



生物多样性 ›› 2025, Vol. 33 ›› Issue (6): 24318. DOI: 10.17520/biods.2024318 cstr: 32101.14.biods.2024318
所属专题: eDNA技术应用
李云翱1,2, 张文富1, 赵桂刚3,4, 杨春燕3,4, 陈向清6, 袁盛东1,5, 曹敏1, 蔡望1,*( ), 杨洁1,5
), 杨洁1,5
收稿日期:2024-07-15
接受日期:2025-04-13
出版日期:2025-06-20
发布日期:2025-07-29
通讯作者:
蔡望
基金资助:
Yun’ao Li1,2, Wenfu Zhang1, Guigang Zhao3,4, Chunyan Yang3,4, Xiangqing Chen6, Shengdong Yuan1,5, Min Cao1, Wang Cai1,*( ), Jie Yang1,5
), Jie Yang1,5
Received:2024-07-15
Accepted:2025-04-13
Online:2025-06-20
Published:2025-07-29
Contact:
Wang Cai
Supported by:摘要:
环境DNA (eDNA)技术为生物多样性保护提供了一种无损伤的监测方法。近年来的研究表明, 从空气中收集eDNA可用于监测森林生态系统中的野生动物。相比其他的eDNA调查方法, 该技术在采样地点选择上具备更高的灵活性, 特别适用于缺乏水体等环境介质的调查区域, 因此, 空气环境DNA在森林生态系统生物多样性监测领域具有广阔的应用潜力。本文选择中国西双版纳20 ha森林动态监测样地, 采用空气环境DNA技术对样地内的陆生脊椎动物多样性展开调查, 并将结果与红外相机监测数据进行对比分析。实验共设置20台空气环境DNA采样器, 在2023年11月的6天内完成了3次采样, 每次采样持续24 h。采集的样本经12SV05引物扩增12S rRNA基因片段, 并在Illumina NovaSeq 6000平台上进行高通量测序, 随后对获得的序列数据进行物种注释, 并评估空气环境DNA与红外相机在物种检测效率上的差异。研究结果表明, 3次的空气环境DNA采样实验共检测到66个可注释到鸟类、哺乳动物、爬行动物和两栖动物的可操作性分类单元(operational taxonomic units, OTUs); 放置于相同位点的20台红外相机, 在总计5,682个有效相机日的监测中, 检测到15种哺乳动物和15种鸟类。分析表明, 相较于红外相机, 空气环境DNA在物种多样性检测方面具有更高的效率。此外, 通过评估α多样性增长曲线发现, 当空气环境DNA样本量达到10个时, 多样性曲线趋于平台期; 在当前实验环境下3天内采集10个样本可实现物种多样性的最大化检测。综上所述, 空气环境DNA是热带雨林陆生脊椎动物多样性监测的有效工具, 并能在短时间内实现一定的物种覆盖。相较于红外相机, 该方法在快速生物多样性调查方面展现出更大的优势。尽管空气环境DNA技术仍处于发展阶段, 其在特定环境条件下的稳定性及检测精度仍需进一步优化, 但随着技术的进步, 空气环境DNA有望成为跨营养级、多物种生物多样性监测的重要工具, 并可为中国大规模标准化的生物多样性监测网络提供科学支撑。
李云翱, 张文富, 赵桂刚, 杨春燕, 陈向清, 袁盛东, 曹敏, 蔡望, 杨洁 (2025) 空气环境DNA在陆生脊椎动物多样性监测上的应用: 以西双版纳20 ha森林动态样地为例. 生物多样性, 33, 24318. DOI: 10.17520/biods.2024318.
Yun’ao Li, Wenfu Zhang, Guigang Zhao, Chunyan Yang, Xiangqing Chen, Shengdong Yuan, Min Cao, Wang Cai, Jie Yang (2025) Application of airborne eDNA for terrestrial animal diversity monitoring: A case study of 20-ha forest dynamics plot in Xishuangbanna, Yunnan, China. Biodiversity Science, 33, 24318. DOI: 10.17520/biods.2024318.
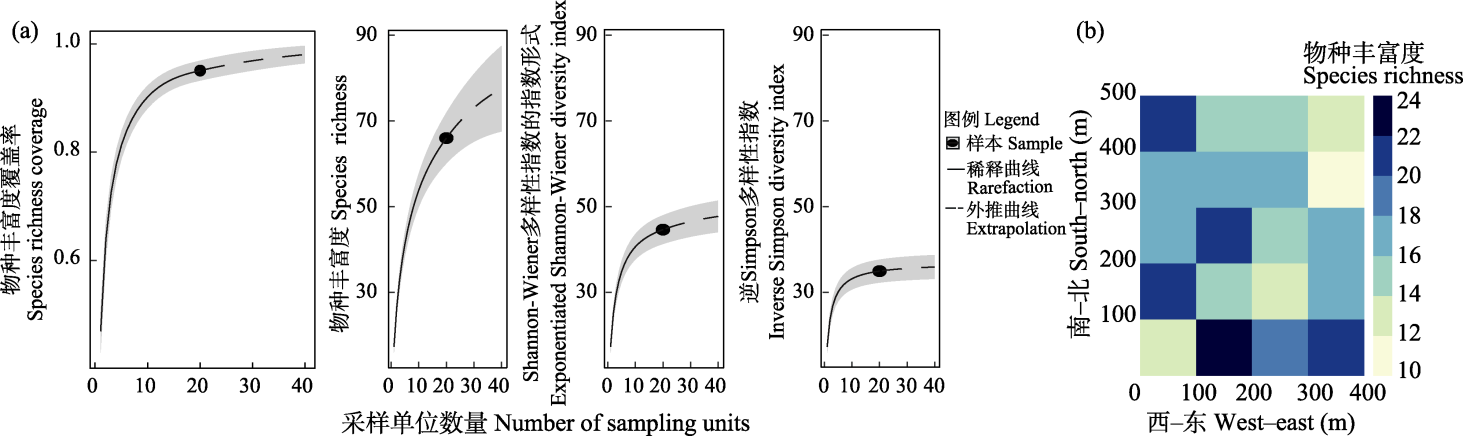
图2 Airborne eDNA监测到的Alpha多样性(a)及样地内物种丰富度分布(b)
Fig. 2 Alpha diversity (a) and distribution of species richness (b) detected by airborne eDNA in the monitoring plot
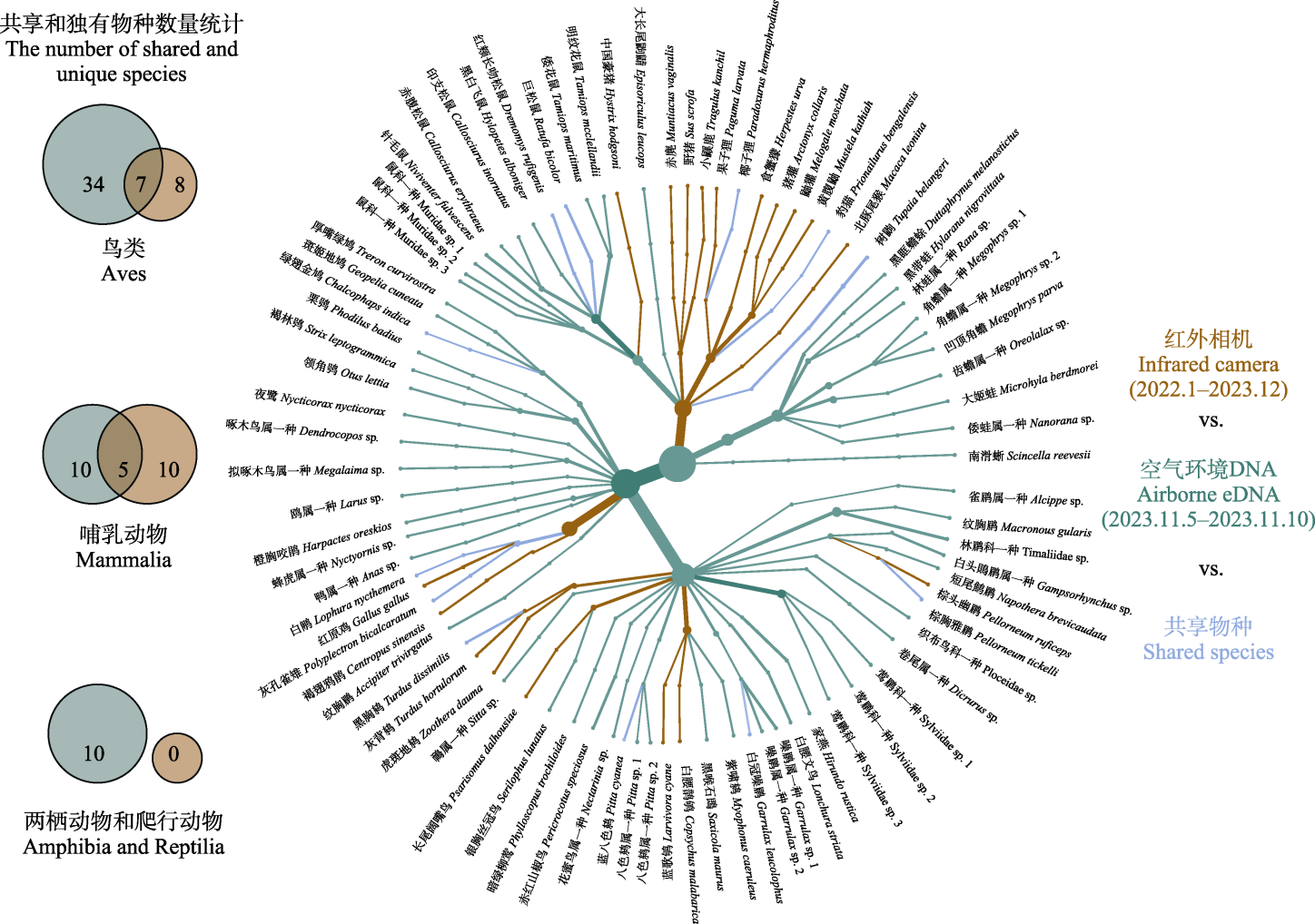
图4 Airborne eDNA (2023.11.5-2023.11.10)和红外相机监测(2022.1-2023.12)结果对比
Fig. 4 Comparison of species composition between airborne eDNA (2023.11.5-2023.11.10) and infrared camera (2022.1-2023.12)
| [1] | Alberdi A, Aizpurua O, Gilbert MTP, Bohmann K (2018) Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods in Ecology and Evolution, 9, 134-147. |
| [2] |
Andersen K, Bird KL, Rasmussen M, Haile J, Breuning-Madsen H, Kjær KH, Orlando L, Gilbert MTP, Willerslev E (2012) Meta-barcoding of ‘dirt’ DNA from soil reflects vertebrate biodiversity. Molecular Ecology, 21, 1966-1979.
DOI PMID |
| [3] | Barlow J, França F, Gardner TA, Hicks CC, Lennox GD, Berenguer E, Castello L, Economo EP, Ferreira J, Guénard B, Gontijo Leal C, Isaac V, Lees AC, Parr CL, Wilson SK, Young PJ, Graham NAJ (2018) The future of hyperdiverse tropical ecosystems. Nature, 559, 517-526. |
| [4] | Barton K (2018) MuMIn: Multi-model Inference, R package version 0.12.0. https://CRAN.R-project.org/package=MuMIn. (accessed on 2024-06-11) |
| [5] | Bates D, Mächler M, Bolker B, Walker S (2014) Package Lme4: Linear Mixed-Effects Models Using Eigen and S4. https://CRAN.R-project.org/package=lme4. (accessed on 2024-06-11) |
| [6] | Bessey C, Gao Y, Truong YB, Miller H, Jarman SN, Berry O (2022) Comparison of materials for rapid passive collection of environmental DNA. Molecular Ecology Resources, 22, 2559-2572. |
| [7] | Bohmann K, Gopalakrishnan S, Nielsen M, dos Santos Bay Nielsen L, Jones G, Streicker DG, Gilbert MTP (2018) Using DNA metabarcoding for simultaneous inference of common vampire bat diet and population structure. Molecular Ecology Resources, 18, 1050-1063. |
| [8] | Bohmann K, Lynggaard C (2023) Transforming terrestrial biodiversity surveys using airborne eDNA. Trends in Ecology & Evolution, 38, 119-121. |
| [9] | Bolte B, Goldsbury J, Huerlimann R, Jerry D, Kingsford M (2021) Validation of eDNA as a viable method of detection for dangerous cubozoan jellyfish. Environmental DNA, 3, 769-779. |
| [10] | Boni MF, Lemey P, Jiang X, Lam TT, Perry BW, Castoe TA, Rambaut A, Robertson DL (2020) Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nature Microbiology, 5, 1408-1417. |
| [11] | Burton AC, Neilson E, Moreira D, Ladle A, Steenweg R, Fisher JT, Bayne E, Boutin S (2015) REVIEW: Wildlife camera trapping: A review and recommendations for linking surveys to ecological processes. Journal of Applied Ecology, 52, 675-685. |
| [12] | Cai W, MacDonald B, Korabik M, Gradin I, Neave EF, Harper LR, Kenchington E, Riesgo A, Whoriskey FG, Mariani S (2024) Biofouling sponges as natural eDNA samplers for marine vertebrate biodiversity monitoring. Science of the Total Environment, 946, 174148. |
| [13] |
Calvignac-Spencer S, Merkel K, Kutzner N, Kühl H, Boesch C, Kappeler PM, Metzger S, Schubert G, Leendertz FH (2013) Carrion fly-derived DNA as a tool for comprehensive and cost-effective assessment of mammalian biodiversity. Molecular Ecology, 22, 915-924.
DOI PMID |
| [14] |
Carraro L, Blackman RC, Altermatt F (2023) Modelling environmental DNA transport in rivers reveals highly resolved spatio-temporal biodiversity patterns. Scientific Reports, 13, 8854.
DOI PMID |
| [15] |
Chen HB, Boutros PC (2011) VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics, 12, 35.
DOI PMID |
| [16] | Cheyne SM, Sastramidjaja WJ, Muhalir, Rayadin Y, MacDonald DW (2016) Mammalian communities as indicators of disturbance across Indonesian Borneo. Global Ecology and Conservation, 7, 157-173. |
| [17] | Clare EL, Economou CK, Bennett FJ, Dyer CE, Adams K, McRobie B, Drinkwater R, Littlefair JE (2022) Measuring biodiversity from DNA in the air. Current Biology, 32, 693-700.e5. |
| [18] | Cristescu ME (2014) From barcoding single individuals to metabarcoding biological communities: Towards an integrative approach to the study of global biodiversity. Trends in Ecology & Evolution, 29, 566-571. |
| [19] | Danabalan R, Planillo A, Butschkau S, Deeg S, Pierre G, Thion C, Calvignac-Spencer S, Kramer-Schadt S, Mazzoni C (2023) Comparison of mosquito and fly derived DNA as a tool for sampling vertebrate biodiversity in suburban forests in Berlin, Germany. Environmental DNA, 5, 476-487. |
| [20] | de Groot GA, Geisen S, Jasper Wubs ER, Meulenbroek L, Laros I, Snoek LB, Lammertsma DR, Hansen LH, Slim PA (2021) The aerobiome uncovered: Multi-marker metabarcoding reveals potential drivers of turn-over in the full microbial community in the air. Environment International, 154, 106551. |
| [21] |
Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, de Vere N, Pfrender ME, Bernatchez L (2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology, 26, 5872-5895.
DOI PMID |
| [22] |
Djurhuus A, Pitz K, Sawaya NA, Rojas-Márquez J, Michaud B, Montes E, Muller-Karger F, Breitbart M (2018) Evaluation of marine zooplankton community structure through environmental DNA metabarcoding. Limnology and Oceanography: Methods, 16, 209-221.
DOI PMID |
| [23] | Dobrowski SZ, Littlefield CE, Lyons DS, Hollenberg C, Carroll C, Parks SA, Abatzoglou JT, Hegewisch K, Gage J (2021) Protected-area targets could be undermined by climate change-driven shifts in ecoregions and biomes. Communications Earth & Environment, 2, 198. |
| [24] | Dong ZY, Chen LL, Zhang NP, Chen L, Sun DB, Ni YM, Li BQ (2023) Response of fish diversity to hydrological connectivity of typical tidal creek system in the Yellow River Delta based on environmental DNA metabarcoding. Biodiversity Science, 31, 23073. (in Chinese with English abstract) |
|
[董志远, 陈琳琳, 张乃鹏, 陈莉, 孙德斌, 倪艳梅, 李宝泉 (2023) 基于环境DNA宏条形码技术研究黄河三角洲典型潮沟系统鱼类多样性及其对水文连通性的响应. 生物多样性, 31, 23073.]
DOI |
|
| [25] |
Drinkwater R, Schnell IB, Bohmann K, Bernard H, Veron G, Clare E, Gilbert MTP, Rossiter SJ (2019) Using metabarcoding to compare the suitability of two blood-feeding leech species for sampling mammalian diversity in North Borneo. Molecular Ecology Resources, 19, 105-117.
DOI PMID |
| [26] |
Ficetola GF, Pansu J, Bonin A, Coissac E, Giguet-Covex C, De Barba M, Gielly L, Lopes CM, Boyer F, Pompanon F, Rayé G, Taberlet P (2015) Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Molecular Ecology Resources, 15, 543-556.
DOI PMID |
| [27] | Foster ZSL, Sharpton TJ, Grünwald NJ (2017) Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Computational Biology, 13, e1005404. |
| [28] | Garrett NR, Watkins J, Simmons NB, Fenton B, Maeda-Obregon A, Sanchez DE, Froehlich EM, Walker FM, Littlefair JE, Clare EL (2023) Airborne eDNA documents a diverse and ecologically complex tropical bat and other mammal community. Environmental DNA, 5, 350-362. |
| [29] | Geml J, Laursen GA, Timling I, Mcfarland JM, Booth MG, Lennon N, Nusbaum C, Taylor DL (2009) Molecular phylogenetic biodiversity assessment of Arctic and boreal ectomycorrhizal Lactarius Pers. (Russulales; Basidiomycota) in Alaska, based on soil and sporocarp DNA. Molecular Ecology, 18, 2213-2227. |
| [30] | Goldberg CS, Turner CR, Deiner K, Klymus KE, Thomsen PF, Murphy MA, Spear SF, McKee A, Oyler-McCance SJ, Cornman RS, Laramie MB, Mahon AR, Lance RF, Pilliod DS, Strickler KM, Waits LP, Fremier AK, Takahara T, Herder JE, Taberlet P (2016) Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods in Ecology and Evolution, 7, 1299-1307. |
| [31] | Hartig F, Abrego N, Bush A, Chase JM, Guillera-Arroita G, Leibold MA, Ovaskainen O, Pellissier L, Pichler M, Poggiato G, Pollock L, Si-Moussi S, Thuiller W, Viana DS, Warton DI, Zurell D, Yu DW (2024) Novel community data in ecology—Properties and prospects. Trends in Ecology & Evolution, 39, 280-293. |
| [32] | Hassan S, Sabreena, Poczai P, Ganai BA, Almalki WH, Gafur A, Sayyed RZ (2022) Environmental DNA metabarcoding: A novel contrivance for documenting terrestrial biodiversity. Biology, 11, 1297. |
| [33] |
He RC, Wang L, Quan RC (2020) Introduction to transboundary animal diversity monitoring platform of southern Yunnan, China and Southeast Asia. Biodiversity Science, 28, 1097-1103. (in Chinese with English abstract)
DOI |
|
[贺如川, 王林, 权锐昌 (2020) 中国滇南-东南亚跨境动物多样性监测平台概述. 生物多样性, 28, 1097-1103.]
DOI |
|
| [34] | Hofreiter M, Mead JI, Martin P, Poinar HN (2003) Molecular caving. Current Biology, 13, R693-R695. |
| [35] | Hsieh TC, Ma KH, Chao A (2016) iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods in Ecology and Evolution, 7, 1451-1456. |
| [36] |
Huang G, Sreekar R, Velho N, Corlett RT, Quan RC, Tomlinson KW (2020) Combining camera-trap surveys and hunter interviews to determine the status of mammals in protected rainforests and rubber plantations of Menglun, Xishuangbanna, SW China. Animal Conservation, 23, 689-699.
DOI |
| [37] |
Ji YQ, Baker CCM, Popescu VD, Wang JX, Wu CY, Wang ZY, Li YH, Wang L, Hua CL, Yang ZX, Yang CY, Xu CCY, Diana A, Wen QZ, Pierce NE, Yu DW (2022) Measuring protected-area effectiveness using vertebrate distributions from leech iDNA. Nature Communications, 13, 1555.
DOI PMID |
| [38] | Johnson M, Barnes MA (2024) Macrobial airborne environmental DNA analysis: A review of progress, challenges, and recommendations for an emerging application. Molecular Ecology Resources, 24, e13998. |
| [39] | Johnson MD, Cox RD, Barnes MA (2019) The detection of a non-anemophilous plant species using airborne eDNA. PLoS ONE, 14, e0225262. |
| [40] | Kampmann ML, Børsting C, Morling N (2017) Decrease DNA contamination in the laboratories. Forensic Science International: Genetics Supplement Series, 6, e577-e578. |
| [41] | Kirtane A, Atkinson JD, Sassoubre L (2020) Design and validation of passive environmental DNA samplers using granular activated carbon and montmorillonite clay. Environmental Science & Technology, 54, 11961-11970. |
| [42] |
Kumar G, Eble JE, Gaither MR (2020) A practical guide to sample preservation and pre-PCR processing of aquatic environmental DNA. Molecular Ecology Resources, 20, 29-39.
DOI PMID |
| [43] | Leempoel K, Hebert T, Hadly EA (2020) A comparison of eDNA to camera trapping for assessment of terrestrial mammal diversity. Proceedings of the Royal Society B: Biological Sciences, 287, 20192353. |
| [44] | Liang J, Zhu H, Wang H, Zhou SS (2007) Changes in species diversity of Parashorea forest in the past 20 years in Xishuangbanna, Yunnan. Chinese Journal of Applied & Environmental Biology, 13, 609-614. (in Chinese with English abstract) |
| [梁娟, 朱华, 王洪, 周仕顺 (2007) 西双版纳补蚌地区望天树林近20a来物种多样性变化研究. 应用与环境生物学报, 13, 609-614.] | |
| [45] | Liu Y, Chen SH (2021) The CNG Field Guide to the Birds of China. Hunan Science and Technology Press, Changsha. (in Chinese) |
| [刘阳, 陈水华 (2021) 中国鸟类观察手册. 湖南科学技术出版社, 长沙.] | |
| [46] |
Lu BY, Li K, Wang CX, Li S (2024) The application and outlook of wildlife tracking using sensor-based tags in China. Biodiversity Science, 32, 23497. (in Chinese with English abstract)
DOI |
|
[鲁彬悦, 李坤, 王晨溪, 李晟 (2024) 基于传感器标记的野生动物追踪技术在中国的应用现状与展望. 生物多样性, 32, 23497.]
DOI |
|
| [47] |
Lyet A, Pellissier L, Valentini A, Dejean T, Hehmeyer A, Naidoo R (2021) eDNA sampled from stream networks correlates with camera trap detection rates of terrestrial mammals. Scientific Reports, 11, 11362.
DOI PMID |
| [48] | Lynggaard C, Bertelsen MF, Jensen CV, Johnson MS, Frøslev TG, Olsen MT, Bohmann K (2022) Airborne environmental DNA for terrestrial vertebrate community monitoring. Current Biology, 32, 701-707.e5. |
| [49] | Lynggaard C, Frøslev TG, Johnson MS, Olsen MT, Bohmann K (2024) Airborne environmental DNA captures terrestrial vertebrate diversity in nature. Molecular Ecology Resources, 24, e13840. |
| [50] |
Ma KP (2011) Assessing progress of biodiversity conservation with monitoring approach. Biodiversity Science, 19, 125-126. (in Chinese)
DOI |
|
[马克平 (2011) 监测是评估生物多样性保护进展的有效途径. 生物多样性, 19, 125-126.]
DOI |
|
| [51] | Mercier C, Boyer F, Bonin A, Coissac E (2013) SUMATRA and SUMACLUST: Fast and exact comparison and clustering of sequences. Abstracts of SeqBio Conference, 25-26 November 2013. |
| [52] | Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature, 403, 853-858. |
| [53] | O’Brien TG, Baillie JEM, Krueger L, Cuke M (2010) The Wildlife Picture Index: Monitoring top trophic levels. Animal Conservation, 13, 335-343. |
| [54] | Pan QH, Wang YX, Yan K (2007) A Field Guide to the Mammals of China. China Forestry Publishing House, Beijing. (in Chinese) |
| [潘清华, 王应祥, 岩崑 (2007) 中国哺乳动物彩色图鉴. 中国林业出版社, 北京.] | |
| [55] | R Core Team (2023) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. https://www.R-project.org/. (accessed on 2024-06-11) |
| [56] | Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E (2011) ecoPrimers: Inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Research, 39, e145. |
| [57] | Rizzoli A, Bolzoni L, Chadwick EA, Capelli G, Montarsi F, Grisenti M, de la Puente JM, Muñoz J, Figuerola J, Soriguer R, Anfora G, Di Luca M, Rosà R (2015) Understanding west Nile virus ecology in Europe: Culex pipiens host feeding preference in a hotspot of virus emergence. Parasites & Vectors, 8, 213. |
| [58] | Roger F, Ghanavi HR, Danielsson N, Wahlberg N, Löndahl J, Pettersson LB, Andersson GKS, Boke Olén N, Clough Y (2022) Airborne environmental DNA metabarcoding for the monitoring of terrestrial insects—A proof of concept from the field. Environmental DNA, 4, 790-807. |
| [59] | Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: A versatile open source tool for metagenomics. PeerJ, 4, e2584. |
| [60] | Sales NG, McKenzie MB, Drake J, Harper LR, Browett SS, Coscia I, Wangensteen OS, Baillie C, Bryce E, Dawson DA, Ochu E, Hänfling B, Handley LL, Mariani S, Lambin X, Sutherland C, McDevitt AD (2020) Fishing for mammals: Landscape-level monitoring of terrestrial and semi-aquatic communities using eDNA from riverine systems. Journal of Applied Ecology, 57, 707-716. |
| [61] | Schnell IB, Bohmann K, Gilbert MT (2015) Tag jumps illuminated-reducing sequence-to-sample misidentifications in metabarcoding studies. Molecular Ecology Resources, 15, 1289-1303. |
| [62] | Thomas AC, Howard J, Nguyen PL, Seimon TA, Goldberg CS (2018) eDNA Sampler: A fully integrated environmental DNA sampling system. Methods in Ecology and Evolution, 9, 1379-1385. |
| [63] | Vélez J, McShea W, Shamon H, Castiblanco-Camacho PJ, Tabak MA, Chalmers C, Fergus P, Fieberg J (2023) An evaluation of platforms for processing camera-trap data using artificial intelligence. Methods in Ecology and Evolution, 14, 459-477. |
| [64] |
Xiao ZS, Li XH, Jiang GS (2014) Applications of camera trapping to wildlife surveys in China. Biodiversity Science, 22, 683-684. (in Chinese)
DOI |
|
[肖治术, 李欣海, 姜广顺 (2014) 红外相机技术在我国野生动物监测研究中的应用. 生物多样性, 22, 683-684.]
DOI |
|
| [65] | Yan B, Wang LF, Liu SQ, Ji K, Mao YN, Zhang ZY (2020) Diversity of animal species on the Chinese side of China-Laos cross-border biodiversity joint protection area. Forest Inventory and Planning, 45(3), 42-46. (in Chinese with English abstract) |
| [岩丙, 王利繁, 刘生强, 吉魁, 毛娅南, 张忠员 (2020) 中老跨境联合保护区域中方一侧动物物种多样性调查. 林业调查规划, 45(3), 42-46.] | |
| [66] | Yang CY, Bohmann K, Wang XY, Cai W, Wales N, Ding ZL, Gopalakrishnan S, Yu DW (2021) Biodiversity Soup II: A bulk-sample metabarcoding pipeline emphasizing error reduction. Methods in Ecology and Evolution, 12, 1252-1264. |
| [67] |
Yoccoz NG, Bråthen KA, Gielly L, Haile J, Edwards ME, Goslar T, Von Stedingk H, Brysting AK, Coissac E, Pompanon F, Sønstebø JH, Miquel C, Valentini A, De Bello F, Chave J, Thuiller W, Wincker P, Cruaud C, Gavory F, Rasmussen M, Gilbert MTP, Orlando L, Brochmann C, Willerslev E, Taberlet P (2012) DNA from soil mirrors plant taxonomic and growth form diversity. Molecular Ecology, 21, 3647-3655.
DOI PMID |
| [68] |
Zepeda-Mendoza ML, Bohmann K, Carmona Baez A, Gilbert MTP (2016) DAMe: A toolkit for the initial processing of datasets with PCR replicates of double-tagged amplicons for DNA metabarcoding analyses. BMC Research Notes, 9, 255.
DOI PMID |
| [69] |
Zhang MX, Cao L, Quan RC, Xiao ZS, Yang XF, Zhang WF, Wang XZ, Deng XB (2014) Camera trap survey of animals in Xishuangbanna forest dynamics plot, Yunnan. Biodiversity Science, 22, 830-832. (in Chinese with English abstract)
DOI |
|
[张明霞, 曹林, 权锐昌, 肖治术, 杨小飞, 张文富, 王学志, 邓晓保 (2014) 利用红外相机监测西双版纳森林动态样地的野生动物多样性. 生物多样性, 22, 830-832.]
DOI |
|
| [70] | Zhao JB, Fan H, Zhao YB, Han LX (2021) Zebra Dove (Geopelia striata) found in Mengla, Yunnan. Chinese Journal of Zoology, 56, 870. (in Chinese) |
| [赵江波, 范欢, 赵夜白, 韩联宪 (2021) 云南勐腊发现斑姬地鸠. 动物学杂志, 56, 870.] | |
| [71] | Zwerts JA, Stephenson PJ, Maisels F, Rowcliffe M, Astaras C, Jansen PA, van der Waarde J, Sterck LEHM, Verweij PA, Bruce T, Brittain S, van Kuijk M (2021) Methods for wildlife monitoring in tropical forests: Comparing human observations, camera traps, and passive acoustic sensors. Conservation Science and Practice, 3, e568. |
| [1] | 钱嘉宁. 履行就地保护义务的投资仲裁风险及应对[J]. 生物多样性, 2025, 33(7): 24333-. |
| [2] | 陈耀辉, 周梓华, 邱洪, 张敬怀. 广东省海岸带牡蛎礁的分布特征、主要威胁及保护建议[J]. 生物多样性, 2025, 33(7): 24414-. |
| [3] | 董雪云, 夏富才, 周繇, 张立秋, 何怀江, 刘冰, 姜润华, 王洪峰. 吉林省野生维管植物名录[J]. 生物多样性, 2025, 33(7): 25120-. |
| [4] | 赵富伟. 遗传资源数字序列信息多边机制的谈判进程及我国的对策[J]. 生物多样性, 2025, 33(6): 24559-. |
| [5] | 彭文, 邓泽帅, 郑文宝, 龚凌轩, 曾玉枫, 孟昊, 陈军, 杨道德. eDNA技术在两栖动物调查中的应用: 以湖南莽山国家级自然保护区为例[J]. 生物多样性, 2025, 33(6): 24552-. |
| [6] | 干靓, 刘巷序, 鲁雪茗, 岳星. 全球生物多样性热点地区中国大城市的保护政策与优化方向[J]. 生物多样性, 2025, 33(5): 24529-. |
| [7] | 曾子轩, 杨锐, 黄越, 陈路遥. 清华大学校园鸟类多样性特征与环境关联[J]. 生物多样性, 2025, 33(5): 24373-. |
| [8] | 臧明月, 刘立, 马月, 徐徐, 胡飞龙, 卢晓强, 李佳琦, 于赐刚, 刘燕. 《昆明-蒙特利尔全球生物多样性框架》下的中国城市生物多样性保护[J]. 生物多样性, 2025, 33(5): 24482-. |
| [9] | 祝晓雨, 王晨灏, 王忠君, 张玉钧. 城市绿地生物多样性研究进展与展望[J]. 生物多样性, 2025, 33(5): 25027-. |
| [10] | 袁琳, 王思琦, 侯静轩. 大都市地区的自然留野: 趋势与展望[J]. 生物多样性, 2025, 33(5): 24481-. |
| [11] | 胡敏, 李彬彬, Coraline Goron. 只绿是不够的: 一个生物多样性友好的城市公园管理框架[J]. 生物多样性, 2025, 33(5): 24483-. |
| [12] | 王欣, 鲍风宇. 基于鸟类多样性提升的南滇池国家湿地公园生态修复效果[J]. 生物多样性, 2025, 33(5): 24531-. |
| [13] | 明玥, 郝培尧, 谭铃千, 郑曦. 基于城市绿色高质量发展理念的中国城市生物多样性保护与提升[J]. 生物多样性, 2025, 33(5): 24524-. |
| [14] | 杨军, 杨旭东, 刘心怡, 周景. 面向《昆蒙框架》目标12的中国城市生物多样性研究展望[J]. 生物多样性, 2025, 33(5): 25104-. |
| [15] | 杨梦婵, 方启年, 戎灿中, 胡思帆, 赵晶晶, 梁智健, 李添明. 公民科学在城市生物多样性调查监测上的应用现状与前景[J]. 生物多样性, 2025, 33(5): 24464-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn