



生物多样性 ›› 2020, Vol. 28 ›› Issue (4): 474-484. DOI: 10.17520/biods.2019290 cstr: 32101.14.biods.2019290
李潮1,金锦锦1,罗锦桢1,2,王春晖1,3,王俊杰1,赵俊1,*( )
)
收稿日期:2019-09-17
接受日期:2019-12-24
出版日期:2020-04-20
发布日期:2020-06-15
通讯作者:
赵俊
基金资助:
Chao Li1,Jinjin Jin1,Jinzhen Luo1,2,Chunhui Wang1,3,Junjie Wang1,Jun Zhao1,*( )
)
Received:2019-09-17
Accepted:2019-12-24
Online:2020-04-20
Published:2020-06-15
Contact:
Jun Zhao
摘要:
唐鱼(Tanichthys albonubes)是为数不多的几种原产中国的世界性观赏鱼类之一。自2003年以来, 多个唐鱼野生种群相继被发现, 其濒危状态和等级由野外灭绝降为极危。为研究唐鱼养殖种群与广州附近野生种群之间的遗传关系, 本文分析了唐鱼3个代表性养殖种群和4个野生种群, 共计186个样本的Cyt b基因、2个核基因(ENC1和RAG1)以及13个微卫星位点数据。基于K2P模型的遗传距离结果显示, 唐鱼野生种群间的遗传距离在0.005-0.015之间, 养殖种群间的遗传距离为0.001-0.009。系统发育分析表明, 唐鱼养殖种群包含4个单倍型谱系分支, 其中2个分别与广州附近2个野生种群聚在一起, 另外2个分别独立成支。单倍型网络亲缘关系分析显示, 清远种群只有1个单倍型且与芳村养殖种群共享, 芳村养殖种群拥有最多的单倍型。基于微卫星数据的STRUCTURE分析表明, 所有种群最佳分簇数为2, 清远种群与养殖种群聚为一簇, 良口和石门种群聚为另一簇。主成分分析结果显示, 养殖种群高度重叠并能与野生种群分开, 清远种群与养殖种群存在部分重叠。利用IMa3的基因流分析表明, 存在清远种群至芳村养殖种群的单向基因流。综合本文结果, 作者认为唐鱼养殖种群应起源于广州附近多个野生种群。清远种群来源于养殖种群中的芳村养殖种群。建议在未来唐鱼的保护策略中, 应禁止不规范的放流活动并且禁止将不同野生种群补充至养殖种群, 同时加强唐鱼养殖种群和野生种群的遗传资源管理和持续监测。
李潮,金锦锦,罗锦桢,王春晖,王俊杰,赵俊 (2020) 唐鱼养殖种群与广州附近4个野生种群的遗传关系. 生物多样性, 28, 474-484. DOI: 10.17520/biods.2019290.
Chao Li,Jinjin Jin,Jinzhen Luo,Chunhui Wang,Junjie Wang,Jun Zhao (2020) Genetic relationships of hatchery populations and wild populations of Tanichthys albonubes near Guangzhou. Biodiversity Science, 28, 474-484. DOI: 10.17520/biods.2019290.
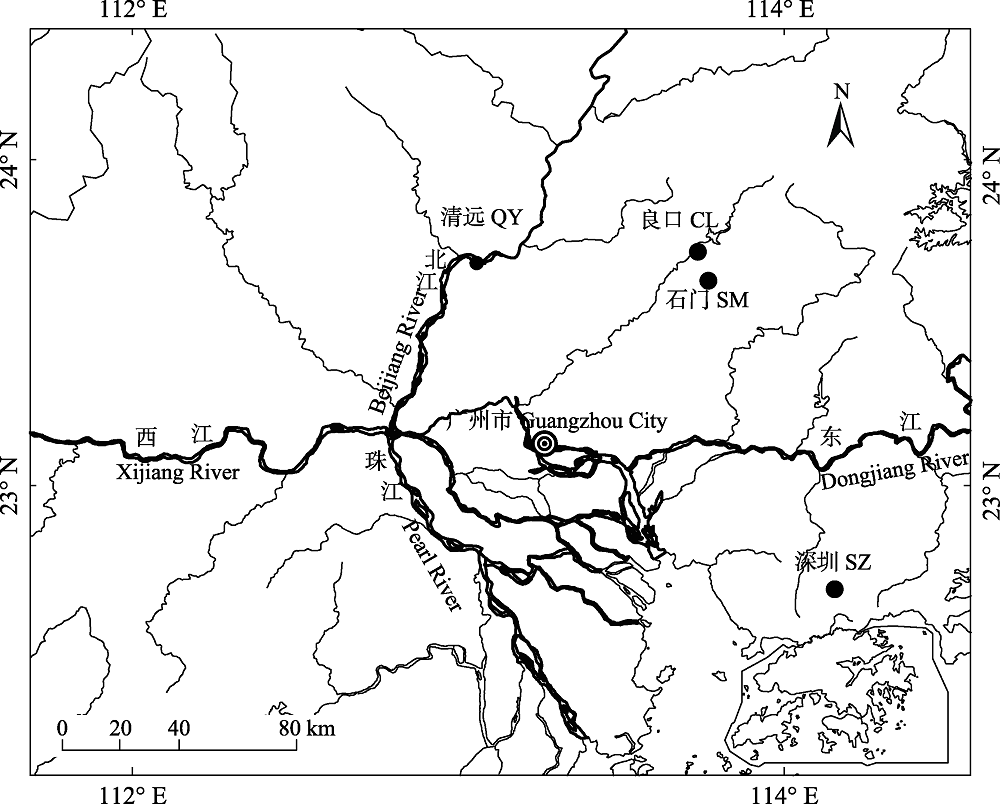
图1 本研究中唐鱼4个野生种群的采样地点(种群代号同表1)
Fig. 1 Sampling location of four wild populations of Tanichthys albonubes in this study. Population codes refer to Table 1.
| 种群 Population | 代号 Code | 所属谱系 Lineage* | 样本量 Sample size* | ||
|---|---|---|---|---|---|
| mtDNA | nuDNA | Microsatellites | |||
| 养殖种群 Hatchery populations | |||||
| 广州芳村 Fangcun, Guangzhou, China | FC | A | 30 | 30 | 30 |
| 新加坡 Singapore | SIG | A | 24 | 24 | 30 |
| 加拿大 Canada | CA | A | 12 | 12 | 12 |
| 野生种群 Wild populations | |||||
| 广州从化良口 Liangkou, Conghua, Guangzhou, China | CL | A | 30 | 30 | 30 |
| 广州从化石门 Shimen, Conghua, Guangzhou, China | SM | A | 30 | 30 | 30 |
| 清远 Qingyuan, China | QY | A | 30 | 30 | 30 |
| 外类群 | |||||
| 深圳 Shenzhen, China | SZ | A | 30 | - | - |
| 总计 Total | 186 | 156 | 162 | ||
表1 本研究的唐鱼样品信息
Table 1 Sampling information of Tanichthys albonubes of the present study
| 种群 Population | 代号 Code | 所属谱系 Lineage* | 样本量 Sample size* | ||
|---|---|---|---|---|---|
| mtDNA | nuDNA | Microsatellites | |||
| 养殖种群 Hatchery populations | |||||
| 广州芳村 Fangcun, Guangzhou, China | FC | A | 30 | 30 | 30 |
| 新加坡 Singapore | SIG | A | 24 | 24 | 30 |
| 加拿大 Canada | CA | A | 12 | 12 | 12 |
| 野生种群 Wild populations | |||||
| 广州从化良口 Liangkou, Conghua, Guangzhou, China | CL | A | 30 | 30 | 30 |
| 广州从化石门 Shimen, Conghua, Guangzhou, China | SM | A | 30 | 30 | 30 |
| 清远 Qingyuan, China | QY | A | 30 | 30 | 30 |
| 外类群 | |||||
| 深圳 Shenzhen, China | SZ | A | 30 | - | - |
| 总计 Total | 186 | 156 | 162 | ||
| 种群Populations | 芳村 FC | 新加坡SIG | 加拿大CA | 良口 CL | 石门 SM | 清远 QY |
|---|---|---|---|---|---|---|
| 新加坡 SIG | 0.008 | |||||
| 加拿大 CA | 0.009 | 0.001 | ||||
| 良口 CL | 0.006 | 0.006 | 0.007 | |||
| 石门 SM | 0.009 | 0.009 | 0.010 | 0.007 | ||
| 清远 QY | 0.007 | 0.001 | 0.002 | 0.005 | 0.008 | |
| 深圳 SZ | 0.013 | 0.014 | 0.015 | 0.012 | 0.015 | 0.014 |
表2 基于线粒体Cyt b基因的唐鱼种群间K2P遗传距离(种群代号同表1)
Table 2 K2P genetic distance among seven populations of Tanichthys albonubes based on Cyt b gene. Population codes refer to Table 1.
| 种群Populations | 芳村 FC | 新加坡SIG | 加拿大CA | 良口 CL | 石门 SM | 清远 QY |
|---|---|---|---|---|---|---|
| 新加坡 SIG | 0.008 | |||||
| 加拿大 CA | 0.009 | 0.001 | ||||
| 良口 CL | 0.006 | 0.006 | 0.007 | |||
| 石门 SM | 0.009 | 0.009 | 0.010 | 0.007 | ||
| 清远 QY | 0.007 | 0.001 | 0.002 | 0.005 | 0.008 | |
| 深圳 SZ | 0.013 | 0.014 | 0.015 | 0.012 | 0.015 | 0.014 |
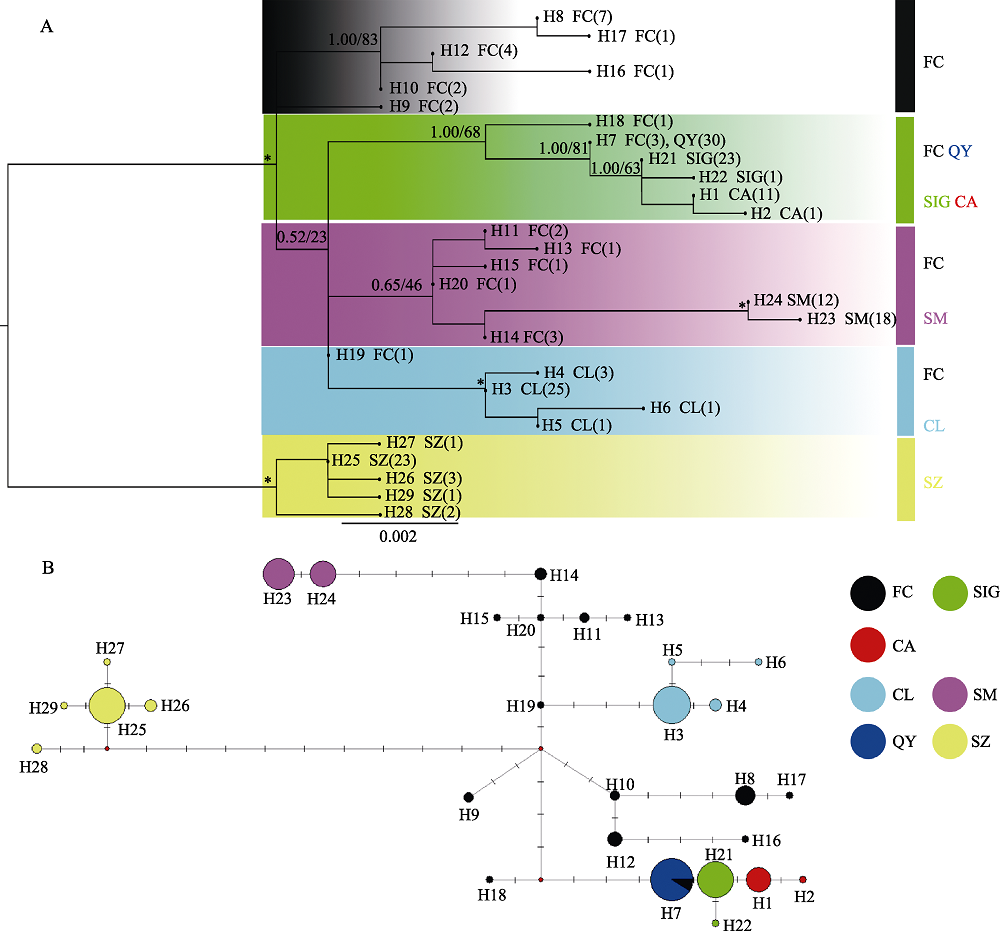
图2 基于Cyt b基因的唐鱼系统发育树和单倍型网络图(种群代号同表1)。(A)基于Cyt b基因的贝叶斯树(BI tree)和最大似然树(ML tree)。*号表示后验概率(posterior probabilities, PP) ≥ 0.90和自举值(bootstrap values, BS) ≥ 90; (B)基于Cyt b基因的单倍型网络图(线上短线表示变异步数)。
Fig. 2 Phylogenetic trees and haplotype networks of Tanichthys albonubes based on Cyt b gene. Population codes refer to Table 1. (A) Bayesian inference (BI) and maximum-likelihood (ML) trees of T. albonubes based on Cyt b gene. The asterisks on the branches indicate posterior probabilities (PP) ≥ 0.90 and bootstrap values (BS) ≥ 90. (B) Haplotype networks of T. albonubes based on Cyt b gene (segments above lines indicate the number of mutations).
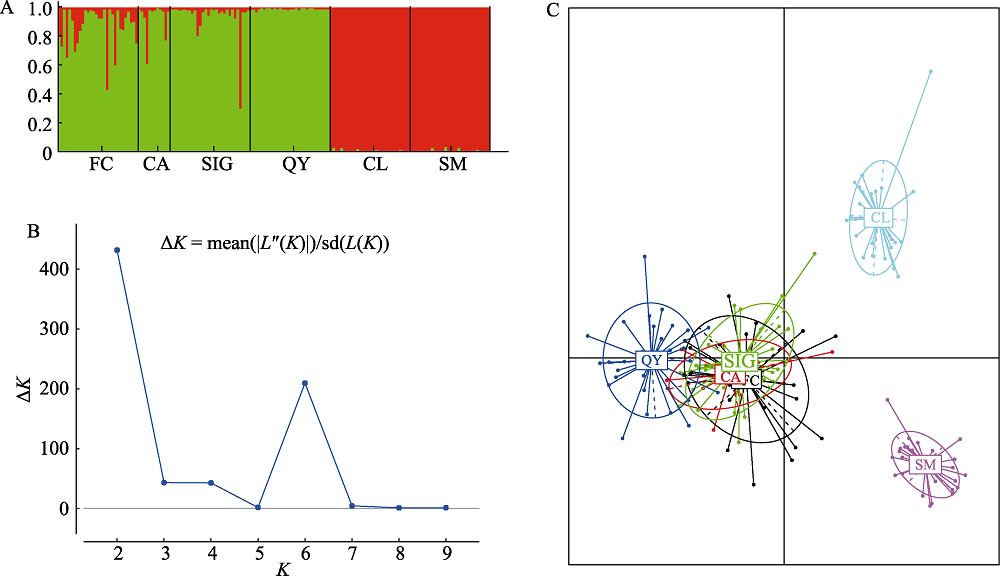
图3 唐鱼养殖种群和野生种群的种群结构分析(种群代号同表1)。(A)基于13个微卫星位点的STRUCTURE结果图(K = 2); (B) K取不同值时的ΔK值; (C)基于13个微卫星位点的主成分分析。
Fig. 3 Population structure analysis of hatchery and wild populations of Tanichthys albonubes. (A) STRUCTURE plots of K = 2 based on 13 microsatellite loci; (B) ΔK values of the different number of clusters (K); (C) PCA analysis based on 13 microsatellite loci. Population codes refer to Table 1.
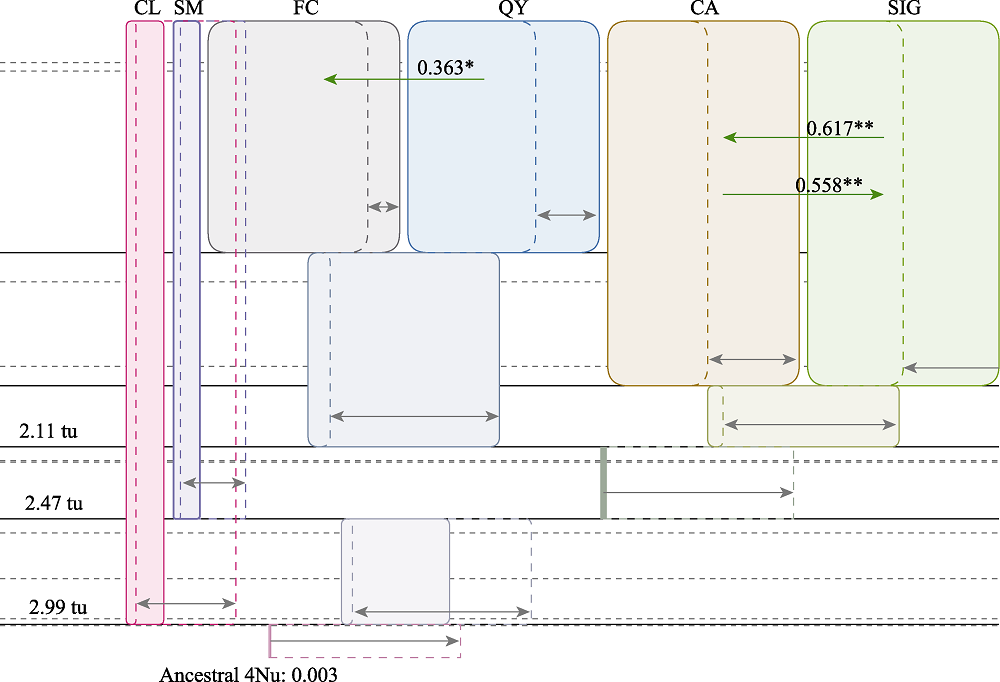
图4 唐鱼养殖种群和野生种群间的基因流估算(种群代号同表1)。灰色箭头和方框代表tu的边界值。tu: 种群分开后所经历的时间。绿色箭头代表基因流的方向, 其上数字表示基因流的大小。图中只显示统计上显著的值, 显著性水平: * P < 0.05; ** P < 0.01。
Fig. 4 Gene flow estimation among hatchery and wild populations of Tanichthys albonubes. Grey arrows and boxes were marginal distribution values in demographic units. tu means time since ancestral population splitting. Green arrows and the numbers above them showed the gene flows (2Nm values) and their directions. Only statistically significant cases of gene flow are presented. * P < 0.05, ** P < 0.01. Population codes refer to Table 1.
| [1] |
Bandelt HJ, Forster P, RŏHL A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48.
DOI URL |
| [2] |
Cao L, Zhang E, Zang CX, Cao WX (2016) Evaluating the status of China’s continental fish and analyzing their causes of endangerment through the red list assessment. Biodiversity Science, 24, 598-609. (in Chinese with English abstract)
DOI URL |
|
[ 曹亮, 张鹗, 臧春鑫, 曹文宣 (2016) 通过红色名录评估研究中国内陆鱼类受威胁现状及其成因. 生物多样性, 24, 598-609.]
DOI URL |
|
| [3] |
Chan B, Chen X (2009) Discovery of Tanichthys albonubes Lin 1932 (Cyprinidae) on Hainan Island and notes on its ecology. Zoological Research, 30, 209-214.
DOI URL |
| [4] | Chen GZ, Fang ZQ, Ma GZ (2004) Embryonic development of Tanichthys albonubes. Journal of Fishery Sciences of China, 11, 489-496. (in Chinese with English abstract) |
| [ 陈国柱, 方展强, 马广智 (2004) 唐鱼胚胎发育观察. 中国水产科学, 11, 489-496.] | |
| [5] | Chen YY (1998) Fauna Sinica • Osteichthyes : Cypriniformes (II). Science Press, Beijing. (in Chinese) |
| [ 陈宜瑜 (1998) 中国动物志•硬骨鱼纲: 鲤形目(中卷). 科学出版社, 北京.] | |
| [6] |
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359-361.
DOI URL |
| [7] | Eastman OR (1938) Tanichthys albonubes Lin, the new wonder fish. The Aquarium, 7, 73-75. |
| [8] |
Ellstrand NC, Rieseberg LH (2016) When gene flow really matters: Gene flow in applied evolutionary biology. Evolutionary Applications, 9, 833-836.
DOI URL |
| [9] |
Evanno GS, Regnaut SJ, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611-2620.
DOI URL |
| [10] | Freyhof J, Herder F (2001) Tanichthys micagemmae, a new miniature cyprinid fish from Central Vietnam (Cypriniformes: Cyprinidae). Ichthyological Exploration of Freshwaters, 12, 215-220. |
| [11] |
Glover KA, Solberg MF, McGinnity P, Hindar K, Verspoor E, Coulson MW, Svåsand T (2017) Half a century of genetic interaction between farmed and wild Atlantic salmon: Status of knowledge and unanswered questions. Fish and Fisheries, 18, 890-927.
DOI URL |
| [12] |
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59, 307-321.
DOI URL |
| [13] | Hey J, Chung Y, Sethuraman A, Lachance J, Tishkoff S, Sousa VC (2018) Phylogeny estimation by integration over isolation with migration models. Molecular Biology and Evolution, 35, 2805-2818. |
| [14] |
Jaisuk C, Senanan W (2018) Effects of landscape features on population genetic variation of a tropical stream fish, stone lapping minnow, Garra cambodgiensis, in the upper Nan River drainage basin, northern Thailand. PeerJ, 6, e4487.
DOI URL |
| [15] |
Jombart T (2008) adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403-1405.
DOI URL |
| [16] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547-1549.
DOI URL |
| [17] |
Larson G, Burger J (2013) A population genetics view of animal domestication. Trends in Genetics, 29, 197-205.
DOI URL |
| [18] |
Lefort V, Longueville JE, Gascuel O (2017) SMS: Smart model selection in PhyML. Molecular Biology and Evolution, 34, 2422-2424.
DOI URL |
| [19] | Li J, Li XH (2011) A new record of fish Tanichthys albonubes (Cypriniformes: Cyprinidae) in Guangxi, China. Chinese Journal of Zoology, 46, 136-140. (in Chinese) |
| [ 李捷, 李新辉 (2011) 广西鱼类一新纪录: 唐鱼(鲤形目: 鲤科). 动物学杂志, 46, 136-140.] | |
| [20] |
Librado P, Rozas J (2009) DNASP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452.
DOI URL |
| [21] | Lin SY (1932) New cyprinid fishes from White Cloud Mountain, Canton. Lingnan Science Journal, 11, 379-383. |
| [22] | Liu HS, Yi ZS, Liang JH, Li WH, Lin XT (2008) Morphological variations between the wild population and hatchery stock of Tanichtys albonubes. Journal of Jinan University, 29, 295-299. (in Chinese with English abstract) |
| [ 刘汉生, 易祖盛, 梁建宏, 李伟华, 林小涛 (2008) 唐鱼野生种群和养殖群体的形态差异分析. 暨南大学学报, 29, 295-299.] | |
| [23] |
Luo JZ, Lin HD, Yang F, Yi ZS, Chan BP, Zhao J (2015) Population genetic structure in wild and hatchery populations of white cloud mountain minnow (Tanichthys albonubes): Recommendations for conservation. Biochemical Systematics and Ecology, 62, 142-150.
DOI URL |
| [24] | Pan JH (1990) Freshwater Fishes of Gongdong Province. Guangdong Science and Technology Press, Guangzhou. (in Chinese) |
| [ 潘炯华 (1990) 广东淡水鱼类志. 广东科技出版社, 广州.] | |
| [25] | Pritchard JK, Stephens P, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959. |
| [26] | R Development Core Team (2015) R: A Language and Environment for Statistical Computing. Foundation for Statistical Computing, Vienna. |
| [27] |
Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, Höhna S, Larget B, Liu L, Suchard M, Huelsenbeck J (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539-542.
DOI URL |
| [28] |
Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312-1313.
DOI URL |
| [29] |
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins, DG (1997) The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882.
DOI URL |
| [30] | Weitzman SH, Chan LL (1966) Identification and relationships of Tanichthys albonubes and Aphyocypris pooni, two cyprinid fishes from South China and Hong Kong. Copeia, 1966, 285-296. |
| [31] |
Yan F, Lv JC, Zhang BL, Yuan ZY, Zhao HP, Huang S, Wei G, Mi X, Zou DH, Xu W, Chen S, Wang J, Xie F, Wu MY, Xiao HB, Liang ZQ, Jin JQ, Wu SF, Xu CS, Tapley B, Turvey ST, Papenfuss TJ, Cunningham AA, Murphy RW, Zhang YP, Che J (2018) The Chinese giant salamander exemplifies the hidden extinction of cryptic species. Current Biology, 28, R590-R592.
DOI URL |
| [32] | Yi ZS, Chen XL, Wu JX, Yu SC, Huang CE (2004) Rediscovering the wild population of White Cloud Mountain minnows (Tanichthys albonubes Lin) in Guangdong Province. Zoological Research, 25, 551-555. (in Chinese) |
| [ 易祖盛, 陈湘粦, 巫锦雄, 余顺昌, 黄慈恩 (2004) 野生唐鱼在广东的再发现. 动物学研究, 25, 551-555.] | |
| [33] | Zhang XX, Zhu QY, Zhao, J (2017) Geometric morphometric analysis of body-form variability in populations of Tanichthys albonubes. Journal of Fisheries of China, 41, 1365-1372. (in Chinese with English abstract) |
| [ 张秀霞, 朱巧莹, 赵俊 (2017) 利用几何形态测量学方法分析唐鱼群体的形态变异. 水产学报, 41, 1365-1373.] | |
| [34] | Zhao J, Hsu CK, Luo JZ, Wang CH, Chan BP, Li J, Kuo PH, Lin HD (2018) Genetic diversity and population history of Tanichthys albonubes (Teleostei: Cyprinidae): Implications for conservation. Aquatic Conversation: Marine and Freshwater Ecosystem, 28, 422-434. |
| [35] | Zhao J, Yi ZS, Zhou XY, Xiao Z (2010) Background Resources of Aquatic Plants and Animals in Guangzhou. Science Press, Beijing. (in Chinese) |
| [ 赵俊, 易祖盛, 周叶先, 肖智 (2010) 广州市水生动植物本底资源. 科学出版社, 北京.] | |
| [36] | Zheng CY (1989) The Fishes of the Zhujiang River. Science Press, Beijing. (in Chinese) |
| [ 郑慈英 (1989) 珠江鱼类志. 科学出版社, 北京.] |
| [1] | 林珍, 向家宝, 蔡何佳奕, 高贝, 杨金涛, 李俊毅, 周青松, 黄晓磊, 邓鋆. 七种半翅目昆虫线粒体基因组数据[J]. 生物多样性, 2025, 33(2): 24434-. |
| [2] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [3] | 吕燕文, 王子韵, 肖钰, 何梓晗, 吴超, 胡新生. 谱系分选理论与检测方法的研究进展[J]. 生物多样性, 2024, 32(4): 23400-. |
| [4] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [5] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [6] | 卢聪聪, 刘倩, 黄晓磊. 三种蚜虫的线粒体基因组数据[J]. 生物多样性, 2022, 30(7): 22204-. |
| [7] | 牛晓锋, 王晓梅, 张研, 赵志鹏, 樊恩源. 鲟鱼分子鉴定方法的整合应用[J]. 生物多样性, 2022, 30(6): 22034-. |
| [8] | 孙琼, 王嵘, 陈小勇. 物种形成过程中的分化基因组岛及其形成机制[J]. 生物多样性, 2022, 30(3): 21383-. |
| [9] | 翟俊杰, 赵慧峰, 商光申, 孙振钧, 张玉峰, 王兴. 蚯蚓基因组学的研究进展: 基于全基因组及线粒体基因组[J]. 生物多样性, 2022, 30(12): 22257-. |
| [10] | 叶俊伟, 田斌. 中国西南地区重要木本油料植物扁核木的遗传结构及成因[J]. 生物多样性, 2021, 29(12): 1629-1637. |
| [11] | 杨计平, 李策, 陈蔚涛, 李跃飞, 李新辉. 西江中下游鳤的遗传多样性与种群动态历史[J]. 生物多样性, 2018, 26(12): 1289-1295. |
| [12] | 武星彤, 陈璐, 王敏求, 张原, 林雪莹, 李鑫玉, 周宏, 文亚峰. 丹霞梧桐群体遗传结构及其遗传分化[J]. 生物多样性, 2018, 26(11): 1168-1179. |
| [13] | 毛建丰, 马永鹏, 周仁超. 结合系统发育与群体遗传学分析检验杂交是否存在的技术策略[J]. 生物多样性, 2017, 25(6): 577-599. |
| [14] | 周秋杰, 蔡亚城, 黄伟伦, 吴伟, 代色平, 王峰, 周仁超. 野牡丹属两个海南特有种与同属广布种自然杂交的分子证据[J]. 生物多样性, 2017, 25(6): 638-646. |
| [15] | 薛建华, 姜莉, 马晓林, 邴艳红, 赵思晨, 马克平. 莲品种DNA指纹图谱的构建[J]. 生物多样性, 2016, 24(1): 3-11. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn