosomal RNAs | rrn4.5(×2), rrn5(×2), rrn16(×2), rrn 23(×2) |
| 转运RNA基因 Transfer RNAs | trnA-UGC(×2), trnC-GCA, trnD-GUG, trnE-UCC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU(×2), trnI-GAU(×2), trnK- UUU, trnL-CAA(×2), trnL-UAA, trnL-UAG, trnM-CAU, trnN-GUU (×2) trnP-UGG, trnQ-UUC, trnR-ACG(×2) trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnV-UAC, trnW-CCA, trnY-GUA | |
| 核糖体小亚基基因 Ribosomal small subunit (SSU) | rps16, rps2, rps14, rps4, rps18, rps11, rps8, rps3, rps19, rps7(×2), rps12, rps15 | |
| 核糖体大亚基基因 Ribosomal large subuni (LSU) | rpl33, rpl20, rpl36, rpl14, rpl16, rpl22, rpl2(×2), rpl23(×2), rpl32 | |
| RNA聚合酶亚基基因 RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| 光合作用相关基因 Genes for photosynthesis | 光合系统I基因 Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| 光合系统II基因 Photosystem II | psbA, psbB, psbK, psbI, psbM, psbD, psbC, psbE, psbJ, psbL, psbT, psbH, psbN, psbF, psbZ, psbJ | |
| 细胞色素复合物基因 Cytochrome b/f complex | petA, petD, petG, petL, petN | |
| ATP合酶基因 ATP synthase | atpA, atpF, atpH, atpI, atpE, atpB | |
| 依赖ATP的蛋白酶单元p基因 ATP-dependent protease subunit p gene | clpP | |
| 二磷酸核酮糖羧化酶大亚基基因 RubiscoCO large subunit | rbcL | |
| NADH脱氢酶基因 NADH dehydrogenase | ndhJ, ndhK, ndhC, ndhB(×2), ndhF, ndhD, ndhE, ndhG, ndhI, ndhA, ndhH | |
| 其他基因 Other genes | 成熟酶基因 Maturase | matK |
| 包裹膜蛋白基因 Envelop membrane protein | cemA | |
| 乙酰辅酶A羧化酶亚基基因 Subunit of acetyl-CoA-carboxylase | accD | |
| c型细胞色素合成基因 c-type cytochrome synthesis ccsA gene | ccsA | |
| 转录起始因子基因 Transcription initiation factor IF-1 | InfA | |
| 未知功能基因 Genes of unknown function | 保守开放阅读框 Conserved open reading frames | ycf1, ycf2(×2), ycf3, ycf4 |
| [1] | Andrews S (2013) Babraham Bioinformatics FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. |
| [2] | Bayly MJ, Rigault P, Spokevicius A, Ladiges PY, Ades PK, Anderson C, Bossinger G, Merchant A, Udovicic F, Woodrow IE (2013) Chloroplast genome analysis of Australian eucalypts—Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Molecular Phylogenetics & Evolution, 69, 704-716. |
| [3] |
Clegg MT, Gaut BS, Learn G, Morton BR (1994) Rates and patterns of chloroplast DNA evolution. Proceedings of the National Academy of Sciences, USA, 91, 6795-6801.
DOI URL |
| [4] |
Daniell H, Lin CS, Ming Y, Chang WJ (2016) Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biology, 17, 134-163.
DOI URL |
| [5] |
Dierckxsens N, Mardulyn P, Smits G (2017) NOVO-Plasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Research, 45, e18.
DOI URL |
| [6] |
Downie SR, Jansen RK (2015) A comparative analysis of whole plastid genomes from the Apiales: Expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Systematic Botany, 40, 336-351.
DOI URL |
| [7] |
Ge L, Shen LQ, Chen QY, Li XM, Zhang L (2017) The complete chloroplast genome sequence of Hydrocotyle sibthorpioides (Apiales: Araliaceae). Mitochondrial DNA Part B, 2, 29-30.
DOI URL |
| [8] |
Jansen RK, Raubeson LA, Boore JL, Depamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ (2005) Methods for obtaining and analyzing whole chloroplast genome sequences. Methods in Enzymology, 395, 348-384.
DOI URL |
| [9] |
Kazutaka K, Kazuharu M, Kei-Ichi K, Takashi M (2002) MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30, 3059-3066.
DOI URL |
| [10] |
Kearse M, Moir R, Wilson A, Stoneshavas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647-1649.
DOI URL |
| [11] | Kim KJ, Lee HL (2005) Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Research, 11, 247-261. |
| [12] | Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology & Evolution, 33, 1870-1874. |
| [13] |
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research, 41, W575.
DOI URL |
| [14] |
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Qi P, Liu Y (2012) SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience, 1, 18.
DOI URL |
| [15] |
McCauley DE, Stevens JE, Peroni PA, Raveill JA (1996) The spatial distribution of chloroplast DNA and allozyme polymorphisms within a population of Silene alba (Caryophyllaceae). American Journal of Botany, 83, 727-731.
DOI URL |
| [16] | Peden JF (1999) CodonW. PhD Dissertation, University of Nottingham, Nottinghamshire, UK. |
| [17] |
Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H (2006) Complete plastid genome sequence of Daucus carota: Implications for biotechnology and phylogeny of angiosperms. BMC Genomics, 7, 222-235.
DOI URL |
| [18] | Shan RH, She ML (1979) Flora Reipublicae Popularis Sinicae, Tomus 14, pp. 12-67. Science Press, Beijing. (in Chinese) |
| [ 单人骅, 佘孟兰 (1979) 中国植物志, 第十四卷, 12-67页. 科学出版社, 北京.] | |
| [19] | Sichuan Provincial Health Department (1979) Sichuan Chinese Herbal Medicine Standard (Trial Draft). Sichuan Provincial Health Department, Chengdu. (in Chinese) |
| [ 四川省卫生局 (1979) 四川省中草药标准(试行稿). 四川省卫生局, 成都.] | |
| [20] |
Small RL, Cronn RC, Wendel JF (2004) Use of nuclear genes for phylogeny reconstruction in plants. Australian Systematic Botany, 17, 145-170.
DOI URL |
| [21] |
Soltis PS, Soltis DE (2000) The role of genetic and genomic attributes in the success of polyploids. Proceedings of the National Academy of Sciences, USA, 97, 7051-7057.
DOI URL |
| [22] | Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theoretical & Applied Genetics, 106, 411-422. |
| [23] | Wang L, Dong WP, Zhou SL (2012) Structural mutations and reorganizations in chloroplast genomes of flowering plants. Acta Botanica Boreali-Occidentalia Sinica. 32, 1282-1288. (in Chinese with English abstract) |
| [ 王玲, 董文攀, 周世良 (2012) 被子植物叶绿体基因组的结构变异研究进展. 西北植物学报, 32, 1282-1288.] | |
| [24] | Xie ZW (1996) The Compilation of Chinese Herbal Medicine. People’s Medical Publishing House, Beijing. (in Chinese) |
| [ 谢宗万 (1996) 全国中草药汇编. 人民卫生出版社, 北京.] | |
| [25] | Xing SC, Clarke JL (2008) Process in chloroplast genome analysis. Progress in Biochemistry and Biophysics, 35, 21-28. (in Chinese with English abstract) |
| [ 邢少辰 , Clarke JL (2008) 叶绿体基因组研究进展. 生物化学与生物物理进展, 35, 21-28.] | |
| [26] | Zhang HY (1994) Annals of Chinese Traditional Medicine Resources. Science Press, Beijing. (in Chinese) |
| [ 张惠源 (1994) 中国中药资源志要. 科学出版社, 北京.] | |
| [27] |
Zhang T, Fang Y, Wang X, Deng X, Zhang X, Hu S, Yu J (2012) The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: Insights into the evolution of plant organellar genomes. PLoS ONE, 7, e30531.
DOI URL |
| [28] | Zhao YB, Yin JL, Guo HY, Zhang YY, Xiao W, Sun C, Wu JY, Qu XB, Yu J, Wang XM, Xiao JF (2015) The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Frontiers in Plant Science, 5, 696-709. |
| [1] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [2] | 罗正明, 刘晋仙, 张变华, 周妍英, 郝爱华, 杨凯, 柴宝峰. 不同退化阶段亚高山草甸土壤原生生物群落多样性特征及驱动因素[J]. 生物多样性, 2023, 31(8): 23136-. |
| [3] | 毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌. 毛竹扩张对杉木林土壤细菌群落的影响[J]. 生物多样性, 2023, 31(6): 22659-. |
| [4] | 赵雯, 王丹丹, 热依拉·木民, 黄开钏, 刘顺, 崔宝凯. 阿尔山地区兴安落叶松林土壤微生物群落结构[J]. 生物多样性, 2023, 31(2): 22258-. |
| [5] | 夏凡, 杨婧, 李建, 史洋, 盖立新, 黄文华, 张经纬, 杨南, 高福利, 韩莹莹, 鲍伟东. 北京地区四个豹猫亚种群肠道菌群的组成[J]. 生物多样性, 2022, 30(9): 22103-. |
| [6] | 孙翌昕, 李英滨, 李玉辉, 李冰, 杜晓芳, 李琪. 高通量测序技术在线虫多样性研究中的应用[J]. 生物多样性, 2022, 30(12): 22266-. |
| [7] | 赵琦, 蒋际宝, 张曾鲁, 金清, 李佳丽, 邱江平. 海南岛蚯蚓物种组成及其系统发育分析[J]. 生物多样性, 2022, 30(12): 22224-. |
| [8] | 高程, 郭良栋. 微生物物种多样性、群落构建与功能性状研究进展[J]. 生物多样性, 2022, 30(10): 22429-. |
| [9] | 夏呈强, 李毅, 党延茹, 察倩倩, 贺晓艳, 秦启龙. 中印度洋与南海西部表层海水细菌多样性[J]. 生物多样性, 2022, 30(1): 21407-. |
| [10] | 栗冬梅, 杨卫红, 李庆多, 韩茜, 宋秀平, 潘虹, 冯云. 巴尔通体在滇西南蝙蝠中高度流行并具有丰富的遗传变异特征[J]. 生物多样性, 2021, 29(9): 1245-1255. |
| [11] | 陆奇丰, 黄至欢, 骆文华. 极小种群濒危植物广西火桐、丹霞梧桐的叶绿体基因组特征[J]. 生物多样性, 2021, 29(5): 586-595. |
| [12] | 王楠, 黄菁华, 霍娜, 杨盼盼, 张欣玥, 赵世伟. 宁南山区不同植被恢复方式下土壤线虫群落特征:形态学鉴定与高通量测序法比较[J]. 生物多样性, 2021, 29(11): 1513-1529. |
| [13] | 靳新影, 张肖冲, 金多, 陈韵, 李靖宇. 腾格里沙漠东南缘不同生物土壤结皮细菌多样性及其季节动态特征[J]. 生物多样性, 2020, 28(6): 718-726. |
| [14] | 韩本凤, 周欣, 张雪. 基因组学技术在病毒鉴定与宿主溯源中的应用[J]. 生物多样性, 2020, 28(5): 587-595. |
| [15] | 张全建, 杨彪, 付强, 王磊, 龚旭, 张远彬. 邛崃山系水鹿的冬季食性[J]. 生物多样性, 2020, 28(10): 1192-1201. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn