



生物多样性 ›› 2024, Vol. 32 ›› Issue (9): 24171. DOI: 10.17520/biods.2024171 cstr: 32101.14.biods.2024171
收稿日期:2024-05-08
接受日期:2024-06-27
出版日期:2024-09-20
发布日期:2024-07-05
通讯作者:
* E-mail: zhangqg@bnu.edu.cn基金资助:
Nan Chen( ), Quan-Guo Zhang*(
), Quan-Guo Zhang*( )(
)( )
)
Received:2024-05-08
Accepted:2024-06-27
Online:2024-09-20
Published:2024-07-05
Contact:
* E-mail: zhangqg@bnu.edu.cnSupported by:摘要:
追踪生物种群在实验室条件下发生的进化, 这种研究途径称为实验进化。不同于大多数“回溯式”的进化研究, 实验进化是一种“前瞻式”的研究途径。这种方法的使用可以追溯到达尔文时代。最近几十年来这一研究手段被用于对现代进化综合理论的若干关键假说进行检验。实验进化的研究材料可以是真实生物种群, 也可以是数字生命形式; 使用真实生物开展的进化实验可以在实验室或野外环境中进行。微生物具有生长繁殖快、易保存等特征, 成为实验进化的主要研究对象。微生物实验进化已经被用于研究包括适应性进化的动态、种群分化规律、种间协同进化模式甚至多细胞生命形式的起源等问题。同时在解决进化的确定性和偶然性、预测全球变化背景下的进化过程等方面有明显的优势。实验进化这一研究方法也有一定的局限性, 特别是其无法完全模拟自然界中的种群及其进化环境。但该方法在未来的科学研究、工业生产和课堂教学中将扮演越来越重要的角色。
陈楠, 张全国 (2024) 实验进化研究途径. 生物多样性, 32, 24171. DOI: 10.17520/biods.2024171.
Nan Chen, Quan-Guo Zhang (2024) The experimental evolution approach. Biodiversity Science, 32, 24171. DOI: 10.17520/biods.2024171.
| 部分参考文献 References | |
|---|---|
| 决定进化的种群遗传过程: 突变、基因流、漂变和选择 Population genetic processes that determine evolution: mutation, gene flow, drift and selection | |
| • 突变速率与适合度效果 Mutation rate and fitness effects | Mukai et al, |
| • 适应进化速率随突变供给的饱和式上升模式 Patterns of diminishing returns in adaptive evolutionary rates from subsequent mutations | Souza et al, |
| • 基因流对种群适应进化的影响 Effects of gene flow on adaptive evolution of population | Souza et al, |
| • 多层级选择以及利他行为进化 Multi-level selection and the evolution of altruistic behavior | Weinig et al, |
| 生物种群适应重要环境因子的规律 Rules for the adaptation of populations to critical environmental factors | |
| • 温度决定进化速率的机制 Mechanisms underlying temperature-determined evolutionary rates | Knies et al, |
| • 温度对代谢等生理过程的限制性及生物的适应 Temperature-dependent physiological processes such as metabolism and thermal adaptation of organisms | Anderson-Teixeira et al, |
| • 生物对全球环境变化的快速适应 Rapid adaptation of organisms to global environmental change | Collins & Bell, |
| 适应的代价与种群间分化 Costs of adaptation and interpopulation differentiation | |
| • 环境间适合度权衡的机制: 拮抗多效应与突变累积 Mechanisms of fitness trade-off across environments: antagonistic pleiotropy and mutations accumulation | Bosshard et al, |
| • 空间异质性和时间异质性环境中种群分化规律 Patterns of population differentiation in spatially and temporally heterogeneous environments | Reboud & Bell, |
| 从种群分化到物种多样性维持 From population differentiation to the maintenance of species diversity | |
| • 适应辐射的影响因素 Factors affecting adaptive radiation | Dodd, |
| • 生物多样性维持中的生态-进化动态 Eco-evolutionary dynamics in biodiversity maintenance | Yoshida et al, |
| 种间协同进化 Interspecific coevolution | |
| • 对抗性种间关系进化的模式和机制 Patterns and mechanisms in the evolution of interspecific antagonistic interactions | Buckling & Rainey, |
| • 红皇后过程与有性过程维持 Red Queen processes and the maintenance of sex | Morran et al, |
| 重大进化转变事件的规律 Patterns of events during major evolutionary transition | |
| • 多细胞属性的进化 Evolution of multicellularity | Ratcliff et al, |
| • 异养属性的进化 Evolution of heterotrophy | Bell, |
| • 真核生物体内共生体的形成 Formation of endosymbionts in eukaryotes | Jeon & Jeon, |
| 进化的确定性和偶然性 Determinism and contingency in evolution | |
| • 进化过程的可重复性 Repeatability of evolutionary processes | Travisano et al, |
| • 进化偶然事件对进化结果的影响 Consequences of contingencies on evolution | Rebolleda-Gomez et al, |
表1 一些经典进化生物学理论主题及对应的实验进化途径解决具体问题的例证。其中加粗的参考文献为正文中介绍的例子。
Table 1 Examples of classical topics in evolutionary biology and related empirical studies using the experimental evolutionary approach. References in bold are examples presented in the main text.
| 部分参考文献 References | |
|---|---|
| 决定进化的种群遗传过程: 突变、基因流、漂变和选择 Population genetic processes that determine evolution: mutation, gene flow, drift and selection | |
| • 突变速率与适合度效果 Mutation rate and fitness effects | Mukai et al, |
| • 适应进化速率随突变供给的饱和式上升模式 Patterns of diminishing returns in adaptive evolutionary rates from subsequent mutations | Souza et al, |
| • 基因流对种群适应进化的影响 Effects of gene flow on adaptive evolution of population | Souza et al, |
| • 多层级选择以及利他行为进化 Multi-level selection and the evolution of altruistic behavior | Weinig et al, |
| 生物种群适应重要环境因子的规律 Rules for the adaptation of populations to critical environmental factors | |
| • 温度决定进化速率的机制 Mechanisms underlying temperature-determined evolutionary rates | Knies et al, |
| • 温度对代谢等生理过程的限制性及生物的适应 Temperature-dependent physiological processes such as metabolism and thermal adaptation of organisms | Anderson-Teixeira et al, |
| • 生物对全球环境变化的快速适应 Rapid adaptation of organisms to global environmental change | Collins & Bell, |
| 适应的代价与种群间分化 Costs of adaptation and interpopulation differentiation | |
| • 环境间适合度权衡的机制: 拮抗多效应与突变累积 Mechanisms of fitness trade-off across environments: antagonistic pleiotropy and mutations accumulation | Bosshard et al, |
| • 空间异质性和时间异质性环境中种群分化规律 Patterns of population differentiation in spatially and temporally heterogeneous environments | Reboud & Bell, |
| 从种群分化到物种多样性维持 From population differentiation to the maintenance of species diversity | |
| • 适应辐射的影响因素 Factors affecting adaptive radiation | Dodd, |
| • 生物多样性维持中的生态-进化动态 Eco-evolutionary dynamics in biodiversity maintenance | Yoshida et al, |
| 种间协同进化 Interspecific coevolution | |
| • 对抗性种间关系进化的模式和机制 Patterns and mechanisms in the evolution of interspecific antagonistic interactions | Buckling & Rainey, |
| • 红皇后过程与有性过程维持 Red Queen processes and the maintenance of sex | Morran et al, |
| 重大进化转变事件的规律 Patterns of events during major evolutionary transition | |
| • 多细胞属性的进化 Evolution of multicellularity | Ratcliff et al, |
| • 异养属性的进化 Evolution of heterotrophy | Bell, |
| • 真核生物体内共生体的形成 Formation of endosymbionts in eukaryotes | Jeon & Jeon, |
| 进化的确定性和偶然性 Determinism and contingency in evolution | |
| • 进化过程的可重复性 Repeatability of evolutionary processes | Travisano et al, |
| • 进化偶然事件对进化结果的影响 Consequences of contingencies on evolution | Rebolleda-Gomez et al, |
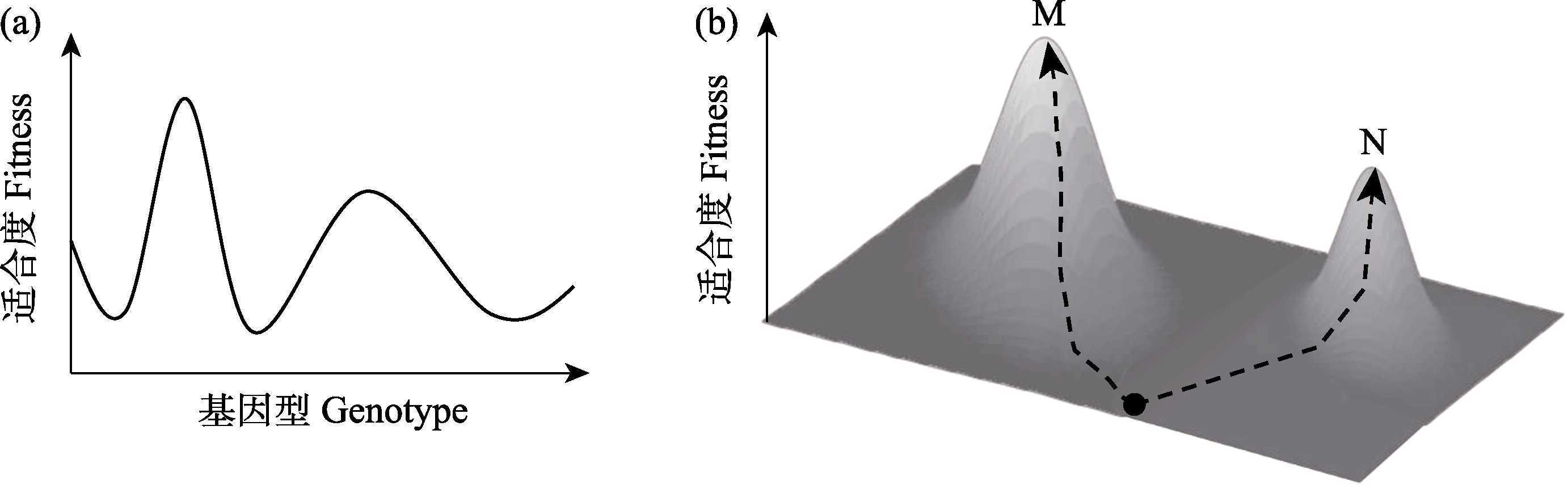
图1 一个二维(a)和一个三维(b)的适合度景观示意图。(b)图中任意一点在水平面上的投影代表特定的基因型。M: 全局适合度最高峰; N: 局域适合度最高峰。
Fig. 1 A graphic illustration of (a) a two- and (b) a three-dimensional fitness landscape. The projection of any point on the horizontal plane in (b) represents a specific genotype. M, Global fitness peak; N, Local fitness peak.
| [1] | Adami C (1998) Introduction to Artificial Life. Springer, New York. |
| [2] |
Alzate A, Bisschop K, Etienne RS, Bonte D (2017) Interspecific competition counteracts negative effects of dispersal on adaptation of an arthropod herbivore to a new host. Journal of Evolutionary Biology, 30, 1966-1977.
DOI PMID |
| [3] | Anderson-Teixeira KJ, Delong JP, Fox AM, Brese DA, Litvak ME (2011) Differential responses of production and respiration to temperature and moisture drive the carbon balance across a climatic gradient in New Mexico. Global Change Biology, 17, 410-424. |
| [4] |
Andersson DI, Hughes D (2010) Antibiotic resistance and its cost: Is it possible to reverse resistance? Nature Reviews Microbiology, 8, 260-271.
DOI PMID |
| [5] |
Andersson DI, Hughes D (2011) Persistence of antibiotic resistance in bacterial populations. FEMS Microbiology Reviews, 35, 901-911.
DOI PMID |
| [6] |
Angert AL, Bradshaw HD Jr, Schemske DW (2008) Using experimental evolution to investigate geographic range limits in monkeyflowers. Evolution, 62, 2660-2675.
DOI PMID |
| [7] | Arrhenius S (1889) Über die Reaktionsgeschwindigkeit Bei der Inversion von Rohrzucker durch Säuren. Zeitschrift Für Physikalische Chemie, 4, 226-248. |
| [8] | Atwood KC, Schneider LK, Ryan FJ (1951) Periodic selection in Escherichia coli. Proceedings of the National Academy of Sciences, USA, 37, 146-155. |
| [9] |
Axelrod R, Hamilton WD (1981) The evolution of cooperation. Science, 211, 1390-1396.
DOI PMID |
| [10] | Bailey SF, Dettman JR, Rainey PB, Kassen R (2013) Competition both drives and impedes diversification in a model adaptive radiation. Proceedings of the Royal Society B, 280, 20131253. |
| [11] | Baym M, Stone LK, Kishony R (2016) Multidrug evolutionary strategies to reverse antibiotic resistance. Science, 351, aad3292. |
| [12] |
Bell G (2013) Experimental evolution of heterotrophy in a green alga. Evolution, 67, 468-476.
DOI PMID |
| [13] | Bell G (2016) Experimental macroevolution. Proceedings of the Royal Society B: Biological Sciences, 283, 20152547. |
| [14] | Berg MP, Kiers ET, Driessen G, van der Heijden M, Kooi BW, Kuenen F, Liefting M, Verhoef HA, Ellers J (2010) Adapt or disperse: Understanding species persistence in a changing world. Global Change Biology, 16, 587-598. |
| [15] | Betts A, Kaltz O, Hochberg ME (2014) Contrasted coevolutionary dynamics between a bacterial pathogen and its bacteriophages. Proceedings of the National Academy of Sciences, USA, 111, 11109-11114. |
| [16] | Blount ZD (2016) A case study in evolutionary contingency. Studies in History and Philosophy of Biological and Biomedical Sciences, 58, 82-92. |
| [17] | Blount ZD, Borland CZ, Lenski RE (2008) Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proceedings of the National Academy of Sciences, USA, 105, 7899-7906. |
| [18] | Blount ZD, Lenski RE, Losos JB (2018) Contingency and determinism in evolution: Replaying life’s tape. Science, 362, eaam5979. |
| [19] |
Bosshard L, Dupanloup I, Tenaillon O, Bruggmann R, Ackermann M, Peischl S, Excoffier L (2017) Accumulation of deleterious mutations during bacterial range expansions. Genetics, 207, 669-684.
DOI PMID |
| [20] | Boyce DG, Lewis MR, Worm B (2010) Global phytoplankton decline over the past century. Nature, 466, 591-596. |
| [21] |
Brakmann S, Grzeszik S (2001) An error-prone T 7 RNA polymerase mutant generated by directed evolution. Chembiochem, 2, 212-219.
PMID |
| [22] | Bratley P, Millo J (1972) Computer recreations. Software: Practice and Experience, 2, 397-400. |
| [23] | Brennan RS, DeMayo JA, Dam HG, Finiguerra M, Baumann H, Buffalo V, Pespeni MH (2022) Experimental evolution reveals the synergistic genomic mechanisms of adaptation to ocean warming and acidification in a marine copepod. Proceedings of the National Academy of Sciences, USA, 119, e2201521119. |
| [24] | Brown JH, Gillooly JF, Allen AP, Savage VM, West GB (2004) Toward a metabolic theory of ecology. Ecology, 85, 1771-1789. |
| [25] | Buckling A, MacLean RC, Brockhurst MA, Colegrave N (2009) The beagle in a bottle. Nature, 457, 824-829. |
| [26] | Buckling A, Rainey PB (2002) Antagonistic coevolution between a bacterium and a bacteriophage. Proceedings of the Royal Society of London, Series B: Biological Sciences, 269, 931-936. |
| [27] |
Bush SE, Villa SM, Altuna JC, Johnson KP, Shapiro MD, Clayton DH (2019) Host defense triggers rapid adaptive radiation in experimentally evolving parasites. Evolution Letters, 3, 120-128.
DOI PMID |
| [28] | Caldeira K, Wickett ME (2003) Anthropogenic carbon and ocean pH. Nature, 425, 365-365. |
| [29] | Cervera H, Lalić J, Elena SF (2016) Efficient escape from local optima in a highly rugged fitness landscape by evolving RNA virus populations. Proceedings of the Royal Society B, 283, 20160984. |
| [30] |
Chavhan Y, Malusare S, Dey S (2020) Larger bacterial populations evolve heavier fitness trade-offs and undergo greater ecological specialization. Heredity, 124, 726-736.
DOI PMID |
| [31] | Chen N, Zhang QG (2023) Linking temperature dependence of fitness effects of mutations to thermal niche adaptation. Journal of Evolutionary Biology, 36, 1517-1524. |
| [32] | Chen N, Zhang QG (2024) Suffering makes you weaker: Limited evolutionary adaptation in competitively inferior populations. Ecology Letters, 27, e14457. |
| [33] | Chen PP, Zhang JZ (2020) Antagonistic pleiotropy conceals molecular adaptations in changing environments. Nature Ecology & Evolution, 4, 461-469. |
| [34] | Chu XL, Zhang DY, Buckling A, Zhang QG (2020) Warmer temperatures enhance beneficial mutation effects. Journal of Evolutionary Biology, 33, 1020-1027. |
| [35] | Clarke L, Pelin A, Phan M, Wong A (2020) The effect of environmental heterogeneity on the fitness of antibiotic resistance mutations in Escherichia coli. Evolutionary Ecology, 34, 379-390. |
| [36] | Clements DR, Jones VL (2021) Rapid evolution of invasive weeds under climate change: Present evidence and future research needs. Frontiers in Agronomy, 3, 664034. |
| [37] |
Colegrave N, Buckling A (2005) Microbial experiments on adaptive landscapes. BioEssays, 27, 1167-1173.
PMID |
| [38] | Collins S, Bell G (2004) Phenotypic consequences of 1,000 generations of selection at elevated CO2 in a green alga. Nature, 431, 566-569. |
| [39] | Condon C, Cooper BS, Yeaman S, Angilletta MJ Jr (2014) Temporal variation favors the evolution of generalists in experimental populations of Drosophila melanogaster. Evolution, 68, 720-728. |
| [40] |
Cooper TF, Lenski RE (2010) Experimental evolution with E. coli in diverse resource environments. I. Fluctuating environments promote divergence of replicate populations. BMC Evolutionary Biology, 10, 11.
DOI PMID |
| [41] | Cooper TF, Ofria C (2002) Evolution of stable ecosystems in populations of digital organisms. In: Artificial Life VIII: Proceedings of the Eighth International Conference on Artificial Life (eds Standish RK, Bedau MA, Abbass HA), pp.227-232. MIT Press, Cambridge, MA. |
| [42] | Cooper VS, Warren TM, Matela AM, Handwork M, Scarponi S (2019) EvolvingSTEM: A microbial evolution-in-action curriculum that enhances learning of evolutionary biology and biotechnology. Evolution, 12, 12. |
| [43] | Dai XF, Shen L (2022) Advances and trends in omics technology development. Frontiers in Medicine, 9, 911861. |
| [44] | Dallinger WH (1887) The president’s address. Journal of the Royal Microscopical Society, 7, 185-199. |
| [45] | Darwin C (1859) The Origin of Species. John Murray, London. |
| [46] |
de Vargas Roditi L, Boyle KE, Xavier JB (2013) Multilevel selection analysis of a microbial social trait. Molecular Systems Biology, 9, 684.
DOI PMID |
| [47] |
Krug J (2014) Empirical fitness landscapes and the predictability of evolution. Nature Reviews Genetics, 15, 480-490.
DOI PMID |
| [48] |
Zeyl CW, Gerrish PJ, Blanchard JL, Lenski RE (1999) Diminishing returns from mutation supply rate in asexual populations. Science, 283, 404-406.
DOI PMID |
| [49] | de Vos MG, Dawid A, Sunderlikova V, Tans SJ (2015) Breaking evolutionary constraint with a tradeoff ratchet. Proceedings of the National Academy of Sciences, USA, 112, 14906-14911. |
| [50] |
Dettman JR, Rodrigue N, Melnyk AH, Wong A, Bailey SF, Kassen R (2012) Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Molecular Ecology, 21, 2058-2077.
DOI PMID |
| [51] | Dewald-Wang EA, Parr N, Tiley K, Lee A, Koskella B (2021) Multiyear time-shift study of bacteria and phage dynamics in the phyllosphere. The American Naturalist, 199, 126-140. |
| [52] | Dewdney A (1984) Computer recreations. Scientific American, 250, 14-23. |
| [53] | Dodd DMB (1989) Reproductive isolation as a consequence of adaptive divergence in Drosophila pseudoobscura. Evolution, 43, 1308-1311. |
| [54] | Dorado-Morales P, Garcillán-Barcia MP, Lasa I, Solano C (2021) Fitness cost evolution of natural plasmids of Staphylococcus aureus. mBio, 12, e03094-03020. |
| [55] |
Dorin A, Stepney S (2024) What is artificial life today, and where should it go? Artificial Life, 30, 1-15.
DOI PMID |
| [56] |
Driscoll WW, Travisano M (2017) Synergistic cooperation promotes multicellular performance and unicellular free-rider persistence. Nature Communications, 8, 15707.
DOI PMID |
| [57] |
Dybdahl MF, Lively CM (1998) Host-parasite coevolution: Evidence for rare advantage and time-lagged selection in a natural population. Evolution, 52, 1057-1066.
DOI PMID |
| [58] |
Eldakar OT, Wilson DS, Dlugos MJ, Pepper JW (2010) The role of multilevel selection in the evolution of sexual conflict in the water strider Aquarius remigis. Evolution, 64, 3183-3189.
DOI PMID |
| [59] | Elser JJ, Dobberfuhl DR, MacKay NA, Schampel JH (1996) Organism size, life history, and N : P stoichiometry: Toward a unified view of cellular and ecosystem processes. Bioscience, 46, 674-684. |
| [60] | Erdos Z, Studholme DJ, Sharma MD, Chandler D, Bass C, Raymond B (2024) Manipulating multi-level selection in a fungal entomopathogen reveals social conflicts and a method for improving biocontrol traits. PLoS Pathogens, 20, e1011775. |
| [61] | Feely RA, Sabine CL, Lee K, Berelson W, Kleypas J, Fabry VJ, Millero FJ (2004) Impact of anthropogenic CO2 on the CaCO3 system in the oceans. Science, 305, 362-366. |
| [62] | Foster BJ, McCulloch GA, Vogel MFS, Ingram T, Waters JM (2021) Anthropogenic evolution in an insect wing polymorphism following widespread deforestation. Biology Letters, 17, 20210069. |
| [63] | Fuhrer J (2003) Agroecosystem responses to combinations of elevated CO2, ozone, and global climate change. Agriculture, Ecosystems & Environment, 97, 1-20. |
| [64] |
Gandon S, Buckling A, Decaestecker E, Day T (2008) Host-parasite coevolution and patterns of adaptation across time and space. Journal of Evolutionary Biology, 21, 1861-1866.
DOI PMID |
| [65] | Garant D, Forde SE, Hendry AP (2007) The multifarious effects of dispersal and gene flow on contemporary adaptation. Functional Ecology, 21, 434-443. |
| [66] | Garland T, Rose MR (2009) Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments. University of California Press, Berkeley, CA. |
| [67] | Gause GF (1934) The Struggle for Existence. Williams and Wilkins, Baltimore, MD. |
| [68] |
Gibson G, Dworkin I (2004) Uncovering cryptic genetic variation. Nature Reviews Genetics, 5, 681-690.
DOI PMID |
| [69] | Goldsmith M, Tawfik DS (2009) Potential role of phenotypic mutations in the evolution of protein expression and stability. Proceedings of the National Academy of Sciences, USA, 106, 6197-6202. |
| [70] |
Gómez-Sanz E, Kadlec K, Feßler AT, Zarazaga M, Torres C, Schwarz S (2013) Novel erm (T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST 398 that also harbor cadmium and copper resistance determinants. Antimicrobial Agents and Chemotherapy, 57, 3275-3282.
DOI PMID |
| [71] |
Gomulkiewicz R, Holt RD, Barfield M (1999) The effects of density dependence and immigration on local adaptation and niche evolution in a black-hole sink environment. Theoretical Population Biology, 55, 283-296.
PMID |
| [72] |
Görke B, Stülke J (2008) Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nature Reviews Microbiology, 6, 613-624.
DOI PMID |
| [73] | Gould SJ (1989) Wonderful Life: The Burgess Shale and the Nature of History. W. W. Norton, New York. |
| [74] |
Gras R, Devaurs D, Wozniak A, Aspinall A (2009) An individual-based evolving predator-prey ecosystem simulation using a fuzzy cognitive map as the behavior model. Artificial Life, 15, 423-463.
DOI PMID |
| [75] | Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI (2011) Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathogens, 7, e1002158. |
| [76] |
Haafke J, Abou Chakra M, Becks L (2016) Eco-evolutionary feedback promotes Red Queen dynamics and selects for sex in predator populations. Evolution, 70, 641-652.
DOI PMID |
| [77] |
Hall AR, Scanlan PD, Morgan AD, Buckling A (2011) Host-parasite coevolutionary arms races give way to fluctuating selection. Ecology Letters, 14, 635-642.
DOI PMID |
| [78] | Halsch CA, Shapiro AM, Fordyce JA, Nice CC, Thorne JH, Waetjen DP, Forister ML (2021) Insects and recent climate change. Proceedings of the National Academy of Sciences, USA, 118, e2002543117. |
| [79] | Hatton IA, Dobson AP, Storch D, Galbraith ED, Loreau M (2019) Linking scaling laws across eukaryotes. Proceedings of the National Academy of Sciences, USA, 116, 21616-21622. |
| [80] | He XL, Liu L (2016) Toward a prospective molecular evolution. Science, 352, 769-770. |
| [81] |
Heilbron K, Toll-Riera M, Kojadinovic M, MacLean RC (2014) Fitness is strongly influenced by rare mutations of large effect in a microbial mutation accumulation experiment. Genetics, 197, 981-990.
DOI PMID |
| [82] | Hosoda K, Habuchi M, Suzuki S, Miyazaki M, Takikawa G, Sakurai T, Kashiwagi A, Sueyoshi M, Matsumoto Y, Kiuchi A, Mori K, Yomo T (2014) Adaptation of a cyanobacterium to a biochemically rich environment in experimental evolution as an initial step toward a chloroplast-like state. PLoS ONE, 9, e98337. |
| [83] | Houle D, Hoffmaster DK, Assimacopoulos S, Charlesworth B (1992) The genomic mutation rate for fitness in Drosophila. Nature, 359, 58-60. |
| [84] |
Huang W, Petrosino J, Hirsch M, Shenkin PS, Palzkill T (1996) Amino acid sequence determinants of beta-lactamase structure and activity. Journal of Molecular Biology, 258, 688-703.
PMID |
| [85] | Huxley JS (1942) Evolution: The Modern Synthesis. Allen and Unwin, London. |
| [86] |
Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. Journal of Molecular Biology, 3, 318-356.
PMID |
| [87] | Jansen G, Barbosa C, Schulenburg H (2013) Experimental evolution as an efficient tool to dissect adaptive paths to antibiotic resistance. Drug Resistance Updates, 16, 96-107. |
| [88] |
Jeon KW, Jeon MS (1976) Endosymbiosis in amoebae: Recently established endosymbionts have become required cytoplasmic components. Journal of Cellular Physiology, 89, 337-344.
PMID |
| [89] | Jin P, Wan J, Zhou Y, Gao K, Beardall J, Lin J, Huang J, Lu Y, Liang S, Wang K, Ma Z, Xia J (2022) Increased genetic diversity loss and genetic differentiation in a model marine diatom adapted to ocean warming compared to high CO2. The ISME Journal, 16, 2587-2598. |
| [90] | Jousset A, Eisenhauer N, Merker M, Mouquet N, Scheu S (2016) High functional diversity stimulates diversification in experimental microbial communities. Science Advances, 2, e1600124. |
| [91] | Kassen R (2014) Experimental Evolution and the Nature of Biodiversity. Roberts and Company, Denver, CO. |
| [92] |
Kassen R, Bataillon T (2006) Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nature Genetics, 38, 484-488.
PMID |
| [93] | Kassen R, Bell G (1998) Experimental evolution in Chlamydomonas. IV. Selection in environments that vary through time at different scales. Heredity, 80, 732-741. |
| [94] | Kassen R, Llewellyn M, Rainey PB (2004) Ecological constraints on diversification in a model adaptive radiation. Nature, 431, 984-988. |
| [95] | Kawecki TJ, Lenski RE, Ebert D, Hollis B, Olivieri I, Whitlock MC (2012) Experimental evolution. Trends in Ecology & Evolution, 27, 547-560. |
| [96] | Kibota TT, Lynch M (1996) Estimate of the genomic mutation rate deleterious to overall fitness in E. coli. Nature, 381, 694-696. |
| [97] |
Knibbe C, Coulon A, Mazet O, Fayard J-M, Beslon G (2007) A long-term evolutionary pressure on the amount of noncoding DNA. Molecular Biology and Evolution, 24, 2344-2353.
DOI PMID |
| [98] |
Knies JL, Kingsolver JG, Burch CL (2009) Hotter is better and broader: Thermal sensitivity of fitness in a population of bacteriophages. The American Naturalist, 173, 419-430.
DOI PMID |
| [99] | Kortright KE, Chan BK, Evans BR, Turner PE (2022) Arms race and fluctuating selection dynamics in Pseudomonas aeruginosa bacteria coevolving with phage OMKO1. Journal of Evolutionary Biology, 35, 1475-1487. |
| [100] |
Kümmerli R, Gardner A, West SA, Griffin AS (2009) Limited dispersal, budding dispersal, and cooperation: An experimental study. Evolution, 63, 939-949.
DOI PMID |
| [101] | Lachapelle J, Reid J, Colegrave N (2015) Repeatability of adaptation in experimental populations of different sizes. Proceedings of the Royal Society B: Biological Sciences, 282, 20143033. |
| [102] | Lee TD, Tjoelker MG, Ellsworth DS, Reich PB (2001) Leaf gas exchange responses of 13 prairie grassland species to elevated CO2 and increased nitrogen supply. New Phytologist, 150, 405-418. |
| [103] |
Lehman J, Clune J, Misevic D, Adami C, Altenberg L, Beaulieu J, Bentley PJ, Bernard S, Beslon G, Bryson DM, Cheney N, Chrabaszcz P, Cully A, Doncieux S, Dyer FC, Ellefsen KO, Feldt R, Fischer S, Forrest S, Fŕenoy A, Gagńe C, Le Goff L, Grabowski LM, Hodjat B, Hutter F, Keller L, Knibbe C, Krcah P, Lenski RE, Lipson H, MacCurdy R, Maestre C, Miikkulainen R, Mitri S, Moriarty DE, Mouret J-B, Nguyen A, Ofria C, Parizeau M, Parsons D, Pennock RT, Punch WF, Ray TS, Schoenauer M, Schulte E, Sims K, Stanley KO, Taddei F, Tarapore D, Thibault S, Watson R, Weimer W, Yosinski J (2020) The surprising creativity of digital evolution: A collection of anecdotes from the evolutionary computation and artificial life research communities. Artificial Life, 26, 274-306.
DOI PMID |
| [104] | Lehmann KDS, Goldman BW, Dworkin I, Bryson DM, Wagner AP (2014) From cues to signals: Evolution of interspecific communication via aposematism and mimicry in a predator-prey system. PLoS ONE, 9, e91783. |
| [105] | Lemmen KD, Zhou L, Papakostas S, Declerck SAJ (2023) An experimental test of the growth rate hypothesis as a predictive framework for microevolutionary adaptation. Ecology, 104, e3853. |
| [106] | Lenski RE, Rose MR, Simpson SC, Tadler SC (1991) Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. The American Naturalist, 138, 1315-1341. |
| [107] | Leónidas Cardoso L, Durão P, Amicone M, Gordo I (2020) Dysbiosis individualizes the fitness effect of antibiotic resistance in the mammalian gut. Nature Ecology & Evolution, 4, 1268-1278. |
| [108] |
Lopez-Pascua LDC, Buckling A (2008) Increasing productivity accelerates host-parasite coevolution. Journal of Evolutionary Biology, 21, 853-860.
DOI PMID |
| [109] |
Lopez-Pascua LDC, Hall AR, Best A, Morgan AD, Boots M, Buckling A (2014) Higher resources decrease fluctuating selection during host-parasite coevolution. Ecology Letters, 17, 1380-1388.
DOI PMID |
| [110] | Lustenhouwer N, Moerman F, Altermatt F, Bassar RD, Bocedi G, Bonte D, Dey S, Fronhofer EA, da Rocha ÉG, Giometto A, Lancaster LT, Robert BP Jr, Saastamoinen M, Travis JMJ, Urquhart CA, Weiss-Lehman C, Williams JL, Börger L, Berger D (2023) Experimental evolution of dispersal: Unifying theory, experiments and natural systems. Journal of Animal Ecology, 92, 1113-1123. |
| [111] |
Maclean CJ, Metzger BPH, Yang J-R, Ho W-C, Moyers B, Zhang J (2017) Deciphering the genic basis of yeast fitness variation by simultaneous forward and reverse genetics. Molecular Biology and Evolution, 34, 2486-2502.
DOI PMID |
| [112] | MacLean RC, Bell G (2003) Divergent evolution during an experimental adaptive radiation. Proceedings of the Royal Society of London, Series B: Biological Sciences, 270, 1645-1650. |
| [113] | Marques DA, Jones FC, Di Palma F, Kingsley DM, Reimchen TE (2018) Experimental evidence for rapid genomic adaptation to a new niche in an adaptive radiation. Nature Ecology & Evolution, 2, 1128-1138. |
| [114] | Marshall DJ, Malerba M, Lines T, Sezmis AL, Hasan CM, Lenski RE, McDonald MJ (2022) Long-term experimental evolution decouples size and production costs in Escherichia coli. Proceedings of the National Academy of Sciences, USA, 119, e2200713119. |
| [115] | McCulloch GA, Waters JM (2023) Rapid adaptation in a fast-changing world: Emerging insights from insect genomics. Global Change Biology, 29, 943-954. |
| [116] | McVeigh I, Hobdy CJ (1952) Development of resistance by Micrococcus pyogenes var. aureus to antibiotics; morphological and physiological changes. American Journal of Botany, 39, 352-359. |
| [117] |
Melnyk AH, Wong A, Kassen R (2015) The fitness costs of antibiotic resistance mutations. Evolutionary Applications, 8, 273-283.
DOI PMID |
| [118] |
Moreno MA, Ofria C (2019) Toward open-ended fraternal transitions in individuality. Artificial Life, 25, 117-133.
DOI PMID |
| [119] | Morgan AD, Gandon S, Buckling A (2005) The effect of migration on local adaptation in a coevolving host-parasite system. Nature, 437, 253-256. |
| [120] |
Morran LT, Schmidt OG, Gelarden IA, Parrish RC, Lively CM (2011) Running with the Red Queen: Host-parasite coevolution selects for biparental sex. Science, 333, 216-218.
DOI PMID |
| [121] | Mu X, Wang N, Li X, Shi K, Zhou Z, Yu Y, Hua X (2016) The effect of colistin resistance-associated mutations on the fitness of Acinetobacter baumannii. Frontiers in Microbiology, 7, 1715. |
| [122] |
Mukai T, Chigusa SI, Mettler LE, Crow JF (1972) Mutation rate and dominance of genes affecting viability in Drosophila melanogaster. Genetics, 72, 335-355.
DOI PMID |
| [123] | Nissan RB, Milshtein E, Pahl V, de Pins B, Jona G, Levi D, Yung H, Nir N, Ezra D, Gleizer S, Link H, Noor E, Milo R (2024) Autotrophic growth of Escherichia coli is achieved by a small number of genetic changes. eLife, 12, RP88793. |
| [124] | Novick A, Szilard L (1950) Experiments with the chemostat on spontaneous mutations of bacteria. Proceedings of the National Academy of Sciences, USA, 36, 708-719. |
| [125] |
O’Donnell DR, Hamman CR, Johnson EC, Kremer CT, Klausmeier CA, Litchman E (2018) Rapid thermal adaptation in a marine diatom reveals constraints and trade-offs. Global Change Biology, 24, 4554-4565.
DOI PMID |
| [126] | O’Neill J (2016) Tackling drug-resistant infections globally: Final report and recommendations. https://apo.org.au/node/63983. (accessed on 2024-11-17) |
| [127] | Orr JA, Luijckx P, Arnoldi J-F, Jackson AL, Piggott JJ (2022) Rapid evolution generates synergism between multiple stressors: Linking theory and an evolution experiment. Global Change Biology, 28, 1740-1752. |
| [128] | Paaby AB, Rockman MV (2014) Cryptic genetic variation: Evolution’s hidden substrate. Nature Reviews Genetics, 15, 247-258. |
| [129] |
Padfield D, Yvon-Durocher G, Buckling A, Jennings S, Yvon-Durocher G (2016) Rapid evolution of metabolic traits explains thermal adaptation in phytoplankton. Ecology Letters, 19, 133-142.
DOI PMID |
| [130] | Papkou A, Garcia-Pastor L, Escudero JA, Wagner A (2023) A rugged yet easily navigable fitness landscape. Science, 382, eadh3860. |
| [131] |
Pargellis AN (2001) Digital life behavior in the amoeba world. Artificial Life, 7, 63-75.
PMID |
| [132] | Payam G, Paula J, Nikolay M, Helena R-M, Jennifer V, Dimitrios K, Simon S, Sergej A, Kristina G, Albert M, Gemma B, Eivind A, Kiran RP, Jonas W (2022) Highly parallelized laboratory evolution of wine yeasts for enhanced metabolic phenotypes. https://www.biorxiv.org/content/10.1101/2022.04.18.488345v3. (accessed on 2024-04-27) |
| [133] | Peck SL, Ellner SP, Gould F (1998) A spatially explicit stochastic model demonstrates the feasibility of Wright’s shifting balance theory. Evolution, 52, 1834-1839. |
| [134] |
Plucain J, Suau A, Cruveiller S, Médigue C, Schneider D, Le Gac M (2016) Contrasting effects of historical contingency on phenotypic and genomic trajectories during a two-step evolution experiment with bacteria. BMC Evolutionary Biology, 16, 86.
DOI PMID |
| [135] |
Qian W, Ma D, Xiao C, Wang Z, Zhang J (2012) The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Reports, 2, 1399-1410.
DOI PMID |
| [136] | Rainey PB, Travisano M (1998) Adaptive radiation in a heterogeneous environment. Nature, 394, 69-72. |
| [137] | Ratcliff WC, Denison RF, Borrello M, Travisano M (2012) Experimental evolution of multicellularity. Proceedings of the National Academy of Sciences, USA, 109, 1595-1600. |
| [138] | Ray TS (1991) An approach to the synthesis of life. In: The Philosophy of Artificial Life (ed. Boden MA), pp. 111-145. Oxford University Press, Oxford. |
| [139] | Rebolleda-Gomez M, Ratcliff W, Travisano M (2012) Adaptation and divergence during experimental evolution of multicellular Saccharomyces cerevisiae. Artificial Life, 13, 99-104. |
| [140] | Reboud X, Bell G (1997) Experimental evolution in Chlamydomonas. III. Evolution of specialist and generalist types in environments that vary in space and time. Heredity, 78, 507-514. |
| [141] | Rocabert C, Knibbe C, Consuegra J, Schneider D, Beslon G (2017) Beware batch culture: Seasonality and niche construction predicted to favor bacterial adaptive diversification. PLoS Computational Biology, 13, e1005459. |
| [142] | Savage VM, Gillooly JF, Brown JH, West GB, Charnov EL (2004) Effects of body size and temperature on population growth. The American Naturalist, 163, 429-441. |
| [143] | Saxe H, Ellsworth DS, Heath J (1998) Tree and forest functioning in an enriched CO2 atmosphere. New Phytologist, 139, 395-436. |
| [144] | Saxer G, Doebeli M, Travisano M (2010) The repeatability of adaptive radiation during long-term experimental evolution of Escherichia coli in a multiple nutrient environment. PLoS ONE, 5, e14184. |
| [145] | Schoustra S, Hwang S, Krug J, de Visser JAGM (2016) Diminishing-returns epistasis among random beneficial mutations in a multicellular fungus. Proceedings of the Royal Society B: Biological Sciences, 283, 20161376. |
| [146] |
Simões P, Santos J, Fragata I, Mueller LD, Rose MR, Matos M (2008) How repeatable is adaptive evolution? The role of geographical origin and founder effects in laboratory adaptation. Evolution, 62, 1817-1829.
DOI PMID |
| [147] | Singh ND, Criscoe DR, Skolfield S, Kohl KP, Keebaugh ES, Schlenke TA (2015) Fruit flies diversify their offspring in response to parasite infection. Science, 349, 747-750. |
| [148] | Sizer IW (1943) Effects of temperature on enzyme kinetics. In: Advances in Enzymology and Related Areas of Molecular Biology (eds Nord FF, Werkman CH), pp. 35-62. Interscience Publishers, New York. |
| [149] |
Slowinski SP, Cho J, Penley MJ, Alexander LW, Greenberg AB, Namburar SR, Morran LT (2023) High parasite virulence necessary for the maintenance of host outcrossing via parasite-mediated selection. Evolution Letters, 7, 371-378.
DOI PMID |
| [150] | Soros L, Stanley K (2014) Identifying necessary conditions for open-ended evolution through the artificial life world of chromaria. Artificial Life Conference Proceedings, 26, 793-800. |
| [151] | Sousa T, Domingos T, Kooijman SALM (2008) From empirical patterns to theory: A formal metabolic theory of life. Philosophical Transactions of the Royal Society B: Biological Sciences, 363, 2453-2464. |
| [152] | Souza V, Turner PE, Lenski RE (1997) Long-term experimental evolution in Escherichia coli. V. Effects of recombination with immigrant genotypes on the rate of bacterial evolution. Journal of Evolutionary Biology, 10, 743-769. |
| [153] | Sun P, Shang Y, Sun R, Tian Y, Heino M (2022) The effects of selective harvest on Japanese Spanish mackerel (Scomberomorus niphonius) phenotypic evolution. Frontiers in Ecology and Evolution, 10, 1-18. |
| [154] |
Thomas MK, Kremer CT, Klausmeier CA, Litchman E (2012) A global pattern of thermal adaptation in marine phytoplankton. Science, 338, 1085-1088.
DOI PMID |
| [155] | Tilman D (1977) Resource competition between plankton algae: An experimental and theoretical approach. Ecology, 58, 338-348. |
| [156] | Tilman D (1982) Resource Competition and Community Structure. Princeton University Press, Princeton, NJ. |
| [157] |
Travisano M, Mongold JA, Bennett AF, Lenski RE (1995) Experimental tests of the roles of adaptation, chance, and history in evolution. Science, 267, 87-90.
PMID |
| [158] | Trindade S, Perfeito L, Gordo I (2010) Rate and effects of spontaneous mutations that affect fitness in mutator Escherichia coli. Philosophical Transactions of the Royal Society B: Biological Sciences, 365, 1177-1186. |
| [159] | Tusso S, Nieuwenhuis BPS, Weissensteiner B, Immler S, Wolf JBW (2021) Experimental evolution of adaptive divergence under varying degrees of gene flow. Nature Ecology & Evolution, 5, 338-349. |
| [160] |
Tyler B, Loomis WF Jr, Magasanik B (1967) Transient repression of the lac operon. Journal of Bacteriology, 94, 2001-2011.
DOI PMID |
| [161] |
Uesugi A, Kessler A (2013) Herbivore exclusion drives the evolution of plant competitiveness via increased allelopathy. New Phytologist, 198, 916-924.
DOI PMID |
| [162] | van Valen LM (1973) A new evolutionary law. Evolutionary Theory, 1, 1-30. |
| [163] |
Venail PA, Kaltz O, Olivieri I, Pommier T, Mouquet N (2011) Diversification in temporally heterogeneous environments: Effect of the grain in experimental bacterial populations. Journal of Evolutionary Biology, 24, 2485-2495.
DOI PMID |
| [164] | Venail PA, MacLean RC, Bouvier T, Brockhurst MA, Hochberg ME, Mouquet N (2008) Diversity and productivity peak at intermediate dispersal rate in evolving metacommunities. Nature, 452, 210-214. |
| [165] |
Vostinar AE, Ofria C (2019) Spatial structure can decrease symbiotic cooperation. Artificial Life, 24, 229-249.
DOI |
| [166] | Wade MJ (2016) Adaptation in Metapopulations. University of Chicago Press, Chicago and London. |
| [167] |
Wagner GP, Zhang J (2011) The pleiotropic structure of the genotype-phenotype map: The evolvability of complex organisms. Nature Reviews Genetics, 12, 204-213.
DOI PMID |
| [168] | Waldvogel AM, Pfenninger M (2021) Temperature dependence of spontaneous mutation rates. Genome Research, 31, 1582-1589. |
| [169] |
Weese DJ, Heath KD, Dentinger BT, Lau JA (2015) Long-term nitrogen addition causes the evolution of less-cooperative mutualists. Evolution, 69, 631-642.
DOI PMID |
| [170] |
Weinig C, Johnston JA, Willis CG, Maloof JN (2007) Antagonistic multilevel selection on size and architecture in variable density settings. Evolution, 61, 58-67.
PMID |
| [171] |
Whitehead DJ, Wilke CO, Vernazobres D, Bornberg-Bauer E (2008) The look-ahead effect of phenotypic mutations. Biology Direct, 3, 18.
DOI PMID |
| [172] | Wilke CO, Adami C (2002) The biology of digital organisms. Trends in Ecology & Evolution, 17, 528-532. |
| [173] |
Wolf JBW, Künstner A, Nam K, Jakobsson M, Ellegren H (2009) Nonlinear dynamics of nonsynonymous (dN) and synonymous (dS) substitution rates affects inference of selection. Genome Biology and Evolution, 1, 308-319.
DOI PMID |
| [174] |
Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR (2002) Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nature Genetics, 32, 569-577.
DOI PMID |
| [175] | Wright S (1932) The roles of mutation, inbreeding, crossbreeding, and selection in evolution, pp.355-366. Brooklyn Botanic Garden, Menasha, WI. |
| [176] | Wünsche A, Dinh DM, Satterwhite RS, Arenas CD, Stoebel DM, Cooper TF (2017) Diminishing-returns epistasis decreases adaptability along an evolutionary trajectory. Nature Ecology & Evolution, 1, 0061. |
| [177] | Xie VC, Pu J, Metzger BPH, Thornton JW, Dickinson BC (2021) Contingency and chance erase necessity in the experimental evolution of ancestral proteins. eLife, 10, e67336. |
| [178] |
Yedid G, Ofria CA, Lenski RE (2008) Historical and contingent factors affect re-evolution of a complex feature lost during mass extinction in communities of digital organisms. Journal of Evolutionary Biology, 21, 1335-1357.
DOI PMID |
| [179] |
Yoshida T, Ellner SP, Jones LE, Bohannan BJM, Lenski RE, Hairston NG Jr (2007) Cryptic population dynamics: Rapid evolution masks trophic interactions. PLoS Biology, 5, e235.
DOI PMID |
| [180] | Yvon-Durocher G, Caffrey JM, Cescatti A, Dossena M, del Giorgio P, Gasol JM, Montoya JM, Pumpanen J, Staehr PA, Trimmer M, Woodward G, Allen AP (2012) Reconciling the temperature dependence of respiration across timescales and ecosystem types. Nature, 487, 472-476. |
| [181] |
Zhang C, Jansen M, de Meester L, Stoks R (2019) Rapid evolution in response to warming does not affect the toxicity of a pollutant: Insights from experimental evolution in heated mesocosms. Evolutionary Applications, 12, 977-988.
DOI PMID |
| [182] | Zhang F, Cheng W (2022) The mechanism of bacterial resistance and potential bacteriostatic strategies. Antibiotics, 11, 1-23. |
| [183] | Zhang QG, Lu HS, Buckling A (2018) Temperature drives diversification in a model adaptive radiation. Proceedings of the Royal Society B: Biological Sciences, 285, 20181515. |
| [184] | Zhao Y, Shu M, Zhang L, Zhong C, Liao N, Wu G (2024) Phage-driven coevolution reveals trade-off between antibiotic and phage resistance in Salmonella anatum. ISME Communications, 4, ycae039. |
| [185] |
Zheng J, Payne JL, Wagner A (2019) Cryptic genetic variation accelerates evolution by opening access to diverse adaptive peaks. Science, 365, 347-353.
DOI PMID |
| [1] | 褚晓琳, 张全国. 演化速率假说的实验验证研究进展[J]. 生物多样性, 2025, 33(4): 25019-. |
| [2] | 宋威, 程才, 王嘉伟, 吴纪华. 土壤微生物对植物多样性–生态系统功能关系的调控作用[J]. 生物多样性, 2025, 33(4): 24579-. |
| [3] | 刘淑琪, 崔东, 江智诚, 刘江慧, 闫江超. 短期氮、水添加和刈割减弱了苦豆子型退化草地土壤生物多样性与生态系统多功能性的联系[J]. 生物多样性, 2025, 33(3): 24305-. |
| [4] | 刘源, 杜剑卿, 马丽媛, 杨刚, 田建卿. 纳木措流域岸边带湿地产甲烷古菌群落多样性与分布特征[J]. 生物多样性, 2025, 33(1): 24247-. |
| [5] | 连佳丽, 陈婧, 杨雪琴, 赵莹, 罗叙, 韩翠, 赵雅欣, 李建平. 荒漠草原植物多样性和微生物多样性对降水变化的响应[J]. 生物多样性, 2024, 32(6): 24044-. |
| [6] | 董云伟, 鲍梦幻, 程娇, 陈义永, 杜建国, 高养春, 胡利莎, 李心诚, 刘春龙, 秦耿, 孙进, 王信, 杨光, 张崇良, 张雄, 张宇洋, 张志新, 战爱斌, 贺强, 孙军, 陈彬, 沙忠利, 林强. 中国海洋生物地理学研究进展和热点: 物种分布模型及其应用[J]. 生物多样性, 2024, 32(5): 23453-. |
| [7] | 郝操, 吴东辉, 莫凌梓, 徐国良. 越冬动物肠道微生物多样性及功能研究进展[J]. 生物多样性, 2024, 32(3): 23407-. |
| [8] | 赵榕江, 吴纪华, 何维明, 赵彩云, 周波, 李博, 杨强. 土壤生物多样性与外来植物入侵: 进展与展望[J]. 生物多样性, 2024, 32(11): 24243-. |
| [9] | 韩丽霞, 王永健, 刘宣. 外来物种入侵与本土物种分布区扩张的异同[J]. 生物多样性, 2024, 32(1): 23396-. |
| [10] | 牛永杰, 马全会, 朱玉, 刘海荣, 吕佳乐, 邹元春, 姜明. 氮沉降对草地昆虫多样性影响的研究进展[J]. 生物多样性, 2023, 31(9): 23130-. |
| [11] | 罗正明, 刘晋仙, 张变华, 周妍英, 郝爱华, 杨凯, 柴宝峰. 不同退化阶段亚高山草甸土壤原生生物群落多样性特征及驱动因素[J]. 生物多样性, 2023, 31(8): 23136-. |
| [12] | 朱晓华, 高程, 王聪, 赵鹏. 尿素对土壤细菌与真菌多样性影响的研究进展[J]. 生物多样性, 2023, 31(6): 22636-. |
| [13] | 曾青, 熊超, 尹梅, 葛安辉, 韩丽丽, 张丽梅. 植物微生物组生态功能与群落构建过程研究进展[J]. 生物多样性, 2023, 31(4): 22667-. |
| [14] | 肖媛媛, 冯薇, 乔艳桂, 张宇清, 秦树高. 固沙灌木林地土壤微生物群落特征对土壤多功能性的影响[J]. 生物多样性, 2023, 31(4): 22585-. |
| [15] | 沈诗韵, 潘远飞, 陈丽茹, 土艳丽, 潘晓云. 喜旱莲子草原产地和入侵地种群的植物-土壤反馈差异[J]. 生物多样性, 2023, 31(3): 22436-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn