



生物多样性 ›› 2016, Vol. 24 ›› Issue (11): 1296-1305. DOI: 10.17520/biods.2016202 cstr: 32101.14.biods.2016202
所属专题: 昆虫多样性与生态功能
金倩1,2, 陈芬1, 罗桂杰1, 蔡卫佳1, 刘旭1, 王昊1, 杨采青2, 郝梦迪2, 张爱兵2,*( )
)
收稿日期:2016-07-22
接受日期:2016-11-02
出版日期:2016-11-20
发布日期:2016-12-14
通讯作者:
张爱兵
基金资助:
Qian Jin1,2, Fen Chen1, Guijie Luo1, Weijia Cai1, Xu Liu1, Hao Wang1, Caiqing Yang2, Mengdi Hao2, Aibing Zhang2,*( )
)
Received:2016-07-22
Accepted:2016-11-02
Online:2016-11-20
Published:2016-12-14
Contact:
Zhang Aibing
摘要:
为了探究基于DNA条形码方法量化物种多样性指标的可行性, 本研究以江苏省宿迁地区蛾类群落为例, 基于DNA条形码方法估计群落物种丰富度并绘制等级多度分布曲线(rank-abundance curves), 同时与基于传统形态学的对应指标进行比较。结果表明: (1)基于DNA条形码的物种丰富度估计与基于形态的物种丰富度估计之间没有显著差异; (2)基于形态和DNA条形码的等级多度分布曲线趋势一致, 通过K-S检测发现二者之间没有显著性差异(P > 0.05)。结果显示, 基于DNA条形码的物种丰富度估计能够在一定程度上补充基于形态学的方法, 可以尝试将其应用于蛾类群落生态学调查研究中。
金倩, 陈芬, 罗桂杰, 蔡卫佳, 刘旭, 王昊, 杨采青, 郝梦迪, 张爱兵 (2016) 基于DNA条形码的物种丰富度估计: 以宿迁地区鳞翅目蛾类为例. 生物多样性, 24, 1296-1305. DOI: 10.17520/biods.2016202.
Qian Jin, Fen Chen, Guijie Luo, Weijia Cai, Xu Liu, Hao Wang, Caiqing Yang, Mengdi Hao, Aibing Zhang (2016) Estimation of species richness of moths (Insecta: Lepidoptera) based on DNA barcoding in Suqian, China. Biodiversity Science, 24, 1296-1305. DOI: 10.17520/biods.2016202.
| MOTU① | 种名 Species② | MOTU | 种名 Species |
|---|---|---|---|
| MOTU01 | Lacanobia contigua | MOTU34 | Parapoynx vittalis |
| MOTU02 | Agrotis segetum 黄地老虎 | MOTU35 | Heliothis assulta 烟青虫 |
| MOTU03 | Conogethes punctiferalis 桃蛀螟 | MOTU36 | Herminia grisealis |
| MOTU04 | Nola cicatricalis | MOTU37 | Spodoptera depravata |
| MOTU05 | Hipoepa fractalis 中影单跗夜蛾 | MOTU38 | Scopula subpunctaria |
| MOTU06 | Oraesia lata 平嘴壶夜蛾 | MOTU39 | Diaphania indica 瓜绢野螟 |
| MOTU07 | Spodoptera litura 斜纹夜蛾 | MOTU40 | Botyodes diniasalis 黄翅缀叶野螟 |
| MOTU08 | Uropyia meticulodina 核桃美舟蛾 | MOTU41 | Emmelia trabealis 谐夜蛾 |
| MOTU09 | Evergestis extimalis | MOTU42 | Termioptycha nigrescens |
| MOTU10 | Mythimna separate | MOTU43 | Ipimorpha subtusa 杨逸色夜蛾 |
| MOTU11 | Ctenoplusia albostriata | MOTU44 | Spilosoma lubricipeda |
| MOTU12 | Peridea lativitta 侧带内斑舟蛾 | MOTU45 | Parapediasia teterrellus |
| MOTU13 | Thyas juno 肖毛翅夜蛾 | MOTU46 | Adoxophyes orana |
| MOTU14 | Nephopterix fumella | MOTU47 | Simplicia rectalis 黑点贫夜蛾 |
| MOTU15 | Spodoptera exigua 甜菜夜蛾 | MOTU48 | Glyptoteles leucacrinella 亮雕斑螟 |
| MOTU16 | Oglasa consanguis | MOTU49 | Somena scintillans |
| MOTU17 | Diaphania perspectalis 黄杨绢野螟 | MOTU50 | Anadevidia peponis 葫芦夜蛾 |
| MOTU18 | Athetis lepigone | MOTU51 | Hypocala subsatura 苹梢鹰夜蛾 |
| MOTU19 | Miyakea raddeella | MOTU52 | Choristoneura diversana 异色卷蛾 |
| MOTU20 | Herpetogramma pseudomagna 狭翅切叶野螟 | MOTU53 | Choristoneura luticostana 棕色卷蛾 |
| MOTU21 | Axylia putris 朽木夜蛾 | MOTU54 | Archips betulana; Archips podana |
| MOTU22 | Mocis ancilla | MOTU55 | Palpita nigropunctalis 白蜡绢须野螟 |
| MOTU23 | Mecyna tricolor | MOTU56 | Epiblema foenella 白钩小卷蛾 |
| MOTU24 | Mamestra brassicae 甘蓝夜蛾 | MOTU57 | Grammodes geometrica 象夜蛾 |
| MOTU25 | Xanthorhoe quadrifasiata | MOTU58; MOTU60 | Plusia nadeja |
| MOTU26 | Euproctis similis | MOTU59 | Sphinx caligineus |
| MOTU27 | Zanclognatha lunalis 朽镰须夜蛾 | MOTU61 | Acronicta rumicis 梨剑纹夜蛾 |
| MOTU28 | Theretra japonica | MOTU62 | Herminia tarsipennalis |
| MOTU29 | Herminia tarsicrinalis | MOTU63 | Pangrapta trimantesalis |
| MOTU30 | Eremodrina morosa | MOTU64 | Eutelia hamulatrix 沟尾夜蛾 |
| MOTU31 | Helicoverpa armigera 棉铃虫 | MOTU65 | Plusilla rosalia |
| MOTU32 | Nomophila noctuella | MOTU66 | Bertula bistrigata |
| MOTU33 | Dysgonia mandschuriana | MOTU67 | Gastropacha populifolia 杨枯叶蛾 |
表1 ABGD方法MOTU分组结果与形态种类对照
Table 1 The comparison of ABGD method with morphospecies
| MOTU① | 种名 Species② | MOTU | 种名 Species |
|---|---|---|---|
| MOTU01 | Lacanobia contigua | MOTU34 | Parapoynx vittalis |
| MOTU02 | Agrotis segetum 黄地老虎 | MOTU35 | Heliothis assulta 烟青虫 |
| MOTU03 | Conogethes punctiferalis 桃蛀螟 | MOTU36 | Herminia grisealis |
| MOTU04 | Nola cicatricalis | MOTU37 | Spodoptera depravata |
| MOTU05 | Hipoepa fractalis 中影单跗夜蛾 | MOTU38 | Scopula subpunctaria |
| MOTU06 | Oraesia lata 平嘴壶夜蛾 | MOTU39 | Diaphania indica 瓜绢野螟 |
| MOTU07 | Spodoptera litura 斜纹夜蛾 | MOTU40 | Botyodes diniasalis 黄翅缀叶野螟 |
| MOTU08 | Uropyia meticulodina 核桃美舟蛾 | MOTU41 | Emmelia trabealis 谐夜蛾 |
| MOTU09 | Evergestis extimalis | MOTU42 | Termioptycha nigrescens |
| MOTU10 | Mythimna separate | MOTU43 | Ipimorpha subtusa 杨逸色夜蛾 |
| MOTU11 | Ctenoplusia albostriata | MOTU44 | Spilosoma lubricipeda |
| MOTU12 | Peridea lativitta 侧带内斑舟蛾 | MOTU45 | Parapediasia teterrellus |
| MOTU13 | Thyas juno 肖毛翅夜蛾 | MOTU46 | Adoxophyes orana |
| MOTU14 | Nephopterix fumella | MOTU47 | Simplicia rectalis 黑点贫夜蛾 |
| MOTU15 | Spodoptera exigua 甜菜夜蛾 | MOTU48 | Glyptoteles leucacrinella 亮雕斑螟 |
| MOTU16 | Oglasa consanguis | MOTU49 | Somena scintillans |
| MOTU17 | Diaphania perspectalis 黄杨绢野螟 | MOTU50 | Anadevidia peponis 葫芦夜蛾 |
| MOTU18 | Athetis lepigone | MOTU51 | Hypocala subsatura 苹梢鹰夜蛾 |
| MOTU19 | Miyakea raddeella | MOTU52 | Choristoneura diversana 异色卷蛾 |
| MOTU20 | Herpetogramma pseudomagna 狭翅切叶野螟 | MOTU53 | Choristoneura luticostana 棕色卷蛾 |
| MOTU21 | Axylia putris 朽木夜蛾 | MOTU54 | Archips betulana; Archips podana |
| MOTU22 | Mocis ancilla | MOTU55 | Palpita nigropunctalis 白蜡绢须野螟 |
| MOTU23 | Mecyna tricolor | MOTU56 | Epiblema foenella 白钩小卷蛾 |
| MOTU24 | Mamestra brassicae 甘蓝夜蛾 | MOTU57 | Grammodes geometrica 象夜蛾 |
| MOTU25 | Xanthorhoe quadrifasiata | MOTU58; MOTU60 | Plusia nadeja |
| MOTU26 | Euproctis similis | MOTU59 | Sphinx caligineus |
| MOTU27 | Zanclognatha lunalis 朽镰须夜蛾 | MOTU61 | Acronicta rumicis 梨剑纹夜蛾 |
| MOTU28 | Theretra japonica | MOTU62 | Herminia tarsipennalis |
| MOTU29 | Herminia tarsicrinalis | MOTU63 | Pangrapta trimantesalis |
| MOTU30 | Eremodrina morosa | MOTU64 | Eutelia hamulatrix 沟尾夜蛾 |
| MOTU31 | Helicoverpa armigera 棉铃虫 | MOTU65 | Plusilla rosalia |
| MOTU32 | Nomophila noctuella | MOTU66 | Bertula bistrigata |
| MOTU33 | Dysgonia mandschuriana | MOTU67 | Gastropacha populifolia 杨枯叶蛾 |
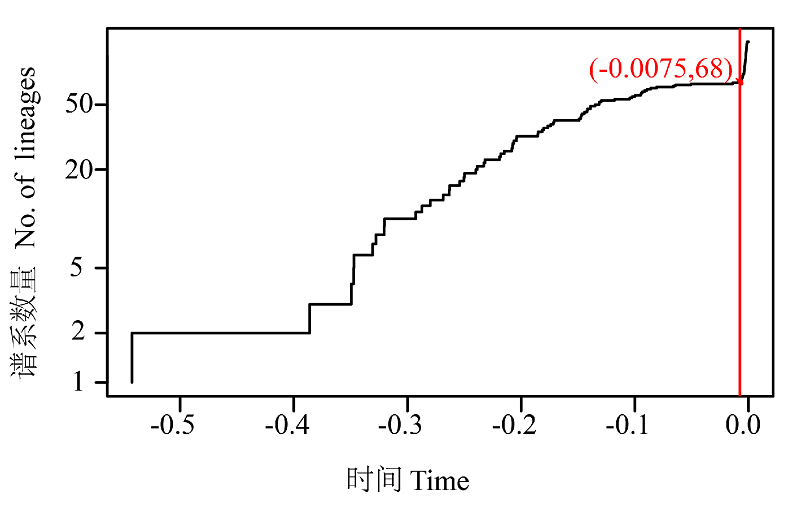
图2 121条单倍型基于超度量树绘制的时间-谱系数量关系图。曲线表示支长速率, 支长速率急剧上升的变化转折点对应物种界定的数量。
Fig. 2 Lineages-through-time plot based on the time calibrated tree obtained from all 121 haplotypes. The sharp increase in branching rate, corresponding to the transition from interspecies to intraspecies branching events.
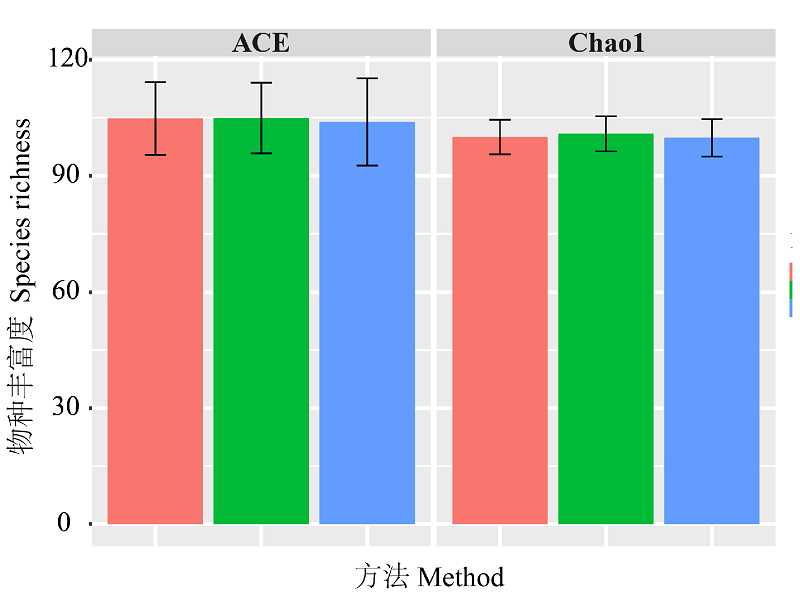
图3 基于不同方法的物种丰富度估计结果。粉红色表示基于ABGD方法的结果, 绿色表示基于GMYC方法的结果, 蓝色表示形态学结果, 黑色误差线表示置信区间。
Fig. 3 Species richness estimation results with different methods. Pink colour represents ABGD-based method. Green colour represents GMYC-based method. Blue colour represents morphology-based method. Black error bar indicates confidence interval.
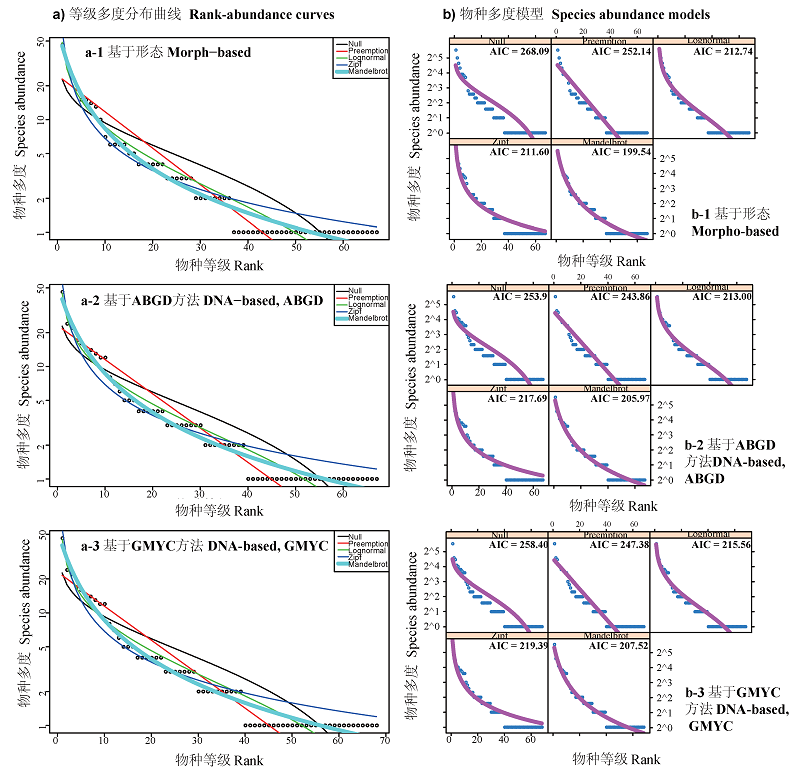
图4 等级多度分布曲线及模型拟合。a-1) 基于形态的等级多度分布曲线, a-2) 基于ABGD方法的等级多度分布曲线, a-3) 基于GMYC方法的等级多度分布曲线; b-1) 基于形态学的物种多度模型, b-2) 基于ABGD方法的物种多度模型, b-3) 基于GMYC方法的物种多度模型。黑点表示实际观测数据, 黑色曲线是零模型断棍模型, 红色曲线是生态位优先占领模型, 绿色曲线是正态分布模型, 深蓝色曲线是Zipf模型, 淡蓝色曲线是Zipf-Mandelbrot模型。
Fig. 4 Rank-abundance curves and fittings of five models. a-1)Species rank-abundance curves based on morphological method, a-2) Species rank-abundance curves based on ABGD method, a-3) Species rank-abundance curves based on GMYC method; b-1) Species abundance models based on morphological method, b-2) Species abundance models based on ABGD method, b-3) Species abundance models based on GMYC method. Black points represent the real abundance data. Black curve represents broken stick model. Red curve represents preemption model. Green curve represents log normal model. Dark blue curve represents Zipf model. Light blue curve represents Zipf-Mandelbrot model.
| 1 | Austerlitz F, David O, Schaeffer B, Bleakley K, Olteanu M, Leblois R, Veuille M, Laredo C (2009) DNA barcode analysis: a comparison of phylogenetic and statistical classification methods. BMC Bioinformatics, 10(Suppl. 14), 1-13. |
| 2 | Blaxter M, Floyd R (2003) Molecular taxonomics for biodiversity surveys: already a reality. Trends in Ecology and Evolution, 18, 268-269. |
| 3 | Boykin LM, Armstrong KF, Kubatko L, Barro PD (2012) Species delimitation and global biosecurity. Evolutionary Bioinformatics, 8, 1-37. |
| 4 | Burnham KP, Anderson DR (2002) Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. Springer-Verlag, New York. |
| 5 | Chao A (1984) Nonparametric estimation of the number of classes in a population. Scandinavian Journal of Statistics, 11, 265-270. |
| 6 | Chao A, Hwang WH, Chen YC, Kuo CY (2000) Estimating the number of shared species in two communities. Statistica Sinica, 10, 227-246. |
| 7 | Colwell RK, Coddington JA (1994) Estimating terrestrial biodiversity through extrapolation. Philosophical Transactions of the Royal Society B: Biological Sciences, 345, 101-118. |
| 8 | Darriba D, Taboada GL, Doallo R, Posada D (2012) jModel- Test 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772. |
| 9 | Decaëns T, Porco D, James SW, Brown G, Da SE, Dupont L, Lapied E, Rougerie R, Taberlet P, Roy V (2015) Dissecting tropical earthworm biodiversity patterns in tropical rainforests through the use of DNA barcoding. Genome, 58, 210. |
| 10 | Dincă V, Zakharov EV, Hebert PDN, Vila R (2011) Complete DNA barcode reference library for a country’s butterfly fauna reveals high performance for temperate Europe. Proceedings of the Royal Society B: Biological Sciences, 278, 347-355. |
| 11 | Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biology, 4, e88. |
| 12 | Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214. |
| 13 | Ebach MC, Holdrege C (2005) DNA barcoding is no substitute for taxonomy. Nature, 434, 697. |
| 14 | Ekrem T, Willassen E, Stur E (2007) A comprehensive DNA sequence library is essential for identification with DNA barcodes. Molecular Phylogenetics and Evolution, 43, 530-542. |
| 15 | Fujisawa T, Barraclough TG (2013) Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent (GMYC) approach: a revised method and evaluation on simulated datasets. Systematic Biology, 62, 707-724. |
| 16 | Gomez-Alvarez V, King GM, Nüsslein K (2007) Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiology Ecology, 60, 60-73. |
| 17 | Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences, 270, 313-321. |
| 18 | Hebert PDN, Humble LM (2011) A comprehensive DNA barcode library for the looper moths (Lepidoptera: Geometridae) of British Columbia, Canada. PLoS ONE, 6, e18290. |
| 19 | Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences, USA, 101, 14812-14817. |
| 20 | Herrera A, Héry M, Stach JEM, Jaffré T, Normand P, Navarro E (2007) Species richness and phylogenetic diversity comparisons of soil microbial communities affected by nickel-mining and revegetation efforts in New Caledonia. European Journal of Soil Biology, 43, 130-139. |
| 21 | Hickerson MJ, Meyer CP, Moritz C (2006) DNA barcoding will often fail to discover new animal species over broad parameter space. Systematic Biology, 55, 729-739. |
| 22 | Jin Q, He LJ, Zhang AB (2012) A simple 2D non-parametric resampling statistical approach to assess confidence in species identification in DNA barcoding—an alternative to Likelihood and Bayesian approaches. PLoS ONE, 7, e50831. |
| 23 | Jin Q, Han HL, Hu XM, Li XH, Zhu CD, Ho SYW, Ward RD, Zhang AB (2013) Quantifying species diversity with a DNA barcoding-based method: Tibetan moth species (Noctuidae) on the Qinghai-Tibetan Plateau. PLoS ONE, 8, e64428. |
| 24 | Jin Q, Zhang AB (2013) Distance-based DNA barcoding methods for insects. Chinese Journal of Applied Entomology, 50, 283-287. (in Chinese) |
| [金倩, 张爱兵 (2013) 昆虫DNA条形码分析中的距离方法. 应用昆虫学报, 50, 283-287.] | |
| 25 | Kempton RA, Taylor LR (1974) Log-series and log-normal parameters as diversity discriminants for the Lepidoptera. The Journal of Animal Ecology, 43, 381-399. |
| 26 | Leray M, Knowlton N (2015) DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity. Proceedings of the National Academy of Sciences, USA, 112, 2076-2081. |
| 27 | Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S (2015) Plant DNA barcoding: from gene to genome. Biological Reviews, 90, 157-166. |
| 28 | MacArthur RH (1957) On the relative abundance of bird species. Proceedings of the National Academy of Sciences, USA, 43, 293-295. |
| 29 | Mandelbrot BB (1983) The Fractal Geometry of Nature. W.H. Freeman and Company, San Francisco. |
| 30 | May RM (1988) How many species are there on earth? Science, 241, 1441-1449. |
| 31 | Meyer CP, Paulay G (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biology, 3, e422. |
| 32 | O’hara RB (2005) Species richness estimators: how many species can dance on the head of a pin? Journal of Animal Ecology, 74, 375-386. |
| 33 | Oline DK (2006) Phylogenetic comparisons of bacterial communities from serpentine and nonserpentine soils. Applied and Environmental Microbiology, 72, 6965-6971. |
| 34 | Ovreas L (2000) Population and community level approaches for analyzing microbial diversity in natural environments. Ecology Letters, 3, 236-251. |
| 35 | Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology, 55, 595-609. |
| 36 | Preston FW (1948) The commonness, and rarity, of species. Ecology, 29, 254-283. |
| 37 | Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology, 21, 1864-1877. |
| 38 | Qin J, Zhang YZ, Zhou X, Kong XB, Wei SJ, Ward RD, Zhang AB (2015) Mitochondrial phylogenomics and genetic relationships of closely related pine moth (Lasiocampidae: Dendrolimus) species in China, using whole mitochondrial genomes. BMC Genomics, 16, 428. |
| 39 | Quicke DLJ, Smith MA, Janzen DH, Hallwachs W, Fernandez-Triana J, Laurenne NM, Zaldivar-Riveron A, Shaw MR, Broad GR, Klopfstein S, Shaw SR, Hrcek J, Hebert PDN, Miller SE, Rodriguez JJ, Whitfield JB, Sharkey MJ, Sharanowski BJ, Jussila R, Gauld ID, Chesters D, Vogler AP (2012) Utility of the DNA barcoding gene fragment for parasitic wasp phylogeny (Hymenoptera: Ichneumonoidea): data release and new measure of taxonomic congruence. Molecular Ecology Resources, 12, 676-685. |
| 40 | Rambaut A, Drummond AJ (2007) Tracer v1.4: MCMC Trace Analyses Tool. . |
| 41 | Ratnasingham S, Hebert PD (2013) A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE, 8, e66213. |
| 42 | Rubinoff D (2006) DNA barcoding evolves into the familiar. Conservation Biology, 20, 1548-1549. |
| 43 | Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406-425. |
| 44 | Schindel DE, Miller SE (2005) DNA barcoding a useful tool for taxonomists. Nature, 435, 17. |
| 45 | Smith MA, Fisher BL, Hebert PDN (2005) DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar. Philosophical Transactions of the Royal Society B: Biological Sciences, 360, 1825-1834. |
| 46 | Tokeshi M (1990) Niche apportionment or random assortment: species abundance patterns revisited. The Journal of Animal Ecology, 10, 1129-1146. |
| 47 | Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294-299. |
| 48 | Ward RD, Hanner R, Hebert PDN (2009) The campaign to DNA barcode all fishes, FISH-BOL. Journal of Fish Biology, 74, 329-356. |
| 49 | Wiemers M, Fiedler K (2007) Does the DNA barcoding gap exist? — a case study in blue butterflies (Lepidoptera: Lycaenidae). Frontiers in Zoology, 4, 8. |
| 50 | Wilson JB (1991) Methods for fitting dominance/diversity curves. Journal of Vegetation Science, 2, 35-46. |
| 51 | Zhang AB, Feng J, Ward RD, Wan P, Gao Q, Wu J, Zhao WZ (2012a) A new method for species identification via protein-coding and non-coding DNA barcodes by combining machine learning with bioinformatic methods. PLoS ONE, 7, e30986. |
| 52 | Zhang AB, Muster C, Liang HB, Zhu CD, Crozier R, Wan P, Feng J, Ward RD (2012b) A fuzzy-set-theory-based approach to analyse species membership in DNA barcoding. Molecular Ecology, 21, 1848-1863. |
| 53 | Zhang AB, Sikes DS, Muster C, Li SQ (2008) Inferring species membership using DNA sequences with back-propagation neural networks. Systematic Biology, 57, 202-215. |
| 54 | Zipf GK (1949) Human Behavior and the Principle of Least Effort. Hafner, New York. |
| [1] | 贾贞妮, 张意岑, 杜彦君, 任海保. 干扰对中亚热带森林群落物种多样性演替动态的影响[J]. 生物多样性, 2025, 33(2): 24078-. |
| [2] | 孟敬慈, 王国栋, 曹光兰, 胡楠林, 赵美玲, 赵延彤, 薛振山, 刘波, 朴文华, 姜明. 中国芦苇沼泽植物物种丰富度分布格局及其驱动因素[J]. 生物多样性, 2024, 32(2): 23194-. |
| [3] | 施国杉, 刘峰, 曹光宏, 陈典, 夏尚文, 邓云, 王彬, 杨效东, 林露湘. 西双版纳热带季节雨林木本植物的beta多样性: 空间、环境与林分结构的作用[J]. 生物多样性, 2024, 32(12): 24285-. |
| [4] | 努日耶·木合太尔, 张秀英, 苏比奴尔·艾力, 李后魂. 中国鳞翅目新物种2023年度报告[J]. 生物多样性, 2024, 32(11): 24428-. |
| [5] | 王丽媛, 胡慧建, 姜杰, 胡一鸣. 南岭哺乳类和鸟类物种丰富度空间分布格局及其影响因子[J]. 生物多样性, 2024, 32(1): 23026-. |
| [6] | 罗小燕, 李强, 黄晓磊. 戴云山国家级自然保护区访花昆虫DNA条形码数据集[J]. 生物多样性, 2023, 31(8): 23236-. |
| [7] | 刘志发, 王新财, 龚粤宁, 陈道剑, 张强. 基于红外相机监测的广东南岭国家级自然保护区鸟兽多样性及其垂直分布特征[J]. 生物多样性, 2023, 31(8): 22689-. |
| [8] | 邢超, 林依, 周智强, 赵联军, 蒋仕伟, 林蓁蓁, 徐基良, 詹祥江. 基于DNA条形码技术构建王朗国家级自然保护区陆生脊椎动物遗传资源数据库及物种鉴定[J]. 生物多样性, 2023, 31(7): 22661-. |
| [9] | 陈声文, 任海保, 童光蓉, 王宁宁, 蓝文超, 薛建华, 米湘成. 钱江源国家公园木本植物物种多样性空间分布格局[J]. 生物多样性, 2023, 31(7): 22587-. |
| [10] | 吴帆, 刘深云, 江虎强, 王茜, 陈开威, 李红亮. 中华蜜蜂和意大利蜜蜂秋冬期传粉植物多样性比较[J]. 生物多样性, 2023, 31(5): 22528-. |
| [11] | 谢艳秋, 黄晖, 王春晓, 何雅琴, 江怡萱, 刘子琳, 邓传远, 郑郁善. 福建海岛滨海特有植物种-面积关系及物种丰富度决定因素[J]. 生物多样性, 2023, 31(5): 22345-. |
| [12] | 张秀英, 苏比奴尔·艾力, 李后魂. 中国鳞翅目新物种2022年度报告[J]. 生物多样性, 2023, 31(10): 23283-. |
| [13] | 古丽扎尔·阿不都克力木, 张秀英, 苏比奴尔·艾力, 李后魂. 中国鳞翅目新物种2021年年度报告[J]. 生物多样性, 2022, 30(8): 22191-. |
| [14] | 杨科, 丁城志, 陈小勇, 丁刘勇, 黄敏睿, 陈晋南, 陶捐. 怒江流域鱼类多样性及空间分布格局[J]. 生物多样性, 2022, 30(7): 21334-. |
| [15] | 田璐嘉, 杨小波, 李东海, 李龙, 陈琳, 梁彩群, 张培春, 李晨笛. 海口和三亚两城市破碎化林地中鸟类群落多样性与嵌套分布格局[J]. 生物多样性, 2022, 30(6): 21424-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()