



生物多样性 ›› 2023, Vol. 31 ›› Issue (4): 22667. DOI: 10.17520/biods.2022667 cstr: 32101.14.biods.2022667
曾青1,2, 熊超1,3, 尹梅4, 葛安辉1,5, 韩丽丽1, 张丽梅1,2,*( )
)
收稿日期:2022-12-02
接受日期:2023-01-28
出版日期:2023-04-20
发布日期:2023-04-20
通讯作者:
*E-mail: 基金资助:
Qing Zeng1,2, Chao Xiong1,3, Mei Yin4, Anhui Ge1,5, Lili Han1, Limei Zhang1,2,*( )
)
Received:2022-12-02
Accepted:2023-01-28
Online:2023-04-20
Published:2023-04-20
Contact:
*E-mail: 摘要:
植物各个器官表面及内部定殖着高度多样化的微生物群落, 这些微生物与植物长期共进化, 作为宿主植物的“共生功能体” (holobiont)在植物生长发育、养分吸收、病害抵御和环境胁迫适应性等方面发挥了重要作用。得益于近10年来多组学技术的发展和应用, 有关植物微生物群落的多样性、组成和功能特征、群落构建的驱动因素和植物-微生物互作机制等方面研究取得了一系列重要进展。然而, 与土壤微生物组相比, 目前对植物微生物组的认识及其应用尚且不足。本文系统总结了植物微生物组的组成特征, 植物微生物在调节植物生长发育、促进养分吸收、提高病害抵御能力及环境胁迫适应性等方面的功能及作用机制, 从宿主选择、环境因子以及生物互作3个方面总结了驱动植物微生物群落构建的因素, 并着重阐述了植物-微生物互作如何塑造植物微生物群落以及如何调节对植物的有益功能。此外, 我们对未来植物微生物组研究和应用面临的挑战进行了展望, 如核心微生物组挖掘和合成群落构建, 植物-微生物互作的分子调控机制, 植物微生物群落水平上的互作机制等。深入理解植物微生物群落特征、生态功能以及构建过程对于精准调控植物微生物组以提高植物适应性和生产力以及维持生态系统健康具有重要意义。
曾青, 熊超, 尹梅, 葛安辉, 韩丽丽, 张丽梅 (2023) 植物微生物组生态功能与群落构建过程研究进展. 生物多样性, 31, 22667. DOI: 10.17520/biods.2022667.
Qing Zeng, Chao Xiong, Mei Yin, Anhui Ge, Lili Han, Limei Zhang (2023) Research progress on ecological functions and community assembly of plant microbiomes. Biodiversity Science, 31, 22667. DOI: 10.17520/biods.2022667.
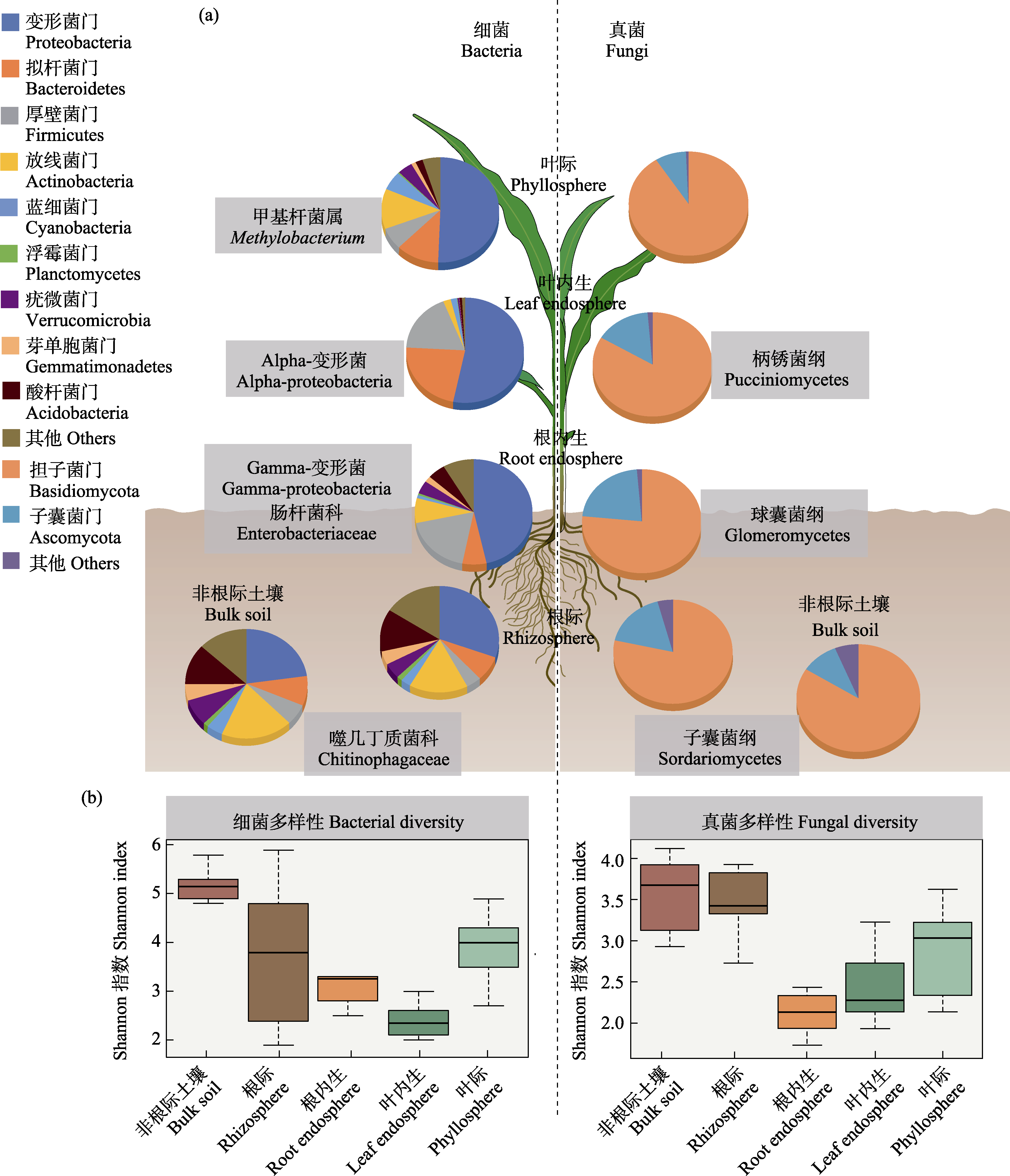
图1 植物不同部位生态位细菌和真菌群落组成(a)和多样性(b)特征。改自Trivedi等(2020)和Xiong等(2021a, b, c)基于玉米、小麦、大麦等植物微生物组研究数据。图a方框中的细菌和真菌类群代表不同部位生态位的指示类群。
Fig. 1 The patterns of plant-associated bacterial and fungal community compositions (a) and diversities (b) across multiple plant compartment niches. Modified from Trivedi et al (2020) and Xiong et al (2021a, b, c) based on data from maize, wheat and barley related studies. The bacterial and fungal taxa in the boxes in Fig. 1a represent biomaker taxa for different compartment niches.
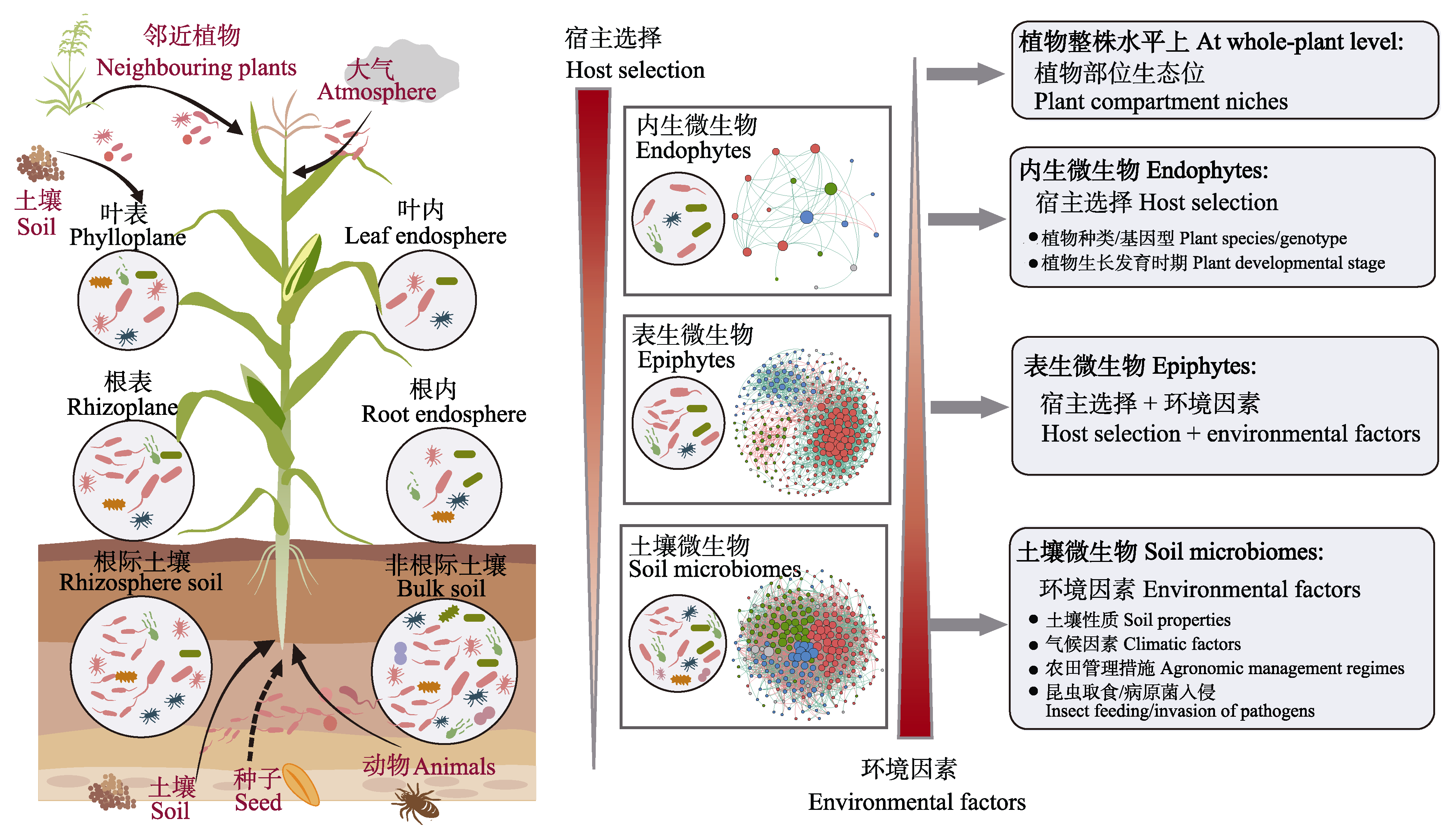
图2 植物微生物组来源以及宿主选择和环境因素共同驱动的土壤-植物连续体微生物群落构建模型
Fig. 2 Sources of plant microbiomes and a conceptual model for microbial community assembly on the soil-plant continuum driven by the interactive effect of host selection and environmental effect
| [1] |
Abdelfattah A, Wisniewski M, Schena L, Tack AJM (2021) Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environmental Microbiology, 23, 2199-2214.
DOI PMID |
| [2] |
Acuña-Rodríguez IS, Newsham KK, Gundel PE, Torres-Díaz C, Molina-Montenegro MA (2020) Functional roles of microbial symbionts in plant cold tolerance. Ecology Letters, 23, 1034-1048.
DOI PMID |
| [3] | Almario J, Jeena G, Wunder J, Langen G, Zuccaro A, Coupland G, Bucher M (2017) Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proceedings of the National Academy of Sciences, USA, 114, E9403-E9412. |
| [4] |
Aydogan EL, Moser G, Müller C, Kämpfer P, Glaeser SP (2018) Long-term warming shifts the composition of bacterial communities in the phyllosphere of Galium album in a permanent grassland field-experiment. Frontiers in Microbiology, 9, 144.
DOI PMID |
| [5] |
Bai B, Liu WD, Qiu XY, Zhang J, Zhang JY, Bai Y (2022) The root microbiome: Community assembly and its contributions to plant fitness. Journal of Integrative Plant Biology, 64, 230-243.
DOI |
| [6] | Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, Hüttel B, McHardy AC, Vorholt JA, Schulze-Lefert P (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature, 528, 364-369. |
| [7] |
Banerjee S, Walder F, Büchi L, Meyer M, Held AY, Gattinger A, Keller T, Charles R, van der Heijden MGA, (2019) Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. The ISME Journal, 13, 1722-1736.
DOI |
| [8] |
Belimov AA, Hontzeas N, Safronova VI, Demchinskaya SV, Piluzza G, Bullitta S, Glick BR (2005) Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biology and Biochemistry, 37, 241-250.
DOI URL |
| [9] |
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends in Plant Science, 17, 478-486.
DOI PMID |
| [10] |
Berg G, Raaijmakers JM (2018) Saving seed microbiomes. The ISME Journal, 12, 1167-1170.
DOI |
| [11] |
Boller T, Felix G (2009) A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annual Review of Plant Biology, 60, 379-406.
DOI PMID |
| [12] |
Brunel C, Pouteau R, Dawson W, Pester M, Ramirez KS, van Kleunen M (2020) Towards unraveling macroecological patterns in rhizosphere microbiomes. Trends in Plant Science, 25, 1017-1029.
DOI PMID |
| [13] | Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host & Microbe, 17, 392-403. |
| [14] |
Bulgari D, Casati P, Quaglino F, Bianco PA (2014) Endophytic bacterial community of grapevine leaves influenced by sampling date and phytoplasma infection process. BMC Microbiology, 14, 1-11.
DOI |
| [15] |
Burch AY, Zeisler V, Yokota K, Schreiber L, Lindow SE (2014) The hygroscopic biosurfactant syringafactin produced by Pseudomonas syringae enhances fitness on leaf surfaces during fluctuating humidity. Environmental Microbiology, 16, 2086-2098.
DOI URL |
| [16] |
Carrión VJ, Perez-Jaramillo J, Cordovez V, Tracanna V, de Hollander M, Ruiz-Buck D, Mendes LW, Gomez-Exposito R, Elsayed SS, Mohanraju P, Arifah A, van der Oost J, Paulson JN, Mendes R, van Wezel GP, Medema MH, Raaijmakers JM (2019) Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science, 366, 606-612.
DOI PMID |
| [17] |
Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM (2015) Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Molecular Plant-Microbe Interactions, 28, 1049-1058.
DOI PMID |
| [18] |
Ceja-Navarro JA, Wang Y, Ning DL, Arellano A, Ramanculova L, Yuan MM, Byer A, Craven KD, Saha MC, Brodie EL, Pett-Ridge J, Firestone MK (2021) Protist diversity and community complexity in the rhizosphere of switchgrass are dynamic as plants develop. Microbiome, 9, 96.
DOI PMID |
| [19] |
Chang WS, van de Mortel M, Nielsen L, Nino de Guzman G, Li XH, Halverson LJ (2007) Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. Journal of Bacteriology, 189, 8290-8299.
DOI URL |
| [20] |
Chaparro JM, Badri DV, Vivanco JM (2014) Rhizosphere microbiome assemblage is affected by plant development. The ISME Journal, 8, 790-803.
DOI |
| [21] |
Chen SM, Waghmode TR, Sun RB, Kuramae EE, Hu CS, Liu BB (2019) Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome, 7, 136.
DOI PMID |
| [22] |
Chen Y, Wang J, Yang N, Wen ZY, Sun XP, Chai YR, Ma ZH (2018) Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nature Communications, 9, 3429.
DOI PMID |
| [23] |
Copeland JK, Yuan LJ, Layeghifard M, Wang PW, Guttman DS (2015) Seasonal community succession of the phyllosphere microbiome. Molecular Plant-Microbe Interactions, 28, 274-285.
DOI PMID |
| [24] |
Cordovez V, Dini-Andreote F, Carrión VJ, Raaijmakers JM (2019) Ecology and evolution of plant microbiomes. Annual Review of Microbiology, 73, 69-88.
DOI PMID |
| [25] |
de Vries FT, Griffiths RI, Knight CG, Nicolitch O, Williams A (2020) Harnessing rhizosphere microbiomes for drought- resilient crop production. Science, 368, 270-274.
DOI URL |
| [26] | Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-gonzález A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N (2018) A global atlas of the dominant bacteria found in soil. Science, 325, 320-325. |
| [27] |
Dini-Andreote F (2020) Endophytes: The second layer of plant defense. Trends in Plant Science, 25, 319-322.
DOI PMID |
| [28] | Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF (2015) Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proceedings of the National Academy of Sciences, USA, 112, E1326-E1332. |
| [29] |
Dodd IC, Pérez-Alfocea F (2012) Microbial amelioration of crop salinity stress. Journal of Experimental Botany, 63, 3415-3428.
DOI PMID |
| [30] |
Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, Hacquard S (2018) Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell, 175, 973-983.
DOI URL |
| [31] | Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V, Jeffery LD (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proceedings of the National Academy of Sciences, USA, 112, E911-E920. |
| [32] |
Edwards JA, Santos-Medellín CM, Liechty ZS, Nguyen B, Lurie E, Eason S, Phillips G, Sundaresan V (2018) Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biology, 16, e2003862.
DOI URL |
| [33] |
Egidi E, Delgado-Baquerizo M, Plett JM, Wang JT, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK (2019) A few Ascomycota taxa dominate soil fungal communities worldwide. Nature Communications, 10, 2369.
DOI PMID |
| [34] |
Finkel OM, Salas-González I, Castrillo G, Conway JM, Law TF, Teixeira PJPL, Wilson ED, Fitzpatrick CR, Jones CD, Dangl JL (2020) A single bacterial genus maintains root growth in a complex microbiome. Nature, 587, 103-108.
DOI |
| [35] |
Gao C, Montoya L, Xu L, Madera M, Hollingsworth J, Purdom E, Singan V, Vogel J, Hutmacher RB, Dahlberg JA, Coleman-Derr D, Lemaux PG, Taylor JW (2020) Fungal community assembly in drought-stressed sorghum shows stochasticity, selection, and universal ecological dynamics. Nature Communications, 11, 34.
DOI PMID |
| [36] |
Gao M, Xiong C, Gao C, Tsui CKM, Wang MM, Zhou X, Zhang AM, Cai L (2021) Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome, 9, 187.
DOI PMID |
| [37] |
Ge AH, Liang ZH, Han LL, Xiao JL, Zhang Y, Zeng Q, Xiang JF, Xiong C, Zhang LM (2022) Rootstock rescues watermelon from Fusarium wilt disease by shaping protective root-associated microbiomes and metabolites in continuous cropping soils. Plant and Soil, 479, 423-442.
DOI |
| [38] |
Ge AH, Liang ZH, Xiao JL, Zhang Y, Zeng Q, Xiong C, Han LL, Wang JT, Zhang LM (2021) Microbial assembly and association network in watermelon rhizosphere after soil fumigation for Fusarium wilt control. Agriculture, Ecosystems & Environment, 312, 107336.
DOI URL |
| [39] |
Gebauer L, Bouffaud ML, Ganther M, Yim B, Vetterlein D, Smalla K, Buscot F, Heintz-Buschart A, Tarkka MT (2021) Soil texture, sampling depth and root hairs shape the structure of ACC deaminase bacterial community composition in maize rhizosphere. Frontiers in Microbiology, 12, 616828.
DOI URL |
| [40] |
Geisen S, Mitchell EAD, Adl S, Bonkowski M, Dunthorn M, Ekelund F, Fernández LD, Jousset A, Krashevska V, Singer D, Spiegel FW, Walochnik J, Lara E (2018) Soil protists: A fertile frontier in soil biology research. FEMS Microbiology Reviews, 42, 293-323.
DOI PMID |
| [41] |
Gong TY, Xin XF (2021) Phyllosphere microbiota: Community dynamics and its interaction with plant hosts. Journal of Integrative Plant Biology, 63, 297-304.
DOI |
| [42] |
Gu SH, Wei Z, Shao ZY, Friman VP, Cao KH, Yang TJ, Kramer J, Wang XF, Li M, Mei XL, Xu YC, Shen QR, Kümmerli R, Jousset A (2020) Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nature Microbiology, 5, 1002-1010.
DOI PMID |
| [43] |
Guo S, Tao CY, Jousset A, Xiong W, Wang Z, Shen ZZ, Wang BB, Xu ZH, Gao ZL, Liu SS, Li R, Ruan YZ, Shen QR, Kowalchuk GA, Geisen S (2022) Trophic interactions between predatory protists and pathogen-suppressive bacteria impact plant health. The ISME Journal, 16, 1932-1943.
DOI |
| [44] |
Hamonts K, Trivedi P, Garg A, Janitz C, Grinyer J, Holford P, Botha FC, Anderson IC, Singh BK (2018) Field study reveals core plant microbiota and relative importance of their drivers. Environmental Microbiology, 20, 124-140.
DOI PMID |
| [45] |
Han GZ (2019) Origin and evolution of the plant immune system. New Phytologist, 222, 70-83.
DOI URL |
| [46] |
Hannula SE, Boschker HTS, de Boer W, van Veen JA (2012) 13C pulse-labeling assessment of the community structure of active fungi in the rhizosphere of a genetically starch-modified potato (Solanum tuberosum) cultivar and its parental isoline. New Phytologist, 194, 784-799.
DOI PMID |
| [47] | Harbort CJ, Hashimoto M, Inoue H, Niu YL, Guan R, Rombolà AD, Kopriva S, Voges MJEEE, Sattely ES, Garrido-Oter R, Schulze-Lefert P (2020) Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host & Microbe, 28, 825-837. |
| [48] |
Hardoim PR, van Overbeek LS, Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends in Microbiology, 16, 463-471.
DOI PMID |
| [49] |
Hassan S, Mathesius U (2012) The role of flavonoids in root-rhizosphere signalling: Opportunities and challenges for improving plant-microbe interactions. Journal of Experimental Botany, 63, 3429-3444.
DOI PMID |
| [50] |
Hassani MA, Durán P, Hacquard S (2018) Microbial interactions within the plant holobiont. Microbiome, 6, 58.
DOI PMID |
| [51] |
Hassani MA, Özkurt E, Franzenburg S, Stukenbrock EH (2020) Ecological assembly processes of the bacterial and fungal microbiota of wild and domesticated wheat species. Phytobiomes Journal, 4, 217-224.
DOI URL |
| [52] |
He XQ, Zhang Q, Li BB, Jin Y, Jiang LB, Wu RL (2021) Network mapping of root-microbe interactions in Arabidopsis thaliana. Npj Biofilms and Microbiomes, 7, 72.
DOI |
| [53] |
Helfer S (2014) Rust fungi and global change. New Phytologist, 201, 770-780.
PMID |
| [54] |
Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O’Connell RJ, Schulze-Lefert P (2016) Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell, 165, 464-474.
DOI PMID |
| [55] |
Hu LF, Robert CAM, Cadot S, Zhang X, Ye M, Li BB, Manzo D, Chervet N, Steinger T, van der Heijden MGA, Schlaeppi K, Erb M (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nature Communications, 9, 2738.
DOI PMID |
| [56] | Humphrey PT, Whiteman NK (2020) Insect herbivory reshapes a native leaf microbiome. Nature Ecology & Evolution, 4, 221-229. |
| [57] | Hussain M, Hamid MI, Tian JQ, Hu JY, Zhang XL, Chen JS, Xiang MC, Liu XZ (2018) Bacterial community assemblages in the rhizosphere soil, root endosphere and cyst of soybean cyst nematode-suppressive soil challenged with nematodes. FEMS Microbiology Ecology, 94, fiy142. |
| [58] |
James MG, Robertson DS, Myers AM (1995) Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell, 7, 417-429.
DOI PMID |
| [59] |
Jia SH, Wang XG, Yuan ZQ, Lin F, Ye J, Lin GG, Hao ZQ, Bagchi R (2020) Tree species traits affect which natural enemies drive the Janzen-Connell effect in a temperate forest. Nature Communications, 11, 286.
DOI PMID |
| [60] |
Ke XB, Lu YH, Conrad R (2014) Different behaviour of methanogenic Archaea and Thaumarchaeota in rice field microcosms. FEMS Microbiology Ecology, 87, 18-29.
DOI PMID |
| [61] | Kembel SW, O’Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL (2014) Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proceedings of the National Academy of Sciences, USA, 111, 13715-13720. |
| [62] |
Kim H, Lee KK, Jeon J, Harris WA, Lee YH (2020) Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome, 8, 20.
DOI URL |
| [63] |
Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. The ISME Journal, 6, 1378-1390.
DOI |
| [64] |
Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ, Jung EJ, Park H, Roy N, Kim H, Lee MM, Rubin EM, Lee SW, Kim JF (2018) Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nature Biotechnology, 36, 1100-1116.
DOI URL |
| [65] |
Laforest-Lapointe I, Messier C, Kembel SW (2016) Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome, 4, 27.
DOI PMID |
| [66] |
Leach JE, Triplett LR, Argueso CT, Trivedi P (2017) Communication in the phytobiome. Cell, 169, 587-596.
DOI PMID |
| [67] |
Lee SM, Kong HG, Song GC, Ryu CM (2021) Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. The ISME Journal, 15, 330-347.
DOI |
| [68] |
Levy A, Gonzalez IS, Mittelviefhaus M, Clingenpeel S, Paredes SH, Miao JM, Wang KR, Devescovi G, Stillman K, Monteiro F, Alvarez BR, Lundberg DS, Lu TY, Lebeis S, Jin Z, McDonald M, Klein AP, Feltcher ME, Rio TG, Grant SR, Doty SL, Ley RE, Zhao BY, Venturi V, Pelletier DA, Vorholt JA, Tringe SG, Woyke T, Dangl JL (2018) Genomic features of bacterial adaptation to plants. Nature Genetics, 50, 138-150.
DOI |
| [69] |
Li PD, Zhu ZR, Zhang YZ, Xu JP, Wang HK, Wang ZY, Li HY (2022a) The phyllosphere microbiome shifts toward combating melanose pathogen. Microbiome, 10, 56.
DOI URL |
| [70] |
Li YB, Yang R, Häggblom MM, Li MY, Guo LF, Li BQ, Kolton M, Cao ZG, Soleimani M, Chen Z, Xu ZM, Gao WL, Yan B, Sun WM (2022b) Characterization of diazotrophic root endophytes in Chinese silvergrass (Miscanthus sinensis). Microbiome, 10, 186.
DOI |
| [71] |
Li ZF, Bai XL, Jiao S, Li YM, Li PR, Yang Y, Zhang H, Wei GH (2021) A simplified synthetic community rescues Astragalus mongholicus from root rot disease by activating plant-induced systemic resistance. Microbiome, 9, 217.
DOI URL |
| [72] |
Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Applied and Environmental Microbiology, 69, 1875-1883.
DOI PMID |
| [73] |
Liu HW, Brettell LE, Singh B (2020) Linking the phyllosphere microbiome to plant health. Trends in Plant Science, 25, 841-844.
DOI PMID |
| [74] |
Liu HW, Li JY, Carvalhais LC, Percy CD, Prakash Verma J, Schenk PM, Singh BK (2021) Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens. New Phytologist, 229, 2873-2885.
DOI URL |
| [75] |
Liu X, Liu M, Xiao Y (2023) The effect of foliar fungal pathogens on plant species coexistence: Progress and challenge. Biodiversity Science, 31, 22525. (in Chinese with English abstract)
DOI URL |
|
[ 刘向, 刘木, 肖瑶 (2023) 叶片病原真菌对植物物种共存的影响: 进展与挑战. 生物多样性, 31, 22525.]
DOI |
|
| [76] |
Madhaiyan M, Alex THH, Ngoh ST, Prithiviraj B, Ji LH (2015) Leaf-residing Methylobacterium species fix nitrogen and promote biomass and seed production in Jatropha curcas. Biotechnology for Biofuels, 8, 222.
DOI PMID |
| [77] |
Mahdi LK, Miyauchi S, Uhlmann C, Garrido-Oter R, Langen G, Wawra S, Niu YL, Guan R, Robertson-Albertyn S, Bulgarelli D, Parker JE, Zuccaro A (2022) The fungal root endophyte Serendipita vermifera displays inter-Kingdom synergistic beneficial effects with the microbiota in Arabidopsis thaliana and barley. The ISME Journal, 16, 876-889.
DOI |
| [78] |
Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, Tsai SM (2018) Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. The ISME Journal, 12, 212-224.
DOI URL |
| [79] |
Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiology Reviews, 37, 634-663.
DOI PMID |
| [80] |
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science, 332, 1097-1100.
DOI PMID |
| [81] |
Moissl-Eichinger C, Pausan M, Taffner J, Berg G, Bang C, Schmitz RA (2018) Archaea are interactive components of complex microbiomes. Trends in Microbiology, 26, 70-85.
DOI PMID |
| [82] |
Morella NM, Gomez AL, Wang G, Leung MS, Koskella B (2018) The impact of bacteriophages on phyllosphere bacterial abundance and composition. Molecular Ecology, 27, 2025-2038.
DOI PMID |
| [83] | Morella NM, Weng FCH, Joubert PM, Metcalf CJE, Lindow S, Koskella B (2020) Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proceedings of the National Academy of Sciences, USA, 117, 1148-1159. |
| [84] | Müller DB, Vogel C, Bai Y, Vorholt JA (2016) The plant microbiota: Systems-level insights and perspectives. Annual Review of Genetics, 50, 211-234. |
| [85] |
Müller H, Berg C, Landa BB, Auerbach A, Moissl-Eichinger C, Berg G (2015) Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Frontiers in Microbiology, 6, 138.
DOI PMID |
| [86] |
Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S (2013) Patterns and processes of microbial community assembly. Microbiology and Molecular Biology Reviews, 77, 342-356.
DOI PMID |
| [87] | Niu B, Paulson JN, Zheng XQ, Kolter R (2017) Simplified and representative bacterial community of maize roots. Proceedings of the National Academy of Sciences, USA, 114, E2450-E2459. |
| [88] | Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proceedings of the National Academy of Sciences, USA, 110, 6548-6553. |
| [89] |
Pérez-Jaramillo JE, Carrión VJ, Bosse M, Ferrão LFV, de Hollander M, Garcia AAF, Ramírez CA, Mendes R, Raaijmakers JM (2017) Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. The ISME Journal, 11, 2244-2257.
DOI URL |
| [90] |
Pérez-Jaramillo JE, Carrión VJ, de Hollander M, Raaijmakers JM (2018) The wild side of plant microbiomes. Microbiome, 6, 143.
DOI PMID |
| [91] |
Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH (2013) Going back to the roots: The microbial ecology of the rhizosphere. Nature Reviews Microbiology, 11, 789-799.
DOI PMID |
| [92] |
Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM (2014) Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology, 52, 347-375.
DOI PMID |
| [93] |
Porter SS, Sachs JL (2020) Agriculture and the disruption of plant-microbial symbiosis. Trends in Ecology and Evolution, 35, 426-439.
DOI PMID |
| [94] |
Pratama AA, van Elsas JD (2018) The ‘neglected’ soil virome—Potential role and impact. Trends in Microbiology, 26, 649-662.
DOI PMID |
| [95] |
Qin Y, Druzhinina IS, Pan XY, Yuan ZL (2016) Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnology Advances, 34, 1245-1259.
DOI PMID |
| [96] |
Raaijmakers JM, Kiers ET (2022) Rewilding plant microbiomes. Science, 378, 599-600.
DOI PMID |
| [97] |
Rastogi G, Coaker GL, Leveau JHJ (2013) New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiology Letters, 348, 1-10.
DOI PMID |
| [98] |
Ravanbakhsh M, Kowalchuk GA, Jousset A (2019) Root-associated microorganisms reprogram plant life history along the growth-stress resistance tradeoff. The ISME Journal, 13, 3093-3101.
DOI |
| [99] |
Remus-Emsermann MNP, Schlechter RO (2018) Phyllosphere microbiology: At the interface between microbial individuals and the plant host. New Phytologist, 218, 1327-1333.
DOI PMID |
| [100] |
Remus-Emsermann MNP, Vorholt JA (2014) Complexities of microbial life on leaf surfaces. Microbe Magazine, 9, 448-452.
DOI URL |
| [101] |
Richardson AE, Barea JM, McNeill AM, Prigent-Combaret C (2009) Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant and Soil, 321, 305-339.
DOI URL |
| [102] |
Rudrappa T, Czymmek KJ, Paré PW, Bais HP (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiology, 148, 1547-1556.
DOI PMID |
| [103] |
Sánchez-Cañizares C, Jorrín B, Poole PS, Tkacz A (2017) Understanding the holobiont: The interdependence of plants and their microbiome. Current Opinion in Microbiology, 38, 188-196.
DOI PMID |
| [104] |
Santos-Medellín C, Liechty Z, Edwards J, Nguyen B, Huang BH, Weimer BC, Sundaresan V (2021) Prolonged drought imparts lasting compositional changes to the rice root microbiome. Nature Plants, 7, 1065-1077.
DOI PMID |
| [105] |
Sapkota R, Knorr K, Jørgensen LN, O’Hanlon KA, Nicolaisen M (2015) Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytologist, 207, 1134-1144.
DOI PMID |
| [106] |
Sapp M, Ploch S, Fiore-Donno AM, Bonkowski M, Rose LE (2018) Protists are an integral part of the Arabidopsis thaliana microbiome. Environmental Microbiology, 20, 30-43.
DOI URL |
| [107] |
Sarkar D, Rovenich H, Jeena G, Nizam S, Tissier A, Balcke GU, Mahdi LK, Bonkowski M, Langen G, Zuccaro A (2019) The inconspicuous gatekeeper: Endophytic Serendipita vermifera acts as extended plant protection barrier in the rhizosphere. New Phytologist, 224, 886-901.
DOI URL |
| [108] |
Sasse J, Martinoia E, Northen T (2018) Feed your friends: Do plant exudates shape the root microbiome? Trends in Plant Science, 23, 25-41.
DOI PMID |
| [109] |
Schmitz L, Yan ZC, Schneijderberg M, de Roij M, Pijnenburg R, Zheng Q, Franken C, Dechesne A, Trindade LM, van Velzen R, Bisseling T, Geurts R, Cheng X (2022) Synthetic bacterial community derived from a desert rhizosphere confers salt stress resilience to tomato in the presence of a soil microbiome. The ISME Journal, 16, 1907-1920.
DOI |
| [110] |
Shade A, Jacques MA, Barret M (2017) Ecological patterns of seed microbiome diversity, transmission, and assembly. Current Opinion in Microbiology, 37, 15-22.
DOI PMID |
| [111] |
Sun AQ, Jiao XY, Chen QL, Trivedi P, Li ZX, Li FF, Zheng Y, Lin YX, Hu HW, He JZ (2021) Fertilization alters protistan consumers and parasites in crop-associated microbiomes. Environmental Microbiology, 23, 2169-2183.
DOI PMID |
| [112] |
Sweeney CJ, de Vries FT, van Dongen BE, Bardgett RD (2021) Root traits explain rhizosphere fungal community composition among temperate grassland plant species. New Phytologist, 229, 1492-1507.
DOI URL |
| [113] | Thiergart T, Durán P, Ellis T, Vannier N, Garrido-Oter R, Kemen E, Roux F, Alonso-Blanco C, Ågren J, Schulze- Lefert P, Hacquard S (2020) Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nature Ecology & Evolution, 4, 122-131. |
| [114] |
Tian BN, Xie JT, Fu YP, Cheng JS, Li B, Chen T, Zhao Y, Gao ZX, Yang PY, Barbetti MJ, Tyler BM, Jiang DH (2020) A cosmopolitan fungal pathogen of dicots adopts an endophytic lifestyle on cereal crops and protects them from major fungal diseases. The ISME Journal, 14, 3120-3135.
DOI |
| [115] |
Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, Fukuda S, Ushio M, Nakaoka S, Onoda Y, Yoshida K, Schlaeppi K, Bai Y, Sugiura R, Ichihashi Y, Minamisawa K, Kiers ET (2018) Core microbiomes for sustainable agroecosystems. Nature Plants, 4, 247-257.
DOI PMID |
| [116] |
Topalović O, Heuer H (2019) Plant-nematode interactions assisted by microbes in the rhizosphere. Current Issues in Molecular Biology, 30, 75-88.
DOI PMID |
| [117] |
Trivedi P, Batista BD, Bazany KE, Singh BK (2022) Plant-microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytologist, 234, 1951-1959.
DOI PMID |
| [118] |
Trivedi P, Leach JE, Tringe SG, Sa TM, Singh BK (2020) Plant-microbiome interactions: From community assembly to plant health. Nature Reviews Microbiology, 18, 607-621.
DOI |
| [119] | van der Heijden MGA, Dombrowski N, Schlaeppi K (2017) Continuum of root-fungal symbioses for plant nutrition. Proceedings of the National Academy of Sciences, USA, 114, 11574-11576. |
| [120] |
van der Heijden MGA, Martin FM, Selosse MA, Sanders IR (2015) Mycorrhizal ecology and evolution: The past, the present, and the future. New Phytologist, 205, 1406-1423.
DOI PMID |
| [121] |
van der Wal A, Leveau JHJ (2011) Modelling sugar diffusion across plant leaf cuticles: the effect of free water on substrate availability to phyllosphere bacteria. Environmental Microbiology, 13, 792-797.
DOI PMID |
| [122] |
Van Deynze A, Zamora P, Delaux PM, Heitmann C, Jayaraman D, Rajasekar S, Graham D, Maeda J, Gibson D, Schwartz KD, Berry AM, Bhatnagar S, Jospin G, Darling A, Jeannotte R, Lopez J, Weimer BC, Eisen JA, Shapiro HY, Ané JM, Bennett AB (2018) Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biology, 16, e2006352.
DOI URL |
| [123] | Vandenkoornhuyse P, Mahé S, Ineson P, Staddon P, Ostle N, Cliquet JB, Francez AJ, Fitter AH, Young JPW (2007) Active root-inhabiting microbes identified by rapid incorporation of plant-derived carbon into RNA. Proceedings of the National Academy of Sciences, USA, 104, 16970-16975. |
| [124] |
Vandenkoornhuyse P, Quaiser A, Duhamel M, Van AL, Dufresne A (2015) The importance of the microbiome of the plant holobiont. New Phytologist, 206, 1196-1206.
DOI PMID |
| [125] |
Vellend M (2010) Conceptual synthesis in community ecology. The Quarterly Review of Biology, 85, 183-206.
DOI URL |
| [126] | Voges MJEEE, Bai Y, Schulze-Lefert P, Sattely ES (2019) Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proceedings of the National Academy of Sciences, USA, 116, 12558-12565. |
| [127] |
Vorholt JA (2012) Microbial life in the phyllosphere. Nature Reviews Microbiology, 10, 828-840.
DOI PMID |
| [128] |
Wagner MR, Lundberg DS, Del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T (2016) Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nature Communications, 7, 12151.
DOI PMID |
| [129] | Walters WA, Jin Z, Youngblut N, Wallace JG, Sutter J, Zhang W, González-Peña A, Peiffer J, Koren O, Shi QJ, Knight R, Glavina Del Rio T, Tringe SG, Buckler ES, Dangl JL, Ley RE (2018) Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proceedings of the National Academy of Sciences, USA, 115, 7368-7373. |
| [130] | Wei Z, Song YQ, Xiong W, Xu YC, Shen QR (2021) Soil Protozoa: Research methods and roles in the biocontrol of soil-borne diseases. Acta Pedologica Sinica, 58, 14-22. (in Chinese with English abstract) |
| [ 韦中, 宋宇琦, 熊武, 徐阳春, 沈其荣 (2021) 土壤原生动物——研究方法及其在土传病害防控中的作用. 土壤学报, 58, 14-22.] | |
| [131] |
Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS (2002) Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annual Review of Phytopathology, 40, 309-348.
PMID |
| [132] |
Xiong C, He JZ, Singh BK, Zhu YG, Wang JT, Li PP, Zhang QB, Han LL, Shen JP, Ge AH, Wu CF, Zhang LM (2021a) Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environmental Microbiology, 23, 1907-1924.
DOI URL |
| [133] |
Xiong C, Singh BK, He JZ, Han YL, Li PP, Wan LH, Meng GZ, Liu SY, Wang JT, Wu CF, Ge AH, Zhang LM (2021b) Plant developmental stage drives the differentiation in ecological role of the maize microbiome. Microbiome, 9, 171.
DOI URL |
| [134] | Xiong C, Zhu YG, Wang JT, Singh B, Han LL, Shen JP, Li PP, Wang GB, Wu CF, Ge AH, Zhang LM, He JZ (2021c) Host selection shapes crop microbiome assembly and network complexity. New Phytologist, 229, 1091-1104. |
| [135] |
Xiong W, Song YQ, Yang KM, Gu YA, Wei Z, Kowalchuk GA, Xu YC, Jousset A, Shen QR, Geisen S (2020) Rhizosphere protists are key determinants of plant health. Microbiome, 8, 27.
DOI PMID |
| [136] |
Xu L, Dong ZB, Chiniquy D, Pierroz G, Deng SW, Gao C, Diamond S, Simmons T, Wipf HML, Caddell D, Varoquaux N, Madera MA, Hutmacher R, Deutschbauer A, Dahlberg JA, Guerinot ML, Purdom E, Banfield JF, Taylor JW, Lemaux PG, Coleman-Derr D (2021) Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nature Communications, 12, 3209.
DOI PMID |
| [137] | Xu L, Naylor D, Dong ZB, Simmons T, Pierroz G, Hixson KK, Kim YM, Zink EM, Engbrecht KM, Wang Y, Gao C, DeGraaf S, Madera MA, Sievert JA, Hollingsworth J, Birdseye D, Scheller HV, Hutmacher R, Dahlberg J, Jansson C, Taylor JW, Lemaux PG, Coleman-Derr D (2018) Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proceedings of the National Academy of Sciences, USA, 115, E4284-E4293. |
| [138] |
Yao BM, Zeng Q, Zhang LM (2022) Research progress on the biodiversity and ecological function of soil protists. Biodiversity Science, 30, 22353. (in Chinese with English abstract)
DOI |
|
[ 姚保民, 曾青, 张丽梅 (2022) 土壤原生生物多样性及其生态功能研究进展. 生物多样性, 30, 22353.]
DOI |
|
| [139] |
Yao H, Sun X, He C, Maitra P, Li XC, Guo LD (2019) Phyllosphere epiphytic and endophytic fungal community and network structures differ in a tropical mangrove ecosystem. Microbiome, 7, 57.
DOI PMID |
| [140] |
Yoshida S, Hiradate S, Koitabashi M, Kamo T, Tsushima S (2017) Phyllosphere Methylobacterium bacteria contain UVA-absorbing compounds. Journal of Photochemistry and Photobiology B: Biology, 167, 168-175.
DOI URL |
| [141] |
Yu P, He XM, Baer M, Beirinckx S, Tian T, Moya YAT, Zhang XC, Deichmann M, Frey FP, Bresgen V, Li CJ, Razavi BS, Schaaf G, von Wirén N, Su Z, Bucher M, Tsuda K, Goormachtig S, Chen XP, Hochholdinger F (2021) Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nature Plants, 7, 481-499.
DOI PMID |
| [142] |
Zhang JY, Liu YX, Zhang N, Hu B, Jin T, Xu HR, Qin Y, Yan PX, Zhang XN, Guo XX, Hui J, Cao SY, Wang X, Wang C, Wang H, Qu BY, Fan GY, Yuan LX, Garrido-Oter R, Chu CC, Bai Y (2019) NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nature Biotechnology, 37, 676-684.
DOI PMID |
| [143] | Zhang LM, He JZ (2012) A novel archaeal phylum: Thaumarchaeota—A review. Acta Microbiologica Sinica, 52, 411-421. (in Chinese with English abstract) |
| [ 张丽梅, 贺纪正 (2012) 一个新的古菌类群——奇古菌门(Thaumarchaeota). 微生物学报, 52, 411-421.] | |
| [144] |
Zhang LY, Zhang ML, Huang SY, Li LJ, Gao Q, Wang Y, Zhang SQ, Huang SM, Yuan L, Wen YC, Liu KL, Yu XC, Li DC, Zhang L, Xu XP, Wei HL, He P, Zhou W, Philippot L, Ai C (2022) A highly conserved core bacterial microbiota with nitrogen-fixation capacity inhabits the xylem sap in maize plants. Nature Communications, 13, 3361.
DOI PMID |
| [145] | Zhang RF, Shen QR (2012) Characterization of the microbial flora and management to induce the disease suppressive soil. Journal of Nanjing Agricultural University, 35, 125-132. (in Chinese with English abstract) |
| [ 张瑞福, 沈其荣 (2012) 抑病型土壤的微生物区系特征及调控. 南京农业大学学报, 35, 125-132.] | |
| [146] |
Zhang RF, Vivanco JM, Shen QR (2017) The unseen rhizosphere root-soil-microbe interactions for crop production. Current Opinion in Microbiology, 37, 8-14.
DOI PMID |
| [147] |
Zhang W, Li XG, Sun K, Tang MJ, Xu FJ, Zhang M, Dai CC (2020) Mycelial network-mediated rhizobial dispersal enhances legume nodulation. The ISME Journal, 14, 1015-1029.
DOI |
| [148] | Zhou JZ, Ning DL (2017) Stochastic community assembly: Does it matter in microbial ecology? Microbiology and Molecular Biology Reviews, 81, e00002-17. |
| [149] |
Zhou ZC, Pan J, Wang FP, Gu JD, Li M (2018) Bathyarchaeota: Globally distributed metabolic generalists in anoxic environments. FEMS Microbiology Reviews, 42, 639-655.
DOI PMID |
| [1] | 罗菲, 汪涯, 曾庆桂, 颜日明, 张志斌, 朱笃. 东乡野生稻根际可培养细菌多样性及其植物促生活性分析[J]. 生物多样性, 2011, 19(4): 476-484. |
| [2] | 刘琳, 孙磊, 张瑞英, 姚娜, 李潞滨. 春兰根中可分泌吲哚乙酸的内生细菌多样性[J]. 生物多样性, 2010, 18(2): 182-187. |
| [3] | 陆凡, 郑小波, 陈志谊, 刘永锋, 王法明, 范永坚. 江苏省稻瘟病菌的毒性多样性及水稻品种的抗病性[J]. 生物多样性, 2001, 09(3): 201-206. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2026 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()