



生物多样性 ›› 2022, Vol. 30 ›› Issue (3): 21383. DOI: 10.17520/biods.2021383 cstr: 32101.14.biods.2021383
所属专题: 物种形成与系统进化
收稿日期:2021-09-22
接受日期:2022-01-18
出版日期:2022-03-20
发布日期:2022-03-10
通讯作者:
陈小勇
作者简介:*E-mail: xychen@des.ecnu.edu.cn基金资助:
Qiong Sun1, Rong Wang1,2, Xiaoyong Chen1,2,*( )
)
Received:2021-09-22
Accepted:2022-01-18
Online:2022-03-20
Published:2022-03-10
Contact:
Xiaoyong Chen
摘要:
理解物种形成机制是生态和进化领域的重要任务。得益于测序技术的快速发展, 越来越多研究发现分化种群(亚种、物种)间的基因组常呈现异质性分化景观, 存在分化基因组岛, 这被认为是基因流存在下的歧化选择引起的, 支持基因流存在下的成种假说。然而, 基因渐渗、祖先多态性的差异分选、连锁选择等其他进化过程也可导致分化基因组岛的形成。现有实证研究在解析分化基因组岛的形成机制时, 往往忽略了上述其他进化过程的作用。为此, 本文在辨析分化基因组岛相关概念的基础上, 总结了利用种群基因组数据鉴定分化基因组岛的方法, 对比了不同进化过程形成分化基因组岛的特征, 指出在区分不同机制时联用基因渐渗程度、绝对分化指数(dXY)、相对节点深度(RND)、重组率等多个指标的必要性, 归纳了物种形成过程中分化基因组岛形成机制解析的研究思路, 并对未来在生殖隔离机制上的深入探索以及实证研究的整合分析等方面进行了展望。
孙琼, 王嵘, 陈小勇 (2022) 物种形成过程中的分化基因组岛及其形成机制. 生物多样性, 30, 21383. DOI: 10.17520/biods.2021383.
Qiong Sun, Rong Wang, Xiaoyong Chen (2022) Genomic island of divergence during speciation and its underlying mechanisms. Biodiversity Science, 30, 21383. DOI: 10.17520/biods.2021383.
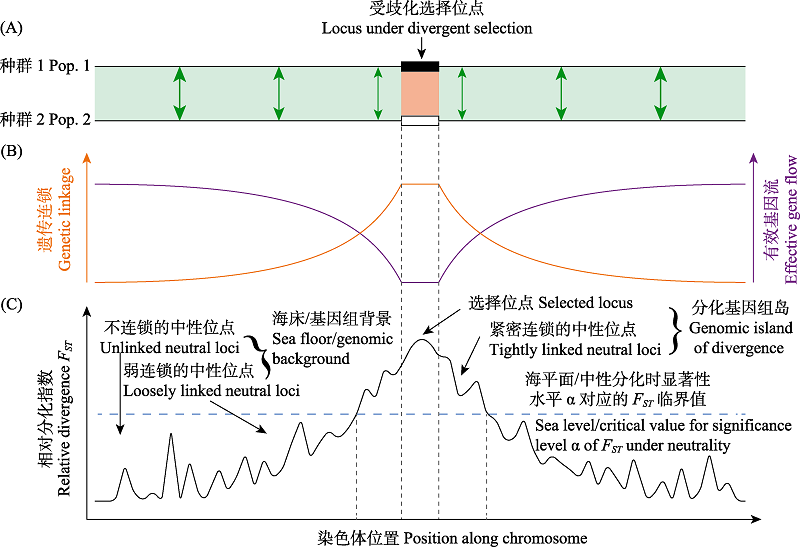
图1 基因流存在下的成种与分化基因组岛。(A)基因流存在下的成种过程初期在基因水平的图示。刚开始分化的两个种群染色体上仅少量位点(此处仅展示1个)受歧化选择, 其等位基因在种群间的交换受限。双向箭头粗细表示有效基因流强弱。(B)中性位点与受歧化选择位点的遗传连锁及中性位点的有效基因流在染色体上的变化。(C)异质性基因组分化景观与分化基因组岛。“海平面”将“分化基因组岛”和“海床”分开来。改自Wu (2001)、Nosil和Feder (2012)和Aeschbacher等(2017)。
Fig. 1 Speciation with gene flow and genomic island of divergence. (A) Conceptual diagram showing the early stage of the process of speciation with gene flow from the genic view. When Pop.1 and Pop.2 start to diverge, only a few loci on the chromosome (here only one is shown) are under divergent selection, and the exchange of the alleles at these loci between populations is restricted. The thickness of the double-headed arrows represents the strength of effective gene flow. (B) Variation of the genetic linkage between neutral loci and divergently selected locus and the effective gene flow at neutral sites along the chromosome. (C) Heterogeneous landscape of genomic divergence and genomic island of divergence. Sea level separates the genomic island of divergence and the sea floor. Adapted from Wu (2001), Nosil & Feder (2012), and Aeschbacher et al (2017).
| 方法 Methods | 优点 Pros | 缺点 Cons | 示例文献 Example references |
|---|---|---|---|
| 基于经验的方法 Empirical approach | |||
| 固定/近固定差异的局部聚集程度 Extent of regional clustering of fixed/nearly fixed differences | 非常简单便捷 Very simple and convenient | 未充分考虑种群统计的影响; 仅使用固定/近固定差异的分子标记进行分析, 忽略了基因组中大量的其他变异信息 The effect of population demography is not fully considered; Only fixed or nearly fixed differences are used in the analysis, with numerous differences in the genome ignored | Turner et al, |
| FST百分位数 Percentile of FST | 非常简单便捷 Very simple and convenient | 未充分考虑种群统计的影响 The effect of population demography is not fully considered | Renaut et al, |
| Z标准化后的FST (ZFST)大小 Value of Z-transformed FST (ZFST) | 非常简单便捷 Very simple and convenient | 未充分考虑种群统计的影响 The effect of population demography is not fully considered | Han et al, |
| 根据实际数据重抽样构造FST零分布检验观测FST的显著性 Significance test on observed FST using the null distribution of FST built by resampling real data | 简单便捷 Simple and convenient | 未充分考虑种群统计的影响 The effect of population demography is not fully considered | Michel et al, |
| 基于模型的方法 Model-based approach | |||
| 隐马尔科夫模型 Hidden Markov model (HMM) | 简单便捷; 无需设置滑窗大小, 考虑了相邻分子标记之间的非独立性 Simple and convenient; No need to set the sliding window size, and the non-independence among neighboring markers is modeled | 未充分考虑种群统计的影响; 每种隐含状态对应观测值的分布、高/低分化水平两种隐含状态互相的转移概率是否为0等方面的假设对鉴定结果有一定影响 The effect of population demography is not fully considered; The identification result is affected by the assumptions such as the distribution of observations corresponding to each hidden state and whether the transition probability between the two hidden states of high/low divergence is 0 | Turner et al, |
| BayeScan | 简单便捷 Simple and convenient | 当研究对象中有经历过严重瓶颈效应的种群时, 鉴定结果存在较高的假阳性率 High false positive rate when studied population underwent a strong bottleneck | Michel et al, |
| 根据假定的种群动态场景构建不同杂合度下FST的零分布检验观测FST的显著性 Significance test on observed FST using the null distribution of FST under different heterozygosity according to the assumed population demographic scenario | 简单便捷; 相关软件内置了不同种群动态场景 Simple and convenient; Different population demographic models are built in related softwares | 假定的种群动态场景可能与现实场景相差甚远, 导致假阳性率较高 The assumed population demographic model may be very different from the real scenario, resulting in a high false positive rate | Michel et al, |
| 根据推断的最优种群统计模型构建FST的零分布检验观测FST的显著性 Significance test on observed FST using the null distribution of FST according to the inferred best-fitting demographic model | 构造FST零分布时使用的种群统计模型更符合现实场景 The demographic model used to build the null distribution of FST is more realistic | 步骤有些繁琐; 未考虑重组率对FST分布的影响 Somewhat complex; The effect of recombination rate on the distribution of FST is ignored | Malinsky et al, |
| 根据推断的最优种群统计模型构建不同重组率下FST的零分布检验观测FST的显著性 Significance test on observed FST using the null distribution of FST under different recombination rate according to the inferred best-fitting demographic model | 构造FST零分布时使用的种群统计模型更符合现实场景; 考虑了重组率对FST分布的影响 The demographic model used to build the null distribution of FST is more realistic, and the effect of recombination rate on distribution of FST is considered | 步骤繁琐 Complicated | 目前尚无示例文献 No example reference is available yet |
表1 不同分化基因组岛鉴定方法的比较
Table 1 Comparison of different methods for identifying genomic island of divergence
| 方法 Methods | 优点 Pros | 缺点 Cons | 示例文献 Example references |
|---|---|---|---|
| 基于经验的方法 Empirical approach | |||
| 固定/近固定差异的局部聚集程度 Extent of regional clustering of fixed/nearly fixed differences | 非常简单便捷 Very simple and convenient | 未充分考虑种群统计的影响; 仅使用固定/近固定差异的分子标记进行分析, 忽略了基因组中大量的其他变异信息 The effect of population demography is not fully considered; Only fixed or nearly fixed differences are used in the analysis, with numerous differences in the genome ignored | Turner et al, |
| FST百分位数 Percentile of FST | 非常简单便捷 Very simple and convenient | 未充分考虑种群统计的影响 The effect of population demography is not fully considered | Renaut et al, |
| Z标准化后的FST (ZFST)大小 Value of Z-transformed FST (ZFST) | 非常简单便捷 Very simple and convenient | 未充分考虑种群统计的影响 The effect of population demography is not fully considered | Han et al, |
| 根据实际数据重抽样构造FST零分布检验观测FST的显著性 Significance test on observed FST using the null distribution of FST built by resampling real data | 简单便捷 Simple and convenient | 未充分考虑种群统计的影响 The effect of population demography is not fully considered | Michel et al, |
| 基于模型的方法 Model-based approach | |||
| 隐马尔科夫模型 Hidden Markov model (HMM) | 简单便捷; 无需设置滑窗大小, 考虑了相邻分子标记之间的非独立性 Simple and convenient; No need to set the sliding window size, and the non-independence among neighboring markers is modeled | 未充分考虑种群统计的影响; 每种隐含状态对应观测值的分布、高/低分化水平两种隐含状态互相的转移概率是否为0等方面的假设对鉴定结果有一定影响 The effect of population demography is not fully considered; The identification result is affected by the assumptions such as the distribution of observations corresponding to each hidden state and whether the transition probability between the two hidden states of high/low divergence is 0 | Turner et al, |
| BayeScan | 简单便捷 Simple and convenient | 当研究对象中有经历过严重瓶颈效应的种群时, 鉴定结果存在较高的假阳性率 High false positive rate when studied population underwent a strong bottleneck | Michel et al, |
| 根据假定的种群动态场景构建不同杂合度下FST的零分布检验观测FST的显著性 Significance test on observed FST using the null distribution of FST under different heterozygosity according to the assumed population demographic scenario | 简单便捷; 相关软件内置了不同种群动态场景 Simple and convenient; Different population demographic models are built in related softwares | 假定的种群动态场景可能与现实场景相差甚远, 导致假阳性率较高 The assumed population demographic model may be very different from the real scenario, resulting in a high false positive rate | Michel et al, |
| 根据推断的最优种群统计模型构建FST的零分布检验观测FST的显著性 Significance test on observed FST using the null distribution of FST according to the inferred best-fitting demographic model | 构造FST零分布时使用的种群统计模型更符合现实场景 The demographic model used to build the null distribution of FST is more realistic | 步骤有些繁琐; 未考虑重组率对FST分布的影响 Somewhat complex; The effect of recombination rate on the distribution of FST is ignored | Malinsky et al, |
| 根据推断的最优种群统计模型构建不同重组率下FST的零分布检验观测FST的显著性 Significance test on observed FST using the null distribution of FST under different recombination rate according to the inferred best-fitting demographic model | 构造FST零分布时使用的种群统计模型更符合现实场景; 考虑了重组率对FST分布的影响 The demographic model used to build the null distribution of FST is more realistic, and the effect of recombination rate on distribution of FST is considered | 步骤繁琐 Complicated | 目前尚无示例文献 No example reference is available yet |
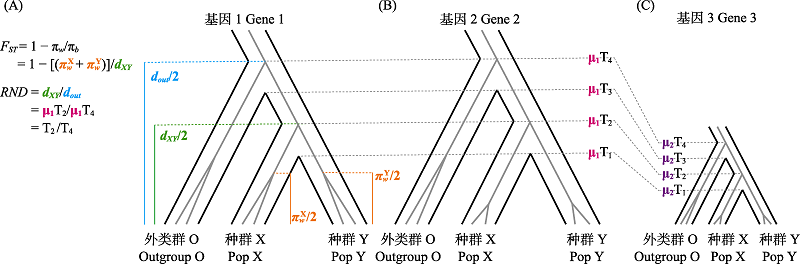
图2 种群内核苷酸多样性(πw)、种群间核苷酸序列分化/绝对分化指标(πb或dXY)、固定指数(FST)和相对节点深度(RND)图示。X、Y为两个正在分化的种群/物种, O为外类群。物种树(黑色)内是示例基因的基因树(灰色)。此处物种树和基因树的大小均与示例基因突变率μ成正比, μ1 > μ2。T1、T3为X与Y、O与XY祖先的种群分化时间/成种时间, T2、T4为X与Y、O与XY祖先的示例基因分化时间。πwX、πwY、dXY (即πb)为X种群内、Y种群内、X与Y种群间两两配对序列的平均核苷酸差异。FST可表示为1-πw/πb或类似形式。RND校正了μ对序列分化的影响。dout是O与X、Y的dOX和dOY的均值。基因1和基因2的dXY相等, 但基因2的πw更小, 因此其FST更高(A和B)。基因2和基因3的FST、T2、T4和RND均相等, 但μ不同, 基因3的dXY更小(B和C)。改自Cruickshank和Hahn (2014)和Rosenzweig等(2016)。
Fig. 2 Illustration of within-population nucleotide diversity (πw), between-population sequence divergence/absolute divergence (πb or dXY), fixation index (FST) and relative node depth (RND). X and Y are two diverging populations or species. The gene tree (grey) of an example gene is contained within the species tree (black). The sizes of species tree and gene tree displayed are both directly proportional to the mutation rate (μ) of the example gene, μ1> μ2. T1 and T3 are the divergence time or speciation time between X and Y, and between O and the ancestor of X and Y, respectively. T2 and T4 are the divergence time of example gene between X and Y, and between O and the ancestor of X and Y, respectively. πwX, πwY, and absolute divergence (dXY) represent the average number of nucleotide differences of pairwise sequences within X, within Y, and between X and Y, respectively. FST is calculated as 1-πw/πb or similar formulas. RND is an estimate of sequence divergence with the effect of μ corrected. dout is the average of absolute divergence between O and X and between O and Y. dXY of gene 1 is the same as that of gene 2, but FST of gene 2 is higher due to lower πw (A and B). There are no differences in FST, T2, T4, and RND between gene 2 and gene 3, but dXY of gene 3 is lower due to lower μ (B and C). Adapted from Cruickshank & Hahn (2014) and Rosenzweig et al (2016).
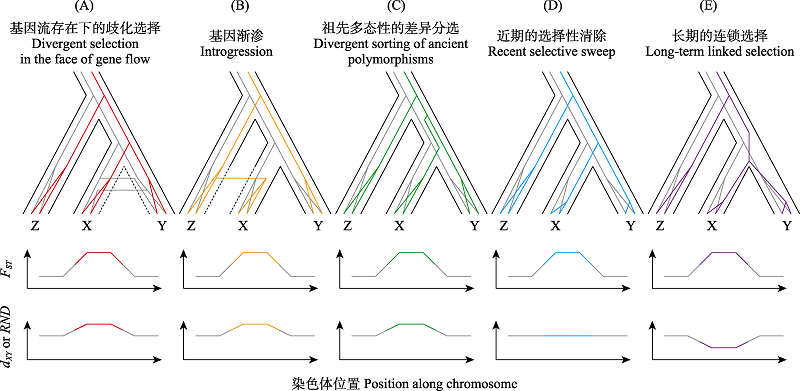
图3 可导致分化基因组岛形成的不同进化过程示意图。X、Y为两个正在分化的种群/物种。第二种机制中(B), 近缘物种Z向X有基因渐渗, 其他机制中Z向X或Y均无基因渐渗(A、C、D、E)。每个物种树内包含两个示例基因的基因树, 彩色基因树表示示例基因是经历了对应进化过程的位点或紧密连锁位点, 灰色基因树表示弱连锁或不连锁的中性位点。物种树下的图表示每种机制两类位点FST、dXY (假设基因组中μ无异质性)和RND (无需对μ的异质性做出假设)的差异。改自Cruickshank和Hahn (2014)和Han等(2017)。
Fig. 3 Schematic diagram illustrating different evolutionary processes that could lead to the formation of genomic island of divergence. X and Y are two diverging populations or species. Z is a related species, from which some genetic materials are introgressed into X in scenario (B) in contrast to the other four scenarios (A, C, D, E). The gene trees of two example genes are contained within the species tree (black). The colored gene tree represents the locus underlying the corresponding evolutionary process or its tightly linked loci, and the grey gene tree represents the loosely linked or unlinked loci. The graphs below species trees show the difference of FST, dXY (on the assumption of no variation of μ), and RND (without the assumption of no variation of μ) between two kinds of loci under various evolutionary processes. Adapted from Cruickshank & Hahn (2014) and Han et al (2017).
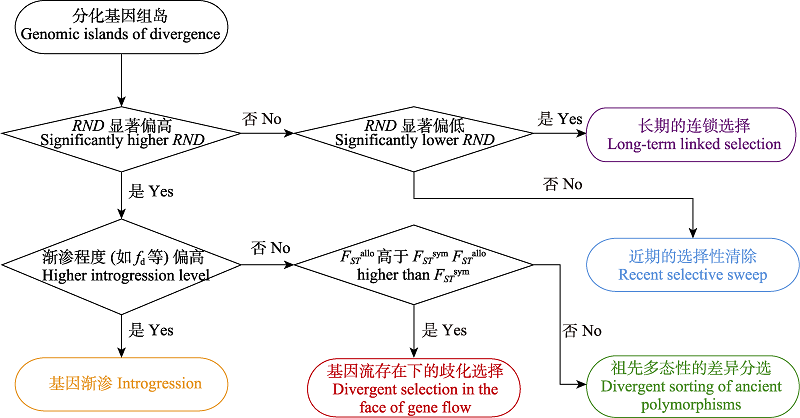
图4 分化基因组岛形成机制解析逻辑流程图。FSTsym和FSTallo分别表示姊妹种同域种群对、异域种群对的FST。此图仅展示了关键证据(辅助性证据见正文)。
Fig. 4 Flow chart illustrating the logic of discerning the mechanism contributing to genomic islands of divergence. FSTsym and FSTallo are abbreviations for FST of sympatric and allopatric populations of focal sister species, respectively. Here, only the key pieces of evidence are listed (see text for complementary evidence).
| [1] |
Ackiss AS, Larson WA, Stott W (2020) Genotyping-by-sequen-cing illuminates high levels of divergence among sympatric forms of coregonines in the Laurentian Great Lakes. Evolutionary Applications, 13, 1037-1054.
DOI URL |
| [2] | Aeschbacher S, Selby JP, Willis JH, Coop G (2017) Population-genomic inference of the strength and timing of selection against gene flow. Proceedings of the National Academy of Sciences, USA, 114, 7061-7066. |
| [3] |
Andrew RL, Rieseberg LH (2013) Divergence is focused on few genomic regions early in speciation: Incipient speciation of sunflower ecotypes. Evolution, 67, 2468-2482.
DOI PMID |
| [4] |
Battey CJ (2020) Evidence of linked selection on the Z chromosome of hybridizing hummingbirds. Evolution, 74, 725- 739.
DOI URL |
| [5] | Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society of London Series B: Biological Sciences, 263, 1619-1626. |
| [6] |
Behrens KA, Girasek QL, Sickler A, Hyde J, Buonaccorsi VP (2021) Regions of genetic divergence in depth-separated Sebastes rockfish species pairs: Depth as a potential driver of speciation. Molecular Ecology, 30, 4259-4275.
DOI URL |
| [7] |
Berg PR, Jentoft S, Star B, Ring KH, Knutsen H, Lien S, Jakobsen KS, André C (2015) Adaptation to low salinity promotes genomic divergence in Atlantic cod (Gadus morhua L.). Genome Biology and Evolution, 7, 1644-1663.
DOI URL |
| [8] |
Booker TR, Yeaman S, Whitlock MC (2020) Variation in recombination rate affects detection of outliers in genome scans under neutrality. Molecular Ecology, 29, 4274-4279.
DOI URL |
| [9] |
Bradbury IR, Hubert S, Higgins B, Bowman S, Borza T, Paterson IG, Snelgrove PVR, Morris CJ, Gregory RS, Hardie D, Hutchings JA, Ruzzante DE, Taggart CT, Bentzen P (2013) Genomic islands of divergence and their consequences for the resolution of spatial structure in an exploited marine fish. Evolutionary Applications, 6, 450-461.
DOI PMID |
| [10] |
Burri R (2017) Interpreting differentiation landscapes in the light of long-term linked selection. Evolution Letters, 1, 118-131.
DOI URL |
| [11] |
Burri R, Nater A, Kawakami T, Mugal CF, Olason PI, Smeds L, Suh A, Dutoit L, Bureš S, Garamszegi LZ, Hogner S, Moreno J, Qvarnström A, Ružić M, Sæther SA, Sætre GP, Török J, Ellegren H (2015) Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Research, 25, 1656-1665.
DOI PMID |
| [12] | Campagna L, Repenning M, Silveira LF, Fontana CS, Tubaro PL, Lovette IJ (2017) Repeated divergent selection on pigmentation genes in a rapid finch radiation. Science Advances, 3, e1602404. |
| [13] |
Charlesworth B (1998) Measures of divergence between populations and the effect of forces that reduce variability. Molecular Biology and Evolution, 15, 538-543.
PMID |
| [14] |
Charlesworth B, Morgan MT, Charlesworth D (1993) The effect of deleterious mutations on neutral molecular variation. Genetics, 134, 1289-1303.
DOI PMID |
| [15] |
Charlesworth B, Nordborg M, Charlesworth D (1997) The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genetical Research, 70, 155-174.
DOI URL |
| [16] | Cheng X, Li LL, Xiao Y, Chen XY, Hu XS (2020) Advances in the methods of detecting interspecific gene introgression and their applications. Scientia Sinica Vita, 50, 1388-1404. (in Chinese with English abstract) |
| [ 程祥, 李玲玲, 肖钰, 陈晓阳, 胡新生 (2020) 种间基因渐渗检测方法及其应用研究进展. 中国科学: 生命科学, 50, 1388-1404.] | |
| [17] |
Choi JY, Purugganan M, Stacy EA (2020) Divergent selection and primary gene flow shape incipient speciation of a riparian tree on Hawaii Island. Molecular Biology and Evolution, 37, 695-710.
DOI URL |
| [18] |
Costello MJ, May RM, Stork NE (2013) Can we name earth’s species before they go extinct? Science, 339, 413-416.
DOI PMID |
| [19] |
Cruickshank TE, Hahn MW (2014) Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Molecular Ecology, 23, 3133-3157.
DOI PMID |
| [20] |
Cutter AD, Payseur BA (2013) Genomic signatures of selection at linked sites: Unifying the disparity among species. Nature Reviews Genetics, 14, 262-274.
DOI PMID |
| [21] |
Ellegren H, Smeds L, Burri R, Olason PI, Backström N, Kawakami T, Künstner A, Mäkinen H, Nadachowska-Brzyska K, Qvarnström A, Uebbing S, Wolf JBW (2012) The genomic landscape of species divergence in Ficedula flycatchers. Nature, 491, 756-760.
DOI URL |
| [22] | Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M (2013) Robust demographic inference from genomic and SNP data. PLoS Genetics, 9, e1003905. |
| [23] |
Excoffier L, Hofer T, Foll M (2009) Detecting loci under selection in a hierarchically structured population. Heredity, 103, 285-298.
DOI PMID |
| [24] |
Feder JL, Egan SP, Nosil P (2012a) The genomics of speciation-with-gene-flow. Trends in Genetics, 28, 342-350.
DOI URL |
| [25] | Feder JL, Gejji R, Yeaman S, Nosil P (2012b) Establishment of new mutations under divergence and genome hitchhiking. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences, 367, 461-474. |
| [26] |
Feder JL, Nosil P (2010) The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution, 64, 1729-1747.
DOI URL |
| [27] | Feder JL, Xie XF, Rull J, Velez S, Forbes A, Leung B, Dambroski H, Filchak KE, Aluja M (2005) Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proceedings of the National Academy of Sciences, USA, 102, 6573-6580. |
| [28] | Feulner PGD, Chain FJJ, Panchal M, Huang Y, Eizaguirre C, Kalbe M, Lenz TL, Samonte IE, Stoll M, Bornberg-Bauer E, Reusch TBH, Milinski M (2015) Genomics of divergence along a continuum of parapatric population differentiation. PLoS Genetics, 11, e1004966. |
| [29] |
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics, 180, 977-993.
DOI URL |
| [30] | Gao F, Li HP (2016) Application of computer simulators in population genetics. Hereditas (Beijing), 38, 707-717. (in Chinese with English abstract) |
| [ 高峰, 李海鹏 (2016) 群体遗传学模拟软件应用现状. 遗传, 38, 707-717.] | |
| [31] |
Guerrero RF, Hahn MW (2017) Speciation as a sieve for ancestral polymorphism. Molecular Ecology, 26, 5362-5368.
DOI PMID |
| [32] | Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD (2009) Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genetics, 5, e1000695. |
| [33] |
Han F, Lamichhaney S, Grant BR, Grant PR, Andersson L, Webster MT (2017) Gene flow, ancient polymorphism, and ecological adaptation shape the genomic landscape of divergence among Darwin’s finches. Genome Research, 27, 1004-1015.
DOI URL |
| [34] |
Harr B (2006) Genomic islands of differentiation between house mouse subspecies. Genome Research, 16, 730-737.
DOI URL |
| [35] |
Harrison RG (2012) The language of speciation. Evolution, 66, 3643-3657.
DOI PMID |
| [36] |
Harrison RG, Larson EL (2014) Hybridization, introgression, and the nature of species boundaries. Journal of Heredity, 105, 795-809.
DOI URL |
| [37] |
Hedrick PW (2013) Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Molecular Ecology, 22, 4606-4618.
DOI PMID |
| [38] |
Hirase S, Yamasaki YY, Sekino M, Nishisako M, Ikeda M, Hara M, Merilä J, Kikuchi K (2021) Genomic evidence for speciation with gene flow in broadcast spawning marine invertebrates. Molecular Biology and Evolution, 38, 4683-4699.
DOI URL |
| [39] |
Hofer T, Foll M, Excoffier L (2012) Evolutionary forces shaping genomic islands of population differentiation in humans. BMC Genomics, 13, 107.
DOI URL |
| [40] |
Hudson AG, Vonlanthen P, Bezault E, Seehausen O (2013) Genomic signatures of relaxed disruptive selection associated with speciation reversal in whitefish. BMC Evolutionary Biology, 13, 108.
DOI PMID |
| [41] |
Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics, 132, 583-589.
DOI PMID |
| [42] |
Irwin DE, Milá B, Toews DPL, Brelsford A, Kenyon HL, Porter AN, Grossen C, Delmore KE, Alcaide M, Irwin JH (2018) A comparison of genomic islands of differentiation across three young avian species pairs. Molecular Ecology, 27, 4839-4855.
DOI URL |
| [43] |
Larson WA, Dann TH, Limborg MT, McKinney GJ, Seeb JE, Seeb LW (2019) Parallel signatures of selection at genomic islands of divergence and the major histocompatibility complex in ecotypes of sockeye salmon across Alaska. Molecular Ecology, 28, 2254-2271.
DOI URL |
| [44] |
Larson WA, Limborg MT, McKinney GJ, Schindler DE, Seeb JE, Seeb LW (2017) Genomic islands of divergence linked to ecotypic variation in sockeye salmon. Molecular Ecology, 26, 554-570.
DOI URL |
| [45] |
Lyu HM, Zhou RC, Shi SH (2015) Recent advances in the study of ecological speciation. Biodiversity Science, 23, 398-407. (in Chinese with English abstract)
DOI URL |
|
[ 吕昊敏, 周仁超, 施苏华 (2015) 生态物种形成及其研究进展. 生物多样性, 23, 398-407.]
DOI |
|
| [46] | Ma T, Wang K, Hu QJ, Xi ZX, Wan DS, Wang Q, Feng JJ, Jiang DC, Ahani H, Abbott RJ, Lascoux M, Nevo E, Liu JQ (2018) Ancient polymorphisms and divergence hitchhiking contribute to genomic islands of divergence within a poplar species complex. Proceedings of the National Academy of Sciences, USA, 115, E236-E243. |
| [47] |
Malinsky M, Challis RJ, Tyers AM, Schiffels S, Terai Y, Ngatunga BP, Miska EA, Durbin R, Genner MJ, Turner GF (2015) Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science, 350, 1493-1498.
DOI PMID |
| [48] |
Malinsky M, Matschiner M, Svardal H (2021) Dsuite - Fast D-statistics and related admixture evidence from VCF files. Molecular Ecology Resources, 21, 584-595.
DOI PMID |
| [49] |
Mallet J, Meyer A, Nosil P, Feder JL (2009) Space, sympatry and speciation. Journal of Evolutionary Biology, 22, 2332- 2341.
DOI PMID |
| [50] |
Mao JF, Ma YP, Zhou RC (2017) Approaches used to detect and test hybridization: Combining phylogenetic and population genetic analyses. Biodiversity Science, 25, 577-599. (in Chinese with English abstract)
DOI URL |
|
[ 毛建丰, 马永鹏, 周仁超 (2017) 结合系统发育与群体遗传学分析检验杂交是否存在的技术策略. 生物多样性, 25, 577-599.]
DOI |
|
| [51] | Marques DA, Lucek K, Meier JI, Mwaiko S, Wagner CE, Excoffier L, Seehausen O (2016) Genomics of rapid incipient speciation in sympatric threespine stickleback. PLoS Genetics, 12, e1005887. |
| [52] |
Martin SH, Davey JW, Jiggins CD (2015) Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Molecular Biology and Evolution, 32, 244-257.
DOI PMID |
| [53] | Mayr E (1942) Systematics and the Origin of Species. Columbia University Press, New York. |
| [54] |
McCulloch GA, Foster BJ, Dutoit L, Harrop TWR, Guhlin J, Dearden PK, Waters JM (2021) Genomics reveals widespread ecological speciation in flightless insects. Systematic Biology, 70, 863-876.
DOI PMID |
| [55] |
Michel AP, Guelbeogo WM, Grushko O, Schemerhorn BJ, Kern M, Willard MB, Sagnon NF, Costantini C, Besansky NJ (2005) Molecular differentiation between chromosomally defined incipient species of Anopheles funestus. Insect Molecular Biology, 14, 375-387.
PMID |
| [56] | Michel AP, Sim S, Powell THQ, Taylor MS, Nosil P, Feder JL (2010) Widespread genomic divergence during sympatric speciation. Proceedings of the National Academy of Sciences, USA, 107, 9724-9729. |
| [57] | Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B (2011) How many species are there on Earth and in the ocean? PLoS Biology, 9, e1001127. |
| [58] | Morales HE, Pavlova A, Amos N, Major R, Kilian A, Greening C, Sunnucks P (2018) Concordant divergence of mitogenomes and a mitonuclear gene cluster in bird lineages inhabiting different climates. Nature Ecology & Evolution, 2, 1258-1267. |
| [59] |
Mořkovský L, Janoušek V, Reif J, Rídl J, Pačes J, Choleva L, Janko K, Nachman MW, Reifová R (2018) Genomic islands of differentiation in two songbird species reveal candidate genes for hybrid female sterility. Molecular Ecology, 27, 949-958.
DOI PMID |
| [60] | Nachman MW, Payseur BA (2012) Recombination rate variation and speciation: Theoretical predictions and empirical results from rabbits and mice. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences, 367, 409-421. |
| [61] | Nadeau NJ, Whibley A, Jones RT, Davey JW, Dasmahapatra KK, Baxter SW, Quail MA, Joron M ffrench-Constant RH, Blaxter ML, Mallet J, Jiggins CD (2012) Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences, 367, 343-353. |
| [62] | Nei M (1987) Molecular Evolutionary Genetics. Columbia University Press, New York. |
| [63] |
Noor MAF, Bennett SM (2009) Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity, 103, 439-444.
DOI PMID |
| [64] |
Nosil P (2008) Speciation with gene flow could be common. Molecular Ecology, 17, 2103-2106.
DOI URL |
| [65] | Nosil P (2012) Ecological Speciation. Oxford University Press, New York. |
| [66] |
Nosil P, Egan SP, Funk DJ (2008) Heterogeneous genomic differentiation between walking-stick ecotypes: “Isolation by adaptation” and multiple roles for divergent selection. Evolution, 62, 316-336.
DOI URL |
| [67] | Nosil P, Feder JL (2012) Genomic divergence during speciation: Causes and consequences. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences, 367, 332-342. |
| [68] |
Nosil P, Funk DJ, Ortiz-Barrientos D (2009) Divergent selection and heterogeneous genomic divergence. Molecular Ecology, 18, 375-402.
DOI URL |
| [69] |
Oziolor EM, Reid NM, Yair S, Lee KM, Guberman VerPloeg S, Bruns PC, Shaw JR, Whitehead A, Matson CW (2019) Adaptive introgression enables evolutionary rescue from extreme environmental pollution. Science, 364, 455-457.
DOI PMID |
| [70] |
Papadopulos AST, Igea J, Dunning LT, Osborne OG, Quan XP, Pellicer J, Turnbull C, Hutton I, Baker WJ, Butlin RK, Savolainen V (2019) Ecological speciation in sympatric palms: 3. Genetic map reveals genomic islands underlying species divergence in Howea. Evolution, 73, 1986-1995.
DOI PMID |
| [71] |
Peñalba JV, Wolf JBW (2020) From molecules to populations: Appreciating and estimating recombination rate variation. Nature Reviews Genetics, 21, 476-492.
DOI PMID |
| [72] |
Pfeifer B, Kapan DD (2019) Estimates of introgression as a function of pairwise distances. BMC Bioinformatics, 20, 207.
DOI URL |
| [73] |
Poelstra JW, Vijay N, Bossu CM, Lantz H, Ryll B, Müller I, Baglione V, Unneberg P, Wikelski M, Grabherr MG, Wolf JBW (2014) The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science, 344, 1410-1414.
DOI PMID |
| [74] |
Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E (2015) Evidence for archaic adaptive introgression in humans. Nature Reviews Genetics, 16, 359-371.
DOI URL |
| [75] |
Ravinet M, Faria R, Butlin RK, Galindo J, Bierne N, Rafajlović M, Noor MAF, Mehlig B, Westram AM (2017) Interpreting the genomic landscape of speciation: A road map for finding barriers to gene flow. Journal of Evolutionary Biology, 30, 1450-1477.
DOI PMID |
| [76] |
Renaut S, Grassa CJ, Yeaman S, Moyers BT, Lai Z, Kane NC, Bowers JE, Burke JM, Rieseberg LH (2013) Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nature Communications, 4, 1827.
DOI PMID |
| [77] |
Rettelbach A, Nater A, Ellegren H (2019) How linked selection shapes the diversity landscape in Ficedula flycatchers. Genetics, 212, 277-285.
DOI PMID |
| [78] |
Richards EJ, Martin CH (2017) Adaptive introgression from distant Caribbean islands contributed to the diversification of a microendemic adaptive radiation of trophic specialist pupfishes. PLoS Genetics, 13, e1006919.
DOI URL |
| [79] |
Rosenzweig BK, Pease JB, Besansky NJ, Hahn MW (2016) Powerful methods for detecting introgressed regions from population genomic data. Molecular Ecology, 25, 2387-2397.
DOI PMID |
| [80] |
Rundle HD, Nosil P (2005) Ecological speciation. Ecology Letters, 8, 336-352.
DOI URL |
| [81] |
Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D (2014) The genomic landscape of Neanderthal ancestry in present-day humans. Nature, 507, 354-357.
DOI URL |
| [82] |
Semenov GA, Safran RJ, Smith CCR, Turbek SP, Mullen SP, Flaxman SM (2019) Unifying theoretical and empirical perspectives on genomic differentiation. Trends in Ecology & Evolution, 34, 987-995.
DOI URL |
| [83] |
Smadja CM, Butlin RK (2011) A framework for comparing processes of speciation in the presence of gene flow. Molecular Ecology, 20, 5123-5140.
DOI URL |
| [84] |
Smith JM, Haigh J (1974) The hitch-hiking effect of a favourable gene. Genetical Research, 23, 23-35.
DOI URL |
| [85] |
Smith NGC, Fearnhead P (2005) A comparison of three estimators of the population-scaled recombination rate: Accuracy and robustness. Genetics, 171, 2051-2062.
PMID |
| [86] |
Soria-Carrasco V, Gompert Z, Comeault AA, Farkas TE, Parchman TL, Johnston JS, Buerkle CA, Feder JL, Bast J, Schwander T, Egan SP, Crespi BJ, Nosil P (2014) Stick insect genomes reveal natural selection’s role in parallel speciation. Science, 344, 738-742.
DOI PMID |
| [87] | Stevison LS, McGaugh SE (2020) It’s time to stop sweeping recombination rate under the genome scan rug. Molecular Ecology, 29, 4249-4253. |
| [88] | Tavares H, Whibley A, Field DL, Bradley D, Couchman M, Copsey L, Elleouet J, Burrus M, Andalo C, Li MM, Li Q, Xue YB, Rebocho AB, Barton NH, Coen E (2018) Selection and gene flow shape genomic islands that control floral guides. Proceedings of the National Academy of Sciences, USA, 115, 11006-11011. |
| [89] |
Teng HJ, Zhang YH, Shi CM, Mao FB, Cai WS, Lu L, Zhao FQ, Sun ZS, Zhang JX (2017) Population genomics reveals speciation and introgression between brown Norway rats and their sibling species. Molecular Biology and Evolution, 34, 2214-2228.
DOI URL |
| [90] |
Todesco M, Owens GL, Bercovich N, Légaré JS, Soudi S, Burge DO, Huang KC, Ostevik KL, Drummond EBM, Imerovski I, Lande K, Pascual-Robles MA, Nanavati M, Jahani M, Cheung W, Staton SE, Muños S, Nielsen R, Donovan LA, Burke JM, Yeaman S, Rieseberg LH (2020) Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature, 584, 602-607.
DOI URL |
| [91] |
Tsumura Y, Uchiyama K, Moriguchi Y, Ueno S, Ihara-Ujino T (2012) Genome scanning for detecting adaptive genes along environmental gradients in the Japanese conifer, Cryptomeria japonica. Heredity, 109, 349-360.
DOI PMID |
| [92] |
Turner TL, Hahn MW, Nuzhdin SV (2005) Genomic islands of speciation in Anopheles gambiae. PLoS Biology, 3, e285.
DOI URL |
| [93] | Via S (2009) Natural selection in action during speciation. Proceedings of the National Academy of Sciences, USA, 106, 9939-9946. |
| [94] |
Vijay N, Bossu CM, Poelstra JW, Weissensteiner MH, Suh A, Kryukov AP, Wolf JBW (2016) Evolution of heterogeneous genome differentiation across multiple contact zones in a crow species complex. Nature Communications, 7, 13195.
DOI URL |
| [95] |
Wang BS, Mojica JP, Perera N, Lee CR, Lovell JT, Sharma A, Adam C, Lipzen A, Barry K, Rokhsar DS, Schmutz J, Mitchell-Olds T (2019) Ancient polymorphisms contribute to genome-wide variation by long-term balancing selection and divergent sorting in Boechera stricta. Genome Biology, 20, 126.
DOI URL |
| [96] |
Wang J, Street NR, Scofield DG, Ingvarsson PK (2016) Variation in linked selection and recombination drive genomic divergence during allopatric speciation of European and American aspens. Molecular Biology and Evolution, 33, 1754-1767.
DOI URL |
| [97] |
Wang ZF, Jiang YZ, Bi H, Lu ZQ, Ma YZ, Yang XY, Chen NN, Tian B, Liu BB, Mao XX, Ma T, DiFazio SP, Hu QJ, Abbott RJ, Liu JQ (2021) Hybrid speciation via inheritance of alternate alleles of parental isolating genes. Molecular Plant, 14, 208-222.
DOI URL |
| [98] |
Wolf JBW, Ellegren H (2017) Making sense of genomic islands of differentiation in light of speciation. Nature Reviews Genetics, 18, 87-100.
DOI URL |
| [99] |
Wu CI (2001) The genic view of the process of speciation. Journal of Evolutionary Biology, 14, 851-865.
DOI URL |
| [100] |
Wu CI, Ting CT (2004) Genes and speciation. Nature Reviews Genetics, 5, 114-122.
DOI URL |
| [101] |
Yeaman S, Aeschbacher S, Bürger R (2016) The evolution of genomic islands by increased establishment probability of linked alleles. Molecular Ecology, 25, 2542-2558.
DOI PMID |
| [1] | 吕燕文, 王子韵, 肖钰, 何梓晗, 吴超, 胡新生. 谱系分选理论与检测方法的研究进展[J]. 生物多样性, 2024, 32(4): 23400-. |
| [2] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [3] | 李潮,金锦锦,罗锦桢,王春晖,王俊杰,赵俊. 唐鱼养殖种群与广州附近4个野生种群的遗传关系[J]. 生物多样性, 2020, 28(4): 474-484. |
| [4] | 毛建丰, 马永鹏, 周仁超. 结合系统发育与群体遗传学分析检验杂交是否存在的技术策略[J]. 生物多样性, 2017, 25(6): 577-599. |
| [5] | 吕昊敏, 周仁超, 施苏华. 生态物种形成及其研究进展[J]. 生物多样性, 2015, 23(3): 398-407. |
| [6] | 苗林, 罗述金. 基因组时代的新视野: 东南亚哺乳动物类群在第四纪冰河时期多样性的起源与分化[J]. 生物多样性, 2014, 22(1): 40-50. |
| [7] | 周伟, 王红. 基于DNA分子标记的花粉流动态分析[J]. 生物多样性, 2014, 22(1): 97-108. |
| [8] | 李忠虎, 刘占林, 王玛丽, 钱增强, 赵鹏, 祝娟, 杨一欣, 阎晓昊, 李银军, 赵桂仿. 基因流存在条件下的物种形成研究述评:生殖隔离机制进化[J]. 生物多样性, 2014, 22(1): 88-96. |
| [9] | 朱璧如, 张大勇. 基于过程的群落生态学理论框架[J]. 生物多样性, 2011, 19(4): 389-399. |
| [10] | 库喜英, 周传江, 何舜平. 中国黄颡鱼的线粒体DNA多样性及其分子系统学[J]. 生物多样性, 2010, 18(3): 262-274. |
| [11] | 张俊红, 黄华宏, 童再康, 程龙军, 梁跃龙, 陈奕良. 光皮桦6个南方天然群体的遗传多样性[J]. 生物多样性, 2010, 18(3): 233-240. |
| [12] | 陈艳, 李宏庆, 刘敏, 陈小勇. 榕-传粉榕小蜂间的专一性与协同进化[J]. 生物多样性, 2010, 18(1): 1-10. |
| [13] | 夏颖哲, 盛岩, 陈宜瑜. 利用线粒体DNA控制区序列分析细鳞鲑种群的遗传结构[J]. 生物多样性, 2006, 14(1): 48-54. |
| [14] | 卢宝荣, 朱有勇, 王云月. 农作物遗传多样性农家保护的现状及前景[J]. 生物多样性, 2002, 10(4): 409-415. |
| [15] | 康明, 黄宏文. 湖北海棠的等位酶变异和遗传多样性研究[J]. 生物多样性, 2002, 10(4): 376-385. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn