



生物多样性 ›› 2013, Vol. 21 ›› Issue (1): 28-37. DOI: 10.3724/SP.J.1003.2013.10097 cstr: 32101.14.SP.J.1003.2013.10097
收稿日期:2012-04-11
接受日期:2012-08-10
出版日期:2013-01-20
发布日期:2013-02-04
通讯作者:
王健鑫
基金资助:
Jianyu He, Xuezhu Liu, Rongtao Zhao, Fangwei Wu, Jianxin Wang*( )
)
Received:2012-04-11
Accepted:2012-08-10
Online:2013-01-20
Published:2013-02-04
Contact:
Wang Jianxin
摘要:
于2011年7月采集了浙江舟山沿岸海底沉积物样品(122°10′41″ E, 29°49′7″ N), 通过埋片原位观察、荧光显微计数、纯培养菌分离以及非培养细菌构建克隆文库的方法, 分析和研究了东海表层沉积物细菌群落结构及其多样性。埋片原位观察和荧光显微计数法的结果表明: 沉积物样品中细菌的细胞数量为(9.30±3.44) ×107 cells/g; 通过分离纯化共获得313株细菌, 分属于20个属, 4种培养基对细菌的分离效果依次为RO>M1>Zobell 2216>MR2A; 常规形态学与生理生化鉴定结果显示芽孢杆菌属(Bacillus)和海球菌属(Marinococcus)从数量和种类上皆为优势菌属, 分别占分离菌株总数的21.08%和17.25%; 对73株典型海洋细菌进行16S rDNA分子鉴定发现, 厚壁菌门(57.5%)、γ-变形杆菌纲(32.9%)、黄杆菌纲(4.1%)和放线菌纲(5.5%)等为主要类群。非培养细菌克隆文库序列分析发现: 细菌克隆子主要属于厚壁菌门和变形杆菌门, 而芽孢杆菌纲和γ-变形杆菌纲是上述两个门的优势类群。综合纯培养与非培养数据得出东海海域表层沉积物细菌多样性丰富, 具有进一步开发研究的价值。
何建瑜, 刘雪珠, 赵荣涛, 吴方伟, 王健鑫 (2013) 东海表层沉积物纯培养与非培养细菌多样性. 生物多样性, 21, 28-37. DOI: 10.3724/SP.J.1003.2013.10097.
Jianyu He,Xuezhu Liu,Rongtao Zhao,Fangwei Wu,Jianxin Wang (2013) Diversity of cultured and uncultured bacteria in surface layer sediment from the East China Sea. Biodiversity Science, 21, 28-37. DOI: 10.3724/SP.J.1003.2013.10097.
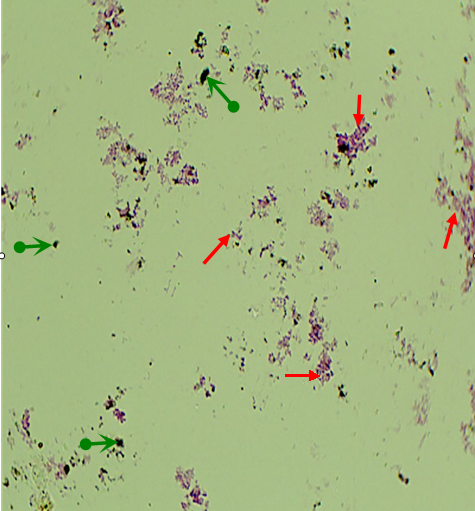
图1 采用埋片观察法得到的东海表层沉积物样品中微生物和有机质的原位分布图。红色箭头为微生物菌体, 绿色箭头为有机杂质。
Fig. 1 Direct observation of the sediment from East China Sea surface by the method of buried piece to find the distribution of microorganisms and organics in situ. Red arrows indicate microorganisms, and green arrows indicate organics.
| 培养基 Medium | 稀释倍数 Dilution multiple | 平板菌落数 No. of plate culture count | 菌落单元数量 Numbers of colony- forming units(CFU/g) |
|---|---|---|---|
| M1 | 1,000 | 73 | (7.30±.0.14)×104 |
| Zobell 2216 | 1,000 | 71 | (7.10±0.08)×104 |
| MR2A | 1,000 | 59 | (5.90±0.09)×104 |
| RO | 10,000 | 49 | (4.90±0.17)×105 |
表1 在4种培养基中利用稀释平板计数的方法统计纯培养的菌落单元总量
Table 1 Statistics of colony-forming units in four kinds of medias by the dilution method of plate counting
| 培养基 Medium | 稀释倍数 Dilution multiple | 平板菌落数 No. of plate culture count | 菌落单元数量 Numbers of colony- forming units(CFU/g) |
|---|---|---|---|
| M1 | 1,000 | 73 | (7.30±.0.14)×104 |
| Zobell 2216 | 1,000 | 71 | (7.10±0.08)×104 |
| MR2A | 1,000 | 59 | (5.90±0.09)×104 |
| RO | 10,000 | 49 | (4.90±0.17)×105 |
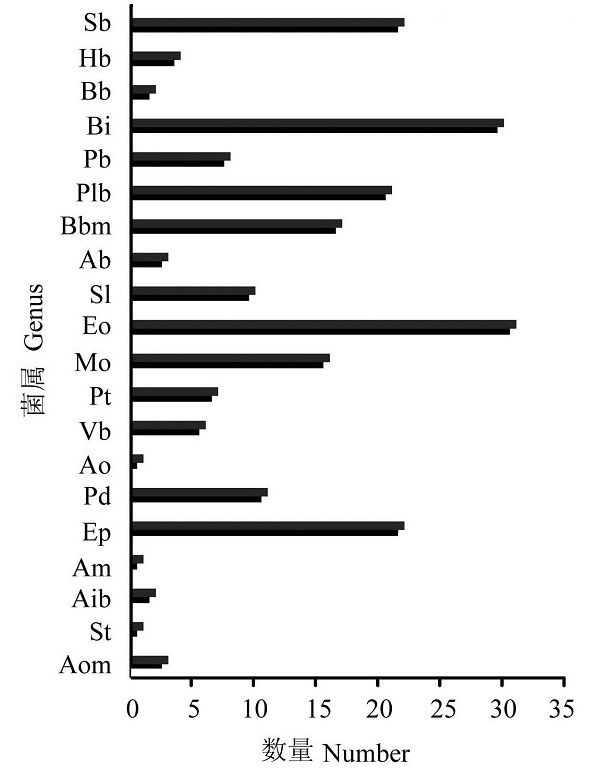
图2 不同菌属菌株数量分布图。Sb: 芽孢乳杆菌属; Hb: 盐芽孢杆菌属; Bb: 短芽孢杆菌属; Bi: 芽孢杆菌属; Pb: 类芽孢杆菌属; Plb: 脂肪杆菌属; Bbm: 短状杆菌属; Ab: 节杆菌属; Sl: 葡萄球菌属; Eo: 肠球菌属; Mo: 海球菌属; Pt: 巴斯德氏菌属; Vb: 弧菌属; Ao: 交替单胞菌属; Pd: 假单胞菌属; Ep: 丹毒丝菌属; Am: 壤霉菌属; Aib: 硫胺素芽孢杆菌属; St: 链球菌属; Aom: 放线菌属
Fig. 2 Distribution of the different genus. Sb, Sporolac- tobacillus; Hb,Halobacillus; Bb, Brevibacillus; Bi, Bacillus; Pb, Paenibacillus; Plb, Pimelobacter; Bbm, Brachybacterium; Ab, Arthrobactor; Sl, Staphylococcus; Eo, Enterococcus; Mo, Marinococcus; Pt, Pasteurella; Vb, Vibrio; Ao, Alteromonas; Pd, Pseudomonas; Ep, Erysipelothrix; Am, Agromyces; Aib, Aneurinibacillus; St, Streptococcus; Aom, Actinomyces.
| 代表菌株 Strain no. | 登录号 Accession no. | 相似菌株(登录号) Closest species (Accession no.) | 相似性 Homology (%) |
|---|---|---|---|
| 芽孢杆菌纲 Bacilli | |||
| DHC01 | JQ904712 | Bacillus pumilus strain PR13 (JQ435673) | 99 |
| DHC02 | JQ904713 | Bacterium YCG4 (JF775413) | 97 |
| DHC03 | JQ904714 | Bacillus tequilensis strain KM35 (JF411314) | 99 |
| DHC04 | JQ904715 | Bacillus sp. M71_S53 (FM992830) | 100 |
| DHC06 | JQ904717 | Bacillus cereus strain CB-10 (JF496530) | 96 |
| DHC07 | JQ904718 | Bacillus subtilis strain B7 (JQ086378) | 100 |
| DHC08 | JQ904719 | Bacillus horikoshii strain NBSL26 (FJ984529) | 99 |
| DHC10 | JQ904720 | Bacillus sp. RHS-str.320 (HE586887) | 97 |
| DHC11 | JQ904721 | Bacillus infantis strain AL10 (JF411305) | 99 |
| DHC12 | JQ904722 | Bacillus aryabhattai isolate PSB59 (HQ242772) | 100 |
| DHC13 | JQ904723 | Bacillus barbaricus strain P2 (GQ200827) | 99 |
| DHC14 | JQ904724 | Bacillus gibsonii strain S-2 (AY737309) | 99 |
| DHC15 | JQ904726 | Paenibacillus sp. GP26-03 (AM162316) | 99 |
| 黄杆菌纲 Flavobacteria | |||
| DHC05 | JQ904716 | Cloacibacterium normanense strain tu33 (FJ544402) | 100 |
| 放线菌纲 Actinobacteria | |||
| DHC09 | JQ904725 | Brevibacterium sp. 210_14 (GQ199716) | 99 |
| γ-变形菌纲 Gamma-proteobacteria | |||
| DHC16 | JQ904727 | Alteromonas sp. SCSWC10 (FJ461444) | 99 |
| DHC17 | JQ904728 | Pseudoalteromonas sp. B296 (FN295786) | 98 |
| DHC18 | JQ904729 | Pseudoalteromonas sp. VS-2 (FJ497709) | 97 |
| DHC19 | JQ904730 | Vibrio algoinfesta strain NBRC 14665 (AB680645) | 99 |
| DHC20 | JQ904731 | Vibrio harveyi EHP8 (FJ227114) | 99 |
| DHC21 | JQ904732 | Vibrio azureus strain CAIM 1457 (JN603238) | 99 |
| DHC22 | JQ904733 | Vibrio parahaemolyticus strain CM12 (EU660326) | 99 |
表2 通过细菌16S rDNA基因序列对东海表层沉积物分离得到的纯培养细菌鉴定结果
Table 2 Identification of pure-cultured bacteria isolated from East China Sea surface sediment by bacterial 16S rDNA gene
| 代表菌株 Strain no. | 登录号 Accession no. | 相似菌株(登录号) Closest species (Accession no.) | 相似性 Homology (%) |
|---|---|---|---|
| 芽孢杆菌纲 Bacilli | |||
| DHC01 | JQ904712 | Bacillus pumilus strain PR13 (JQ435673) | 99 |
| DHC02 | JQ904713 | Bacterium YCG4 (JF775413) | 97 |
| DHC03 | JQ904714 | Bacillus tequilensis strain KM35 (JF411314) | 99 |
| DHC04 | JQ904715 | Bacillus sp. M71_S53 (FM992830) | 100 |
| DHC06 | JQ904717 | Bacillus cereus strain CB-10 (JF496530) | 96 |
| DHC07 | JQ904718 | Bacillus subtilis strain B7 (JQ086378) | 100 |
| DHC08 | JQ904719 | Bacillus horikoshii strain NBSL26 (FJ984529) | 99 |
| DHC10 | JQ904720 | Bacillus sp. RHS-str.320 (HE586887) | 97 |
| DHC11 | JQ904721 | Bacillus infantis strain AL10 (JF411305) | 99 |
| DHC12 | JQ904722 | Bacillus aryabhattai isolate PSB59 (HQ242772) | 100 |
| DHC13 | JQ904723 | Bacillus barbaricus strain P2 (GQ200827) | 99 |
| DHC14 | JQ904724 | Bacillus gibsonii strain S-2 (AY737309) | 99 |
| DHC15 | JQ904726 | Paenibacillus sp. GP26-03 (AM162316) | 99 |
| 黄杆菌纲 Flavobacteria | |||
| DHC05 | JQ904716 | Cloacibacterium normanense strain tu33 (FJ544402) | 100 |
| 放线菌纲 Actinobacteria | |||
| DHC09 | JQ904725 | Brevibacterium sp. 210_14 (GQ199716) | 99 |
| γ-变形菌纲 Gamma-proteobacteria | |||
| DHC16 | JQ904727 | Alteromonas sp. SCSWC10 (FJ461444) | 99 |
| DHC17 | JQ904728 | Pseudoalteromonas sp. B296 (FN295786) | 98 |
| DHC18 | JQ904729 | Pseudoalteromonas sp. VS-2 (FJ497709) | 97 |
| DHC19 | JQ904730 | Vibrio algoinfesta strain NBRC 14665 (AB680645) | 99 |
| DHC20 | JQ904731 | Vibrio harveyi EHP8 (FJ227114) | 99 |
| DHC21 | JQ904732 | Vibrio azureus strain CAIM 1457 (JN603238) | 99 |
| DHC22 | JQ904733 | Vibrio parahaemolyticus strain CM12 (EU660326) | 99 |

图3 根据东海表层沉积物中纯培养得到的细菌菌株16S rDNA基因序列构建的系统发育树
Fig. 3 Phylogenetic tree based on 16S rDNA sequences of pure-cultured clones from East China Sea surface sediment
| 代表克隆子 Clones no. | 登录号 Accession no. | 相似菌株(登录号) Closest species (Accession no.) | 相似菌株分离源 Closest species origin | 相似性 Homology (%) |
|---|---|---|---|---|
| 芽孢杆菌纲 Bacilli | ||||
| DHUC01 | JQ904734 | Bacillus cereus strain E22(GU826149 ) | 巴西土壤 Brazilian soils | 99 |
| DHUC02 | JQ904735 | Bacillus sp. C1(JN571045) | 人造草坪土壤 Chinampa soils | 98 |
| DHUC03 | JQ904736 | Bacillus aquimaris strain T172 (HQ647277) | 东乡野生水稻 Dongxiang wild rice | 99 |
| DHUC04 | JQ904737 | Bacillus subtilis strain MSI-9 (JN993708) | 人参土壤 Ginseng soil | 98 |
| DHUC05 | JQ904738 | Bacillus megaterium strain XAS5-1 (JF496312) | 生物土壤结皮 Biological soil crusts | 100 |
| DHUC06 | JQ904739 | Bacillus subterraneus strain DSM13966T (FR733689) | 普通土壤 Normal soil | 97 |
| DHUC07 | JQ904746 | Bacillus barbaricus strain P2 (GQ200827) | 盐田 Solar salterns | 94 |
| DHUC08 | JQ904740 | Uncultured Bacillus sp. (HQ397100) | 普通土壤 Normal soil | 100 |
| DHUC09 | JQ904741 | Geobacillus stearothermophilus strain DoB50 (JQ359102) | 铁皮石斛 Dendrobium officinale | 99 |
| DHUC10 | JQ904747 | Brevibacillus sp. 100-5 (FJ959373) | 麦田土壤 Wheat field soil | 94 |
| α-变形菌纲 Alpha-proteobacteria | ||||
| DHUC11 | JQ904742 | Uncultured alpha-proteobacterium (FJ205181) | 不活动的热液区沉积物 Inactive hydrothermal field sediments | 95 |
| γ-变形菌纲 Gamma-proteobacteria | ||||
| DHUC12 | JQ904743 | Uncultured Alteromonas sp. (HQ012278) | 海洋 Marine | 100 |
| DHUC13 | JQ904744 | Vibrio harveyi strain EHP8(FJ227114) | 枪乌贼发光器官 Loliginid squid light organs | 100 |
| DHUC14 | JQ904745 | Vibrio hepatarius (NR025491) | 水生动物和海洋环境 Aquatic animals and the marine environment | 98 |
表3 东海表层沉积物非培养细菌的16S rDNA序列在数据库中的同源分类比较以及相似菌株的分离源分析
Table 3 Taxonomic affiliation of clones recovered in the bacterial clone libraries from East China Sea surface sediment and analysis of closest species origin
| 代表克隆子 Clones no. | 登录号 Accession no. | 相似菌株(登录号) Closest species (Accession no.) | 相似菌株分离源 Closest species origin | 相似性 Homology (%) |
|---|---|---|---|---|
| 芽孢杆菌纲 Bacilli | ||||
| DHUC01 | JQ904734 | Bacillus cereus strain E22(GU826149 ) | 巴西土壤 Brazilian soils | 99 |
| DHUC02 | JQ904735 | Bacillus sp. C1(JN571045) | 人造草坪土壤 Chinampa soils | 98 |
| DHUC03 | JQ904736 | Bacillus aquimaris strain T172 (HQ647277) | 东乡野生水稻 Dongxiang wild rice | 99 |
| DHUC04 | JQ904737 | Bacillus subtilis strain MSI-9 (JN993708) | 人参土壤 Ginseng soil | 98 |
| DHUC05 | JQ904738 | Bacillus megaterium strain XAS5-1 (JF496312) | 生物土壤结皮 Biological soil crusts | 100 |
| DHUC06 | JQ904739 | Bacillus subterraneus strain DSM13966T (FR733689) | 普通土壤 Normal soil | 97 |
| DHUC07 | JQ904746 | Bacillus barbaricus strain P2 (GQ200827) | 盐田 Solar salterns | 94 |
| DHUC08 | JQ904740 | Uncultured Bacillus sp. (HQ397100) | 普通土壤 Normal soil | 100 |
| DHUC09 | JQ904741 | Geobacillus stearothermophilus strain DoB50 (JQ359102) | 铁皮石斛 Dendrobium officinale | 99 |
| DHUC10 | JQ904747 | Brevibacillus sp. 100-5 (FJ959373) | 麦田土壤 Wheat field soil | 94 |
| α-变形菌纲 Alpha-proteobacteria | ||||
| DHUC11 | JQ904742 | Uncultured alpha-proteobacterium (FJ205181) | 不活动的热液区沉积物 Inactive hydrothermal field sediments | 95 |
| γ-变形菌纲 Gamma-proteobacteria | ||||
| DHUC12 | JQ904743 | Uncultured Alteromonas sp. (HQ012278) | 海洋 Marine | 100 |
| DHUC13 | JQ904744 | Vibrio harveyi strain EHP8(FJ227114) | 枪乌贼发光器官 Loliginid squid light organs | 100 |
| DHUC14 | JQ904745 | Vibrio hepatarius (NR025491) | 水生动物和海洋环境 Aquatic animals and the marine environment | 98 |
| 1 | Bowman JP, McCuaig RD (2003) Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Applied and Environmental Microbiology, 69, 2463-2483. |
| 2 | Chen C (陈琛), Huang S (黄珊), Wu QH (吴群河), Li RY (李锐仪), Zhang RD (张仁铎) (2010) Factors affecting the DAPI fluorescence direct count in the tidal river sediment. Environmental Science(环境科学), 31, 1918-1925. (in Chinese with English abstract) |
| 3 | Dong HL, Zhang GX, Jiang HC, Yu BS, Chapman LR, Lucas CR, Fields MW (2006) Microbial diversity in sediments of saline Qinghai Lake, China: linking geochemical controls to microbial ecology. Microbial Ecology, 51, 65-82. |
| 4 | Dong XZ (东秀珠), Cai MY (蔡妙英) (2001) Manual for Identification of Common Bacterial System (常见细菌系统鉴定手册). Science Press, Beijing. (in Chinese) |
| 5 | Feng BW, Li XR, Wang JH, Hu ZY, Meng H, Xiang LY, Quan ZX (2009) Bacterial diversity of water and sediment in the Changjiang estuary and coastal area of the East China Sea. FEMS Microbiology Ecology, 70, 236-248. |
| 6 | Gontang EA, Fenical W, Jensen PR (2007) Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Applied and Environmental Microbiology, 73, 3272-3282. |
| 7 | Gray JP, Herwig RP (1996) Phylogenetic analysis of the bacterial communities in marine sediments. Applied and Environmental Microbiology, 62, 4049-4059. |
| 8 | Guo F (郭非), Geng XY (耿绪云), Li X (李翔), Wei JL (魏俊利), Sun JS (孙金生) (2009) Preliminary investigation on the culturable bacteria diversity in Tianjin Coastal Waters. Marine Sciences(海洋科学), 33, 35-40. (in Chinese with English abstract) |
| 9 | Haglund AL, Lantz P, Törnblom E, Tranvik L (2003) Depth distribution of active bacteria and bacterial activity in lake sediment. FEMS Microbiology Ecology, 46, 31-38. |
| 10 | Hill TCJ, Walsh KA, Harris JA, Moffett BF (2003) Using ecological diversity measures with bacterial communities. FEMS Microbiology Ecology, 43, 1-11. |
| 11 | Huo YY (霍颖异), Xu XW (许学伟), Wang CS (王春生), Yang JY (杨俊毅), Wu M (吴敏) (2008) Bacterial diversity of the sediment from Cangnan Large Fishing Bay. Acta Ecologica Sinica(生态学报), 28, 5166-5172. (in Chinese with English abstract) |
| 12 | Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K, Nealson KH, Horikoshi K (2003) Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Applied and Environmental Microbiology, 69, 7224-7235. |
| 13 | Jiao NZ (焦念志) (2006) Marine Microbial Ecology (海洋微型生物生态学). Science Press, Beijing. (in Chinese) |
| 14 | Kepner RL Jr, Pratt JR (1994) Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiological Reviews, 58, 603-615. |
| 15 | Li JB (李家彪) (2008) Donghai Regional Geology (东海区域地质). China Ocean Press, Beijing. (in Chinese) |
| 16 | Li JL, Bai J, Gao HW, Liu GX (2011) Distribution of ammonia-oxidizing Beta-proteobacteria community in surface sediment off the Changjiang River Estuary in summer. Acta Oceanologica Sinica, 30, 92-99. |
| 17 | Li L, Kato C, Horikoshi K (1999) Microbial diversity in sediments collected from the deepest cold-seep area, the Japan Trench. Marine Biotechnology, 1, 391-400. |
| 18 | Li T (李涛), Wang P (王鹏), Wang PX (汪品先) (2008) Microbial diversity in surface sediments of the Xisha Trough, the South China Sea. Acta Ecologica Sinica(生态学报), 28, 1166-1173. (in Chinese with English abstract) |
| 19 | Li ZG (李振高), Luo YM (骆永明), Teng Y (滕应) (2008) Research Methods for Soil and Environmental Microbiology (土壤与环境微生物研究法). Science Press, Beijing. (in Chinese) |
| 20 | Ludwig JA, Reynolds JF (1988) Statistical Ecology. John Wiley & Sons, New York. |
| 21 | Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics, 5, 150-163. |
| 22 | Köster M, Wardenga R, Blume M (2008) Microscale investigations of microbial communities in coastal surficial sediments. Marine Ecology, 29, 89-105. |
| 23 | Neto M, Ohannessian A, Delolme C, Bedell JP (2007) Towards an optimized protocol for measuring global dehydrogenase activity in storm-water sediments. Journal of Soils and Sediments, 7, 101-110. |
| 24 | Park SJ, Park BJ, Pham VH, Yoon DN, Kim SK, Rhee SK (2008) Microeukaryotic diversity in marine environments, an analysis of surface layer sediments from the East Sea. The Journal of Microbiology, 46, 244-249. |
| 25 | Polymenakou PN, Fragkioudaki G, Tselepides A (2007) Bacterial and organic matter distribution in the sediments of the Thracian Sea (NE Aegean Sea). Continental Shelf Research, 27, 2187-2197. |
| 26 | Ramalho R, Cunha J, Teixeira P, Gibbs PA (2001) Improved methods for the enumeration of heterotrophic bacteria in bottled mineral waters. Journal of Microbiological Methods, 44, 97-103. |
| 27 | Ravenschlag K, Sahm K, Amann R (2001) Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard). Applied and Environmental Microbiology, 67, 387-395. |
| 28 | Saha P, Chakrabarti T (2006) Emticicia oligotrophica gen. nov., sp. nov., a new member of the family 'Flexibac- teraceae', phylum Bacteroidetes. International Journal of Systematic and Evolutionary Microbiology, 56, 991-995. |
| 29 | Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Applied and Environmental Microbiology, 71, 1501-1506. |
| 30 | Song JM (宋金明) (2000) Roles of marine sediments diversity in biogenic element. Marine Sciences(海洋科学), 24, 22-26. (in Chinese with English abstract) |
| 31 | Song ZG (宋志刚), Xu QZ (许强芝), Lu XA (鲁心安), Jiao BH (焦炳华), Ai F (艾峰), Yang Y (杨妤), Liu XY (刘小宇), Shi XQ (施晓琼) (2006) A primary study on population biodiversity of marine microorganisms from East China Sea. Microbiology(微生物学通报), 33, 63-67. (in Chinese with English abstract) |
| 32 | Sun FQ (孙风芹), Wang BJ (汪保江), Li GY (李光玉), Liu XP (刘秀片), Du YP (杜雅萍), Lai QL (赖其良), Shao ZZ (邵宗泽) (2008) Diversity of bacteria isolated from the South China Sea sediments. Acta Microbiologica Sinica(微生物学报), 48, 1578-1587. (in Chinese with English abstract) |
| 33 | Sun HM (孙慧敏), Dai SK (戴世鲲), Wang GH (王广华), Xie LW (谢练武), Li X (李翔) (2010) Phylogenetic diversity analysis of bacteria in the deep-sea sediments from the Bashi Channel by 16S rDNA BLAST. Journal of Tropical Oceanography(热带海洋学报), 29, 41-46. (in Chinese with English abstract) |
| 34 | Urakawa H, Kita-Tsukamota K, Ohwada K (1999) Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology, 145, 3305-3315. |
| 35 | Wang JX (王健鑫), Liu XZ (刘雪珠), Li P (李鹏), Chen XL (陈小龙) (2008) Microbes isolated from the intertidal zone of Zhoushan sea area and their antimicrobial activity. Journal of Shanghai Fisheries University(上海水产大学学报), 17, 507-511. (in Chinese with English abstract) |
| 36 | Wang JX (王健鑫), Xu XE (许贤恩), Zhou LL (周链链), Tao S (陶诗), Liu XZ (刘雪珠), Shi G (石戈) (2012) A preliminary study of microbial diversity of the surface layer sediments from the East China Sea. Oceanologia et Limnologia Sinica(海洋与湖沼), 43, 805-813. (in Chinese with English abstract) |
| 37 | Wang SJ (王书锦), Hu JC (胡江春), Xue DL (薛德林), Ma CX (马成新), Xie QH (谢秋宏), Liu QY (刘全永) (2001) Study of marine microbial resources in Huanghai, Bohai and Liaoning offshore of China. Journal of Jinzhou Normal College (Natural Sciences Edition) (锦州师范学院学报(自然科学版)), 22, 1-5. (in Chinese with English abstract) |
| 38 | Xu F (许飞), Dai X (戴欣), Chen YQ (陈月琴), Zhou H (周惠), Cai JH (蔡剑华), Qu LH (屈良鹄) (2004) Phylogenetic diversity of bacteria and archaea in the Nansha marine sediments, as determined by 16S rDNA analysis. Oceanologia et Limnologia Sinica(海洋与湖沼), 35, 89-94. (in Chinese with English abstract) |
| 39 | Xu MX, Wang P, Wang FP, Xiao X (2005) Microbial diversity at a deep-sea station of the Pacific Nodule Province. Biodiversity and Conservation, 14, 3363-3380. |
| 40 | Zeng YH (曾永辉) (2008) Microbial Diversity and Environ- mental Adaptation Mechanisms in Typical Marine Envir- onments (典型海洋环境中浮游细菌多样性及环境适应机制的研究). PhD dissertation, Xiamen University, Xiamen. (in Chinese with English abstract) |
| 41 | Zhang LB (张林宝), Li TG (李铁刚), Dang HY (党宏月), Sharen GW (萨仁高娃), Zhang W (张伟) (2010) Vertical distribution of Archaea in mud wedge sediments located at inner continental shelf of the East China Sea. Earth Science (Journal of China University of Geosciences) (地球科学(中国地质大学学报)), 35, 254-260. (in Chinese with English abstract) |
| 42 | Zheng ZX (郑兆祥), Yuan Y (袁扬), Chen MM (陈蒙蒙), Du C (杜翠), Liu XZ (刘雪珠) (2011) Isolation of marine bacteria from marine mud of the sea floor in Zhoushan Sea area and screening of marine bacteria against Vibrio spp., the pathogens in aquacultue. Journal of Zhejiang Ocean University (Natural Science) (浙江海洋学院学报(自然科学版)), 30, 51-55, 90. (in Chinese with English abstract) |
| 43 | ZoBell CE (1941) Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. Journal of Marine Research, 4, 42-75. |
| [1] | 吴晓晴 张美惠 葛苏婷 李漫淑 宋坤 沈国春 达良俊 张健. 上海近自然林重建过程中木本植物物种多样性与地上生物量的时空动态——以闵行区生态岛为例[J]. 生物多样性, 2025, 33(5): 24444-. |
| [2] | 干靓 刘巷序 鲁雪茗 岳星. 全球生物多样性热点地区大城市的保护政策与优化方向[J]. 生物多样性, 2025, 33(5): 24529-. |
| [3] | 曾子轩 杨锐 黄越 陈路遥. 清华大学校园鸟类多样性特征与环境关联[J]. 生物多样性, 2025, 33(5): 24373-. |
| [4] | 周昊, 王茗毅, 张楚格, 肖治术, 欧阳芳. 昆虫旅馆在独栖蜂多样性保护中的现状与挑战[J]. 生物多样性, 2025, 33(5): 24472-. |
| [5] | 臧明月, 刘立, 马月, 徐徐, 胡飞龙, 卢晓强, 李佳琦, 于赐刚, 刘燕. 《昆明-蒙特利尔全球生物多样性框架》下的中国城市生物多样性保护[J]. 生物多样性, 2025, 33(5): 24482-. |
| [6] | 祝晓雨, 王晨灏, 王忠君, 张玉钧. 城市绿地生物多样性研究进展与展望[J]. 生物多样性, 2025, 33(5): 25027-. |
| [7] | 袁琳, 王思琦, 侯静轩. 大都市地区的自然留野:趋势与展望[J]. 生物多样性, 2025, 33(5): 24481-. |
| [8] | 胡敏, 李彬彬, Coraline Goron. 只绿是不够的: 一个生物多样性友好的城市公园管理框架[J]. 生物多样性, 2025, 33(5): 24483-. |
| [9] | 王欣, 鲍风宇. 基于鸟类多样性提升的南滇池国家湿地公园生态修复效果分析[J]. 生物多样性, 2025, 33(5): 24531-. |
| [10] | 明玥, 郝培尧, 谭铃千, 郑曦. 基于城市绿色高质量发展理念的中国城市生物多样性保护与提升研究[J]. 生物多样性, 2025, 33(5): 24524-. |
| [11] | 徐欢, 辛凤飞, 施宏亮, 袁琳, 薄顺奇, 赵欣怡, 邓帅涛, 潘婷婷, 余婧, 孙赛赛, 薛程. 生态修复技术集成应用对长江口北支生境与鸟类多样性提升效果评估[J]. 生物多样性, 2025, 33(5): 24478-. |
| [12] | 谢淦, 宣晶, 付其迪, 魏泽, 薛凯, 雒海瑞, 高吉喜, 李敏. 草地植物多样性无人机调查的物种智能识别模型构建[J]. 生物多样性, 2025, 33(4): 24236-. |
| [13] | 王太, 宋福俊, 张永胜, 娄忠玉, 张艳萍, 杜岩岩. 河西走廊内陆河水系鱼类多样性及资源现状[J]. 生物多样性, 2025, 33(4): 24387-. |
| [14] | 褚晓琳, 张全国. 演化速率假说的实验验证研究进展[J]. 生物多样性, 2025, 33(4): 25019-. |
| [15] | 张浩斌, 肖路, 刘艳杰. 夜间灯光对外来入侵植物和本地植物群落多样性和生长的影响[J]. 生物多样性, 2025, 33(4): 24553-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn