



生物多样性 ›› 2025, Vol. 33 ›› Issue (8): 25022. DOI: 10.17520/biods.2025022 cstr: 32101.14.biods.2025022
收稿日期:2025-01-13
接受日期:2025-05-08
出版日期:2025-08-20
发布日期:2025-09-17
通讯作者:
*E-mail: songgang@ioz.ac.cn
基金资助:
Ping Fan1,2( ), Zhixin Wen2(
), Zhixin Wen2( ), Gang Song2,*(
), Gang Song2,*( )(
)( )
)
Received:2025-01-13
Accepted:2025-05-08
Online:2025-08-20
Published:2025-09-17
Contact:
*E-mail: songgang@ioz.ac.cn
Supported by:摘要:
气候变化与人类活动已成为威胁全球生物多样性的首要因素。遗传多样性作为生物多样性的核心组成部分, 在物种适应环境变化的过程中发挥关键作用。作为陆生脊椎动物的两大类群, 两栖动物和哺乳动物在演化背景、生理功能、生态行为等方面有明显不同, 尤其是体温调节和活动能力方面的显著差异使得两栖动物较哺乳动物对气候变化及人类活动更为敏感, 但我们尚不清楚这些差异是否会导致不同的遗传多样性水平响应模式。同时, 单倍型多样性和核苷酸多样性作为量化遗传多样性的重要指标, 二者对气候与人类活动的响应模式的异同也值得探究。本研究以两栖动物和哺乳动物为研究对象, 选取细胞色素c氧化酶亚基I (COI)基因片段, 探讨气候因子与人类活动对两个脊椎动物类群单倍型多样性和核苷酸多样性的影响。结果表明, 两栖动物与哺乳动物的总体核苷酸多样性(D = 0.230, P < 0.01)和单倍型多样性(D = 0.211, P < 0.05)均存在显著差异。气候因子和人类影响因子对两栖动物和哺乳动物核苷酸多样性和单倍型多样性的影响呈现出不同的模式。对于两栖动物的单倍型多样性而言, 降水季节性范围与其呈显著正相关(β = 0.467, P < 0.05), 而年均温范围则与其呈显著负相关(β = −0.223, P < 0.05), 同时人类影响因子与核苷酸多样性呈显著正相关(β = 0.035, P < 0.05); 对于哺乳动物的单倍型多样性而言, 人类影响因子与其呈显著负相关(β = ‒0.018, P < 0.05), 而年均温则与核苷酸多样性呈显著正相关(β = 0.002, P < 0.05)。上述结果揭示了遗传多样性响应气候和人类活动影响的复杂性, 我们建议应综合不同多样性指标探讨遗传多样性分布格局及其驱动因素。未来研究需针对不同动物类群, 深入探究人类活动和气候因子对遗传多样性的影响机制, 以制定更具针对性的生物多样性保护策略。
范平, 温知新, 宋刚 (2025) 气候因子和人类活动对两栖及哺乳动物不同遗传多样性指标的影响. 生物多样性, 33, 25022. DOI: 10.17520/biods.2025022.
Ping Fan, Zhixin Wen, Gang Song (2025) The effect of climatic factors and anthropogenic activities on different genetic diversity indicators of amphibians and mammals. Biodiversity Science, 33, 25022. DOI: 10.17520/biods.2025022.
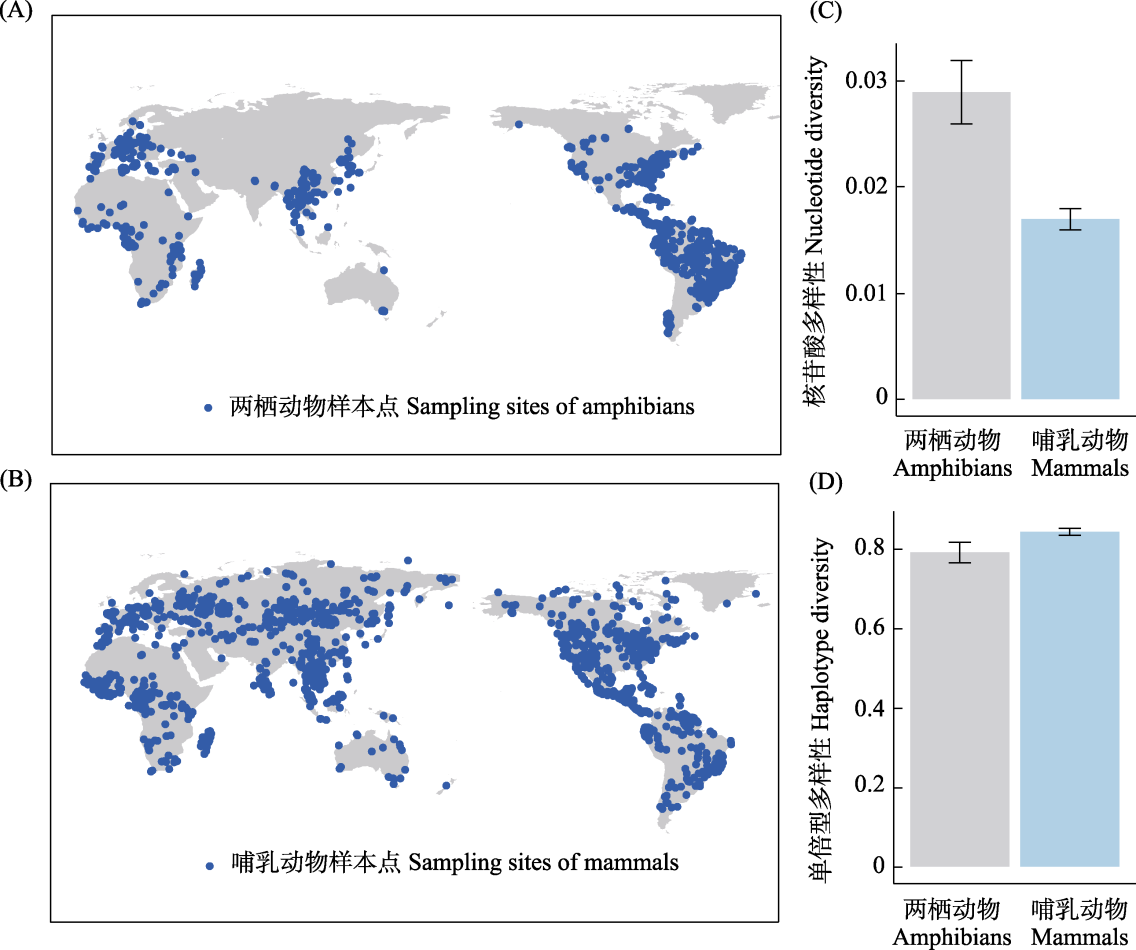
图1 两栖动物和哺乳动物采样点及遗传多样性概述。两栖动物采样点(A)、哺乳动物采样点(B)及两类脊椎动物对应的核苷酸多样性(C)和单倍型多样性(D)。
Fig. 1 Overview of sampling points and genetic diversity for amphibians and mammals. This figure illustrates the sampling points for amphibians (A) and mammals (B), along with the corresponding nucleotide diversity (C) and haplotype diversity (D) for these two vertebrate groups.
| 遗传多样性 Genetic diversity | 分布面积 Area size | 年均温 AT | 年均温范围 ATR | 温度季节 性范围 STR | 降水季 节性 SP | 降水季节 性范围 SPR | 人类影响 因子 HII | Radj2 | AIC | P | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 两栖动物 Amphibians | 单倍型多样性 Haplotype diversity | 0.014 | ‒0.223 | 0.467 | 0.073 | 225.269 | < 0.01 | ||||
| 核苷酸多样性 Nucleotide diversity | 0.046 | 0.004 | 0.008 | 0.035 | 0.123 | 389.035 | < 0.001 | ||||
| 哺乳动物 Mammals | 单倍型多样性 Haplotype diversity | 0.040 | 0.132 | −0.018 | 0.042 | 857.587 | < 0.05 | ||||
| 核苷酸多样性 Nucleotide diversity | 0.071 | 0.002 | 0.181 | 0.006 | 0.073 | 999.273 | < 0.001 |
表1 解释遗传多样性变异程度的最佳多元回归模型
Table 1 Multivariate regression models with variables and genetic diversity best suited for amphibians and mammals
| 遗传多样性 Genetic diversity | 分布面积 Area size | 年均温 AT | 年均温范围 ATR | 温度季节 性范围 STR | 降水季 节性 SP | 降水季节 性范围 SPR | 人类影响 因子 HII | Radj2 | AIC | P | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 两栖动物 Amphibians | 单倍型多样性 Haplotype diversity | 0.014 | ‒0.223 | 0.467 | 0.073 | 225.269 | < 0.01 | ||||
| 核苷酸多样性 Nucleotide diversity | 0.046 | 0.004 | 0.008 | 0.035 | 0.123 | 389.035 | < 0.001 | ||||
| 哺乳动物 Mammals | 单倍型多样性 Haplotype diversity | 0.040 | 0.132 | −0.018 | 0.042 | 857.587 | < 0.05 | ||||
| 核苷酸多样性 Nucleotide diversity | 0.071 | 0.002 | 0.181 | 0.006 | 0.073 | 999.273 | < 0.001 |
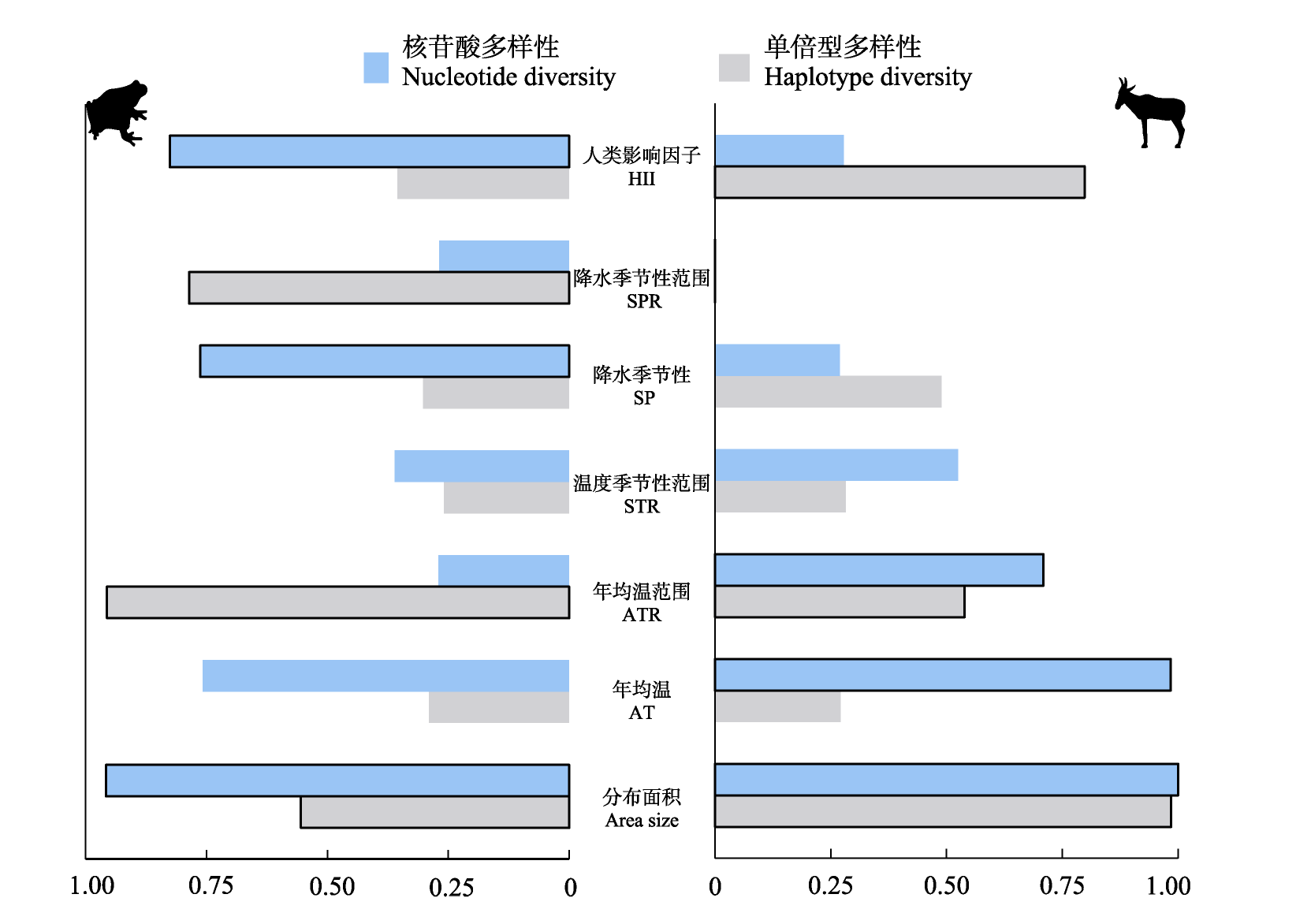
图2 基于Akaike权重的各解释变量对两栖动物(左)和哺乳动物(右)遗传多样性的相对重要程度。黑框标出了对遗传多样性变异相对重要的前3个因子。
Fig. 2 Relative importance of each explanatory variable for amphibians (left panel) and mammals (right panel) based on the Akaike weights. The top three of the relative importance of variables for genetic diversity are marked by black border. Area size, The distribution area size of species; AT, Annual temperature; ATR, Annual temperature range; SP, Precipitation seasonality; SPR, Precipitation seasonality range; STR, Temperature seasonality range; HII, Human influence index.
| [1] | Agneray AC, Parchman TL, Leger EA (2022) Phenotypes and environment predict seedling survival for seven co-occurring Great Basin plant taxa growing with invasive grass. Ecology and Evolution, 12, e8870. |
| [2] |
Almeida-Rocha JM, Soares LASS, Andrade ER, Gaiotto FA, Cazetta E (2020) The impact of anthropogenic disturbances on the genetic diversity of terrestrial species: A global meta-analysis. Molecular Ecology, 29, 4812-4822.
DOI PMID |
| [3] | Bell G (1982) The Masterpiece of Nature:The Evolution and Genetics of Sexuality. University of California Press, Berkeley. |
| [4] |
Blomberg SP, Garland TJ, Ives AR (2003) Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution, 57, 717-745.
DOI PMID |
| [5] |
Brand AB, Snodgrass JW (2010) Value of artificial habitats for amphibian reproduction in altered landscapes. Conservation Biology, 24, 295-301.
DOI PMID |
| [6] |
Brown SC, Wigley TML, Otto-Bliesner BL, Rahbek C, Fordham DA (2020) Persistent Quaternary climate refugia are hospices for biodiversity in the Anthropocene. Nature Climate Change, 10, 244-248.
DOI |
| [7] | Cagan A, Baez-Ortega A, Brzozowska N, Abascal F, Coorens THH, Sanders MA, Lawson ARJ, Harvey LMR, Bhosle S, Jones D, Alcantara RE, Butler TM, Hooks Y, Roberts K, Anderson E, Lunn S, Flach E, Spiro S, Januszczak I, Wrigglesworth E, Jenkins H, Dallas T, Masters N, Perkins MW, Deaville R, Druce M, Bogeska R, Milsom MD, Neumann B, Gorman F, Constantino-Casas F, Peachey L, Bochynska D, St John Smith E, Gerstung M, Campbell PJ, Murchison EP, Stratton MR, Martincorena I (2022) Somatic mutation rates scale with lifespan across mammals. Nature, 604, 517-524. |
| [8] |
Camus MF, Wolff JN, Sgrò CM, Dowling DK (2017) Experimental support that natural selection has shaped the latitudinal distribution of mitochondrial haplotypes in Australian Drosophila melanogaster. Molecular Biology and Evolution, 34, 2600-2612.
DOI URL |
| [9] |
Caruso NM, Staudhammer CL, Rissler LJ (2020) A demographic approach to understanding the effects of climate on population growth. Oecologia, 193, 889-901.
DOI PMID |
| [10] | Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM (2015) Accelerated modern human-induced species losses: Entering the sixth mass extinction. Science Advances, 1, e1400253. |
| [11] |
Chapin III FS, Zavaleta ES, Eviner VT, Naylor RL, Vitousek PM, Reynolds HL, Hooper DU, Lavorel S, Sala OE, Hobbie SE, Mack MC, Díaz S (2000) Consequences of changing biodiversity. Nature, 405, 234-242.
DOI |
| [12] |
Cramer MJ, Willig MR (2005) Habitat heterogeneity, species diversity and null models. Oikos, 108, 209-218.
DOI URL |
| [13] | Daversa DR, Monsalve-Carcaño C, Carrascal LM, Bosch J (2018) Seasonal migrations, body temperature fluctuations, and infection dynamics in adult amphibians. PeerJ, 6, e4698. |
| [14] |
Deflem IS, Calboli FCF, Christiansen H, Hellemans B, Raeymaekers JAM, Volckaert FAM (2022) Contrasting population genetic responses to migration barriers in two native and an invasive freshwater fish. Evolutionary Applications, 15, 2010-2027.
DOI PMID |
| [15] |
Ding CC, Liang DN, Xin WP, Li CW, Jiang ZG (2022) A dataset on the morphological, life-history and ecological traits of the mammals in China. Biodiversity Science, 30, 21520. (in Chinese with English abstract)
DOI |
|
[丁晨晨, 梁冬妮, 信文培, 李春旺, 蒋志刚 (2022) 中国哺乳动物形态、生活史和生态学特征数据集. 生物多样性, 30, 21520.]
DOI |
|
| [16] |
Dirzo R, Young HS, Galetti M, Ceballos G, Isaac NJB, Collen B (2014) Defaunation in the anthropocene. Science, 345, 401-406.
DOI PMID |
| [17] |
Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, Marquéz JRG, Gruber B, Lafourcade B, Leitão PJ, Münkemüller T, McClean C, Osborne PE, Reineking B, Schröder B, Skidmore AK, Zurell D, Lautenbach S (2013) Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography, 36, 27-46.
DOI URL |
| [18] |
Drakulić S, Feldhaar H, Lisičić D, Mioč M, Cizelj I, Seiler M, Spatz T, Rödel MO (2016) Population-specific effects of developmental temperature on body condition and jumping performance of a widespread European frog. Ecology and Evolution, 6, 3115-3128.
DOI PMID |
| [19] |
Ellegren H, Galtier N (2016) Determinants of genetic diversity. Nature Reviews Genetics, 17, 422-433.
DOI PMID |
| [20] |
Epps CW, Keyghobadi N (2015) Landscape genetics in a changing world: Disentangling historical and contemporary influences and inferring change. Molecular Ecology, 24, 6021-6040.
DOI PMID |
| [21] |
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annual Review of Ecology, Evolution, and Systematics, 34, 487-515.
DOI URL |
| [22] |
Fan P, Fjeldså J, Liu X, Dong YF, Chang YB, Qu YH, Song G, Lei FM (2021) An approach for estimating haplotype diversity from sequences with unequal lengths. Methods in Ecology and Evolution, 12, 1658-1667.
DOI URL |
| [23] | Fan P, Song G, Qiao HJ, Zhang DZ, Ji YZ, Qu YH, Fjeldså J, Lei FM (2025) Revaluation of the genetic diversity-area relationship by integrating nucleotide and haplotype diversity. Current Zoology, doi: 10.1093/cz/zoae078. |
| [24] |
Ficetola GF, Maiorano L (2016) Contrasting effects of temperature and precipitation change on amphibian phenology, abundance and performance. Oecologia, 181, 683-693.
DOI PMID |
| [25] | Figueroa-Corona L, Moreno-Letelier A, Ortega-Del Vecchyo D, Peláez P, Gernandt DS, Eguiarte LE, Wegrzyn J, Piñero D (2022) Changes in demography and geographic distribution in the weeping pinyon pine (Pinus pinceana) during the Pleistocene. Ecology and Evolution, 12, e9369. |
| [26] | Garg RK, Mishra V (2018) Molecular insights into the genetic and haplotype diversity among four populations of Catla catla from Madhya Pradesh revealed through mtDNA cyto b gene sequences. Journal of Genetic Engineering & Biotechnology, 16, 169-174. |
| [27] |
Grant W, Bowen B (1998) Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89, 415-426.
DOI URL |
| [28] |
Gratton P, Marta S, Bocksberger G, Winter M, Keil P, Trucchi E, Kuhl H (2017a) Which latitudinal gradients for genetic diversity? Trends in Ecology & Evolution, 32, 724-726.
DOI URL |
| [29] |
Gratton P, Marta S, Bocksberger G, Winter M, Trucchi E, Kühl H (2017b) A world of sequences: Can we use georeferenced nucleotide databases for a robust automated phylogeography? Journal of Biogeography, 44, 475-486.
DOI URL |
| [30] | Guevara EE, Webster TH, Lawler RR, Bradley BJ, Greene LK, Ranaivonasy J, Ratsirarson J, Harris RA, Liu Y, Murali S, Raveendran M, Hughes DST, Muzny DM, Yoder AD, Worley KC, Rogers J (2021) Comparative genomic analysis of sifakas (Propithecus) reveals selection for folivory and high heterozygosity despite endangered status. Science Advances, 7, eabd2274. |
| [31] | Haberl H, Erb KH, Krausmann F, Gaube V, Bondeau A, Plutzar C, Gingrich S, Lucht W, Fischer-Kowalski M (2007) Quantifying and mapping the human appropriation of net primary production in earth’s terrestrial ecosystems. Proceedings of the National Academy of Sciences, USA, 104, 12942-12947. |
| [32] | Haugen H, Dervo BK, Østbye K, Heggenes J, Devineau O, Linløkken A (2024) Genetic diversity, gene flow, and landscape resistance in a pond-breeding amphibian in agricultural and natural forested landscapes in Norway. Evolutionary Applications, 17, e13633. |
| [33] | Hazwan M, Samantha LD, Tee SL, Kamarudin N, Norhisham AR, Lechner AM, Azhar B (2022) Habitat fragmentation and logging affect the occurrence of lesser mouse-deer in tropical forest reserves. Ecology and Evolution, 12, e8745. |
| [34] |
He K, Hu NQ, Chen X, Li JT, Jiang XL (2016) Interglacial refugia preserved high genetic diversity of the Chinese mole shrew in the mountains of southwest China. Heredity, 116, 23-32.
DOI PMID |
| [35] |
Hewitt GM (1999) Post-glacial re-colonization of European biota. Biological Journal of the Linnean Society, 68, 87-112.
DOI URL |
| [36] | Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society Series B: Biological Sciences, 359, 183-195. |
| [37] |
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965-1978.
DOI URL |
| [38] |
Hoban S, Bruford M, D’Urban Jackson J, Lopes-Fernandes M, Heuertz M, Hohenlohe PA, Paz-Vinas I, Sjögren-Gulve P, Segelbacher G, Vernesi C, Aitken S, Bertola LD, Bloomer P, Breed M, Rodríguez-Correa H, Funk WC, Grueber CE, Hunter ME, Jaffe R, Liggins L, Mergeay J, Moharrek F, O’Brien D, Ogden R, Palma-Silva C, Pierson J, Ramakrishnan U, Simo-Droissart M, Tani N, Waits L, Laikre L (2020) Genetic diversity targets and indicators in the CBD post-2020 Global Biodiversity Framework must be improved. Biological Conservation, 248, 108654.
DOI URL |
| [39] | Hung C-M, Shaner P-JL, Zink RM, Liu W-C, Chu T-C, Huang W-S, Li S-H (2014) Drastic population fluctuations explain the rapid extinction of the passenger pigeon. Proceedings of the National Academy of Sciences, USA, 111, 10636-10641. |
| [40] | Hung C-M, Zink RM, Li S-H (2018) Can genomic variation explain the extinction of the passenger pigeon? Journal of Avian Biology, 49, e01858. |
| [41] |
Irisarri I, San Mauro D, Abascal F, Ohler A, Vences M, Zardoya R (2012) The origin of modern frogs (Neobatrachia) was accompanied by acceleration in mitochondrial and nuclear substitution rates. BMC Genomics, 13, 626.
DOI PMID |
| [42] | Jetz W, Pyron RA (2018) The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nature Ecology & Evolution, 2, 850-858. |
| [43] |
Lawrence ER, Fraser DJ (2020) Latitudinal biodiversity gradients at three levels: Linking species richness, population richness and genetic diversity. Global Ecology and Biogeography, 29, 770-788.
DOI URL |
| [44] |
Lei FM, Qu YH, Song G (2014) Species diversification and phylogeographical patterns of birds in response to the uplift of the Qinghai-Tibet Plateau and Quaternary glaciations. Current Zoology, 60, 149-161.
DOI URL |
| [45] |
Leitwein M, Duranton M, Rougemont Q, Gagnaire PA, Bernatchez L (2020) Using haplotype information for conservation genomics. Trends in Ecology & Evolution, 35, 245-258.
DOI URL |
| [46] |
Li XL, Dong F, Lei FM, Alström P, Zhang RY, Ödeen A, Fjeldså J, Ericson PGP, Zou FS, Yang XJ (2016) Shaped by uneven Pleistocene climate: Mitochondrial phylogeographic pattern and population history of white wagtail Motacilla alba (Aves: Passeriformes). Journal of Avian Biology, 47, 263-274.
DOI URL |
| [47] |
Li YM, Wang SQ, Cheng CY, Zhang JQ, Wang SP, Hou XL, Liu X, Yang XJ, Li XP (2021) Latitudinal gradients in genetic diversity and natural selection at a highly adaptive gene in terrestrial mammals. Ecography, 44, 206-218.
DOI URL |
| [48] | Mastretta-Yanes A, da Silva JM, Grueber CE, Castillo-Reina L, Köppä V, Forester BR, Funk WC, Heuertz M, Ishihama F, Jordan R, Mergeay J, Paz-Vinas I, Rincon-Parra VJ, Rodriguez-Morales MA, Arredondo-Amezcua L, Brahy G, DeSaix M, Durkee L, Hamilton A, Hunter ME, Koontz A, Lang I, Latorre-Cárdenas MC, Latty T, Llanes-Quevedo A, MacDonald AJ, Mahoney M, Miller C, Ornelas JF, Ramírez-Barahona S, Robertson E, Russo IM, Santiago MA, Shaw RE, Shea GM, Sjögren-Gulve P, Spence ES, Stack T, Suárez S, Takenaka A, Thurfjell H, Turbek S, van der Merwe M, Visser F, Wegier A, Wood G, Zarza E, Laikre L, Hoban S (2024) Multinational evaluation of genetic diversity indicators for the Kunming-Montreal Global Biodiversity Framework. Ecology Letters, 27, e14461. |
| [49] |
Millette KL, Fugère V, Debyser C, Greiner A, Chain FJJ, Gonzalez A (2020) No consistent effects of humans on animal genetic diversity worldwide. Ecology Letters, 23, 55-67.
DOI PMID |
| [50] |
Miraldo A, Li S, Borregaard MK, Flórez-Rodríguez A, Gopalakrishnan S, Rizvanovic M, Wang ZH, Rahbek C, Marske KA, Nogués-Bravo D (2016) An Anthropocene map of genetic diversity. Science, 353, 1532-1535.
PMID |
| [51] |
Mitrovski P, Hoffmann AA, Heinze DA, Weeks AR (2008) Rapid loss of genetic variation in an endangered possum. Biology Letters, 4, 134-138.
DOI PMID |
| [52] |
Newbold T, Hudson LN, Hill SLL, Contu S, Lysenko I, Senior RA, Börger L, Bennett DJ, Choimes A, Collen B, Day J, De Palma A, Díaz S, Echeverria-Londoño S, Edgar MJ, Feldman A, Garon M, Harrison MLK, Alhusseini T, Ingram DJ, Itescu Y, Kattge J, Kemp V, Kirkpatrick L, Kleyer M, Correia DLP, Martin CD, Shai MR, Novosolov M, Yuan P, Phillips HRP, Purves DW, Robinson A, Simpson J, Tuck SL, Weiher E, White HJ, Ewers RM, Mace GM, Scharlemann JPW, Purvis A (2015) Global effects of land use on local terrestrial biodiversity. Nature, 520, 45-50.
DOI |
| [53] | Orkin JD, Kuderna LFK, Hermosilla-Albala N, Fontsere C, Aylward ML, Janiak MC, Andriaholinirina N, Balaresque P, Blair ME, Fausser J-L, Gut IG, Gut M, Hahn MW, Harris RA, Horvath JE, Keyser C, Kitchener AC, Le MD, Lizano E, Merker S, Nadler T, Perry GH, Rabarivola CJ, Rasmussen L, Raveendran M, Roos C, Wu DD, Zaramody A, Zhang GJ, Zinner D, Pozzi L, Rogers J, Farh KK-H, Marques Bonet T (2025) Ecological and anthropogenic effects on the genomic diversity of lemurs in Madagascar. Nature Ecology & Evolution, 9, 42-56. |
| [54] |
Patenković A, Tanasković M, Erić P, Erić K, Mihajlović M, Stanisavljević L, Davidović S (2022) Urban ecosystem drives genetic diversity in feral honey bee. Scientific Reports, 12, 17692.
DOI PMID |
| [55] |
Pauls SU, Nowak C, Bálint M, Pfenninger M (2013) The impact of global climate change on genetic diversity within populations and species. Molecular Ecology, 22, 925-946.
DOI PMID |
| [56] | Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO (2014) The biodiversity of species and their rates of extinction, distribution, and protection. Science, 344, 1246752. |
| [57] |
Rohde K (1992) Latitudinal gradients in species diversity: The search for the primary cause. Oikos, 65, 514-527.
DOI URL |
| [58] |
Romiguier J, Gayral P, Ballenghien M, Bernard A, Cahais V, Chenuil A, Chiari Y, Dernat R, Duret L, Faivre N, Loire E, Lourenco JM, Nabholz B, Roux C, Tsagkogeorga G, Weber AAT, Weinert LA, Belkhir K, Bierne N, Glémin S, Galtier N (2014) Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature, 515, 261-263.
DOI |
| [59] |
Santos JC (2012) Fast molecular evolution associated with high active metabolic rates in poison frogs. Molecular Biology and Evolution, 29, 2001-2018.
DOI PMID |
| [60] |
Sexton JP, Hangartner SB, Hoffmann AA (2014) Genetic isolation by environment or distance: Which pattern of gene flow is most common? Evolution, 68, 1-15.
DOI PMID |
| [61] |
Shafer ABA, Wolf JBW (2013) Widespread evidence for incipient ecological speciation: A meta-analysis of isolation-by-ecology. Ecology Letters, 16, 940-950.
DOI PMID |
| [62] |
Sills J, Ceballos G, Ehrlich PR (2018) The misunderstood sixth mass extinction. Science, 360, 1080-1081.
DOI PMID |
| [63] |
Song G, Yu LJ, Gao B, Zhang RY, Qu YH, Lambert DM, Li S, Zhou TL, Lei FM (2013) Gene flow maintains genetic diversity and colonization potential in recently range-expanded populations of an Oriental bird, the Light-vented Bulbul (Pycnonotus sinensis, Aves: Pycnonotidae). Diversity and Distributions, 19, 1248-1262.
DOI URL |
| [64] |
Song YF, Chen CW, Wang YP (2022) A dataset on the life-history and ecological traits of Chinese amphibians. Biodiversity Science, 30, 22053. (in Chinese with English abstract)
DOI |
|
[宋云枫, 陈传武, 王彦平 (2022) 中国两栖类生活史和生态学特征数据集. 生物多样性, 30, 22053.]
DOI |
|
| [65] | Suárez-Montes P, Chávez-Pesqueira M, Núñez-Farfán J (2016) Life history and past demography maintain genetic structure, outcrossing rate, contemporary pollen gene flow of an understory herb in a highly fragmented rainforest. PeerJ, 4, e2764. |
| [66] |
Symonds MRE, Moussalli A (2011) A brief guide to model selection, multimodel inference and model averaging in behavioural ecology using Akaike’s information criterion. Behavioral Ecology and Sociobiology, 65, 13-21.
DOI URL |
| [67] |
Tajima F (1993) Statistical analysis of DNA polymorphism. The Japanese Journal of Genetics, 68, 567-595.
DOI URL |
| [68] | Teixeira JC, Huber CD (2021) The inflated significance of neutral genetic diversity in conservation genetics. Proceedings of the National Academy of Sciences, USA, 118, e2015096118. |
| [69] |
Theodoridis S, Fordham DA, Brown SC, Li S, Rahbek C, Nogues-Bravo D (2020) Evolutionary history and past climate change shape the distribution of genetic diversity in terrestrial mammals. Nature Communications, 11, 2557.
DOI PMID |
| [70] | Upham NS, Esselstyn JA, Jetz W (2019) Inferring the mammal tree: Species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biology, 17, e3000494. |
| [71] |
Waldvogel AM, Pfenninger M (2021) Temperature dependence of spontaneous mutation rates. Genome Research, 31, 1582-1589.
DOI URL |
| [72] | Wang RJ, Peña-Garcia Y, Bibby MG, Raveendran M, Harris RA, Jansen HT, Robbins CT, Rogers J, Kelley JL, Hahn MW (2022) Examining the effects of hibernation on germline mutation rates in grizzly bears. Genome Biology and Evolution, 14, evac148. |
| [73] |
Watterson GA (1986) The homozygosity test after a change in population size. Genetics, 112, 899-907.
DOI PMID |
| [74] |
Wei FW, Ma TX, Hu YB (2021) Research advances and perspectives of conservation genetics of threatened mammals in China. Acta Theriologica Sinica, 41, 571-580. (in Chinese with English abstract)
DOI |
|
[魏辅文, 马天笑, 胡义波 (2021) 中国濒危兽类保护遗传学研究进展与展望. 兽类学报, 41, 571-580.]
DOI |
|
| [75] |
Wiens JJ, Graham CH (2005) Niche conservatism: Integrating evolution, ecology, and conservation biology. Annual Review of Ecology Evolution and Systematics, 36, 519-539.
DOI URL |
| [76] | Xia WC, Zhang C, Zhuang HF, Ren BP, Zhou J, Shen J, Krzton A, Luan XF, Li DY (2020) The potential distribution and disappearing of Yunnan snub-nosed monkey: Influences of habitat fragmentation. Global Ecology and Conservation, 21, e00835. |
| [77] | Xu XR, Lv XY, He XT, Wang YY (2019) Analysis of factors affecting animal phylogeographic patterns in China. Chinese Bulletin of Life Sciences, 31, 968-974. (in Chinese with English abstract) |
| [徐小蓉, 吕小艳, 何小涛, 王野影 (2019) 中国动物谱系地理格局的影响因素分析. 生命科学, 31, 968-974.] | |
| [78] |
Yang DD, Jiang ZG, Ma JZ, Hu HJ, Li PF (2005) Causes of endangerment or extinction of some mammals and its relevance to the reintroduction of Père David’s deer in the Dongting Lake drainage area. Biodiversity Science, 13, 451-461. (in Chinese with English abstract)
DOI URL |
|
[杨道德, 蒋志刚, 马建章, 胡慧建, 李鹏飞 (2005) 洞庭湖流域麋鹿等哺乳动物濒危灭绝原因的分析及其对麋鹿重引入的启示. 生物多样性, 13, 451-461.]
DOI |
|
| [79] |
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends in Ecology & Evolution, 11, 413-418.
DOI URL |
| [80] | Zhai DD, Li WJ, Liu HZ, Cao WX, Gao X (2019) Genetic diversity and temporal changes of an endemic cyprinid fish species, Ancherythroculter nigrocauda, from the upper reaches of Yangtze River. Zoological Research, 40, 427-438. |
| [81] |
Zhu LF, Hu YB, Qi DW, Wu H, Zhan XJ, Zhang ZJ, Bruford MW, Wang JL, Yang XY, Gu XD, Zhang L, Zhang BW, Zhang SN, Wei FW (2013) Genetic consequences of historical anthropogenic and ecological events on giant pandas. Ecology, 94, 2346-2357.
PMID |
| [1] | 薛瑞翔, 马雪蓉, 吴炯文, 刘爱君, 张细权, 季从亮, 殷颖珊, 朱炜健, 罗庆斌. 中山麻鸭群体遗传多样性与遗传结构[J]. 生物多样性, 2025, 33(8): 24592-. |
| [2] | 范平, 王欢, 温知新, 宋刚, 雷富民. 气候因子对鸟类遗传多样性与物种分布面积关系的影响[J]. 生物多样性, 2025, 33(8): 25072-. |
| [3] | 杨洋, 邹睿, 谯娅琴, 孟想, 涂飞云. 海南省陆生哺乳动物物种多样性[J]. 生物多样性, 2025, 33(8): 25044-. |
| [4] | 周智成, 曹天玲, 刘如垚, 丁琪琪, 马轲, 杨丽萍, 周传江, 聂国兴, 汤永涛. 基于线粒体COI基因的黄河流域麦穗鱼种群遗传多样性与遗传结构[J]. 生物多样性, 2025, 33(8): 24501-. |
| [5] | 王儒晓, 史博洋, 潘达, 孙红英. 中国特有华溪蟹属淡水蟹多样性格局及其保护空缺[J]. 生物多样性, 2025, 33(8): 25123-. |
| [6] | 徐进博, 崔雅倩, 王渊, 王伟波, 刘锋, 王广龙, 扈晶晶, 普布顿珠, 边巴多吉, 旦增, 胡开, 王小川, 宋刚, 吕永磊, 温知新. 西藏雅鲁藏布大峡谷国家级自然保护区内白颊猕猴的栖息地适宜性评价[J]. 生物多样性, 2025, 33(7): 24493-. |
| [7] | 彭文, 邓泽帅, 郑文宝, 龚凌轩, 曾玉枫, 孟昊, 陈军, 杨道德. eDNA技术在两栖动物调查中的应用: 以湖南莽山国家级自然保护区为例[J]. 生物多样性, 2025, 33(6): 24552-. |
| [8] | 张彤云, 胡自民. 二裂墨角藻谱系多样性模式显示纽芬兰大浅滩存在一个海洋冰期避难所[J]. 生物多样性, 2025, 33(6): 24416-. |
| [9] | 罗敏, 杨永川, 靳程, 周礼华, 龙宇潇. 重庆中心城区城市森林兽类组成特征及其对人类活动的响应[J]. 生物多样性, 2025, 33(5): 24402-. |
| [10] | 顾婧婧, 刘宜卓, 苏杨. 基层地方政府在完成《昆蒙框架》中的作用和难点: 基于《联合国气候变化框架公约》任务的比较[J]. 生物多样性, 2025, 33(3): 24585-. |
| [11] | 李华亮, 张明军, 张熙斌, 谭荣, 李诗川, 冯尔辉, 林雪云, 陈珉, 颜文博, 曾治高. 海南东寨港国家级自然保护区两栖类群落组成及影响因素[J]. 生物多样性, 2025, 33(2): 24350-. |
| [12] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [13] | 卢佳玉, 石小亿, 多立安, 王天明, 李治霖. 基于红外相机技术的天津城市地栖哺乳动物昼夜活动节律评价[J]. 生物多样性, 2024, 32(8): 23369-. |
| [14] | 吴琪, 张晓青, 杨雨婷, 周艺博, 马毅, 许大明, 斯幸峰, 王健. 浙江钱江源-百山祖国家公园庆元片区叶附生苔多样性及其时空变化[J]. 生物多样性, 2024, 32(4): 24010-. |
| [15] | 曹可欣, 王敬雯, 郑国, 武鹏峰, 李英滨, 崔淑艳. 降水格局改变及氮沉降对北方典型草原土壤线虫多样性的影响[J]. 生物多样性, 2024, 32(3): 23491-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn