



生物多样性 ›› 2013, Vol. 21 ›› Issue (1): 71-79. DOI: 10.3724/SP.J.1003.2013.09138 cstr: 32101.14.SP.J.1003.2013.09138
所属专题: 物种形成与系统进化
收稿日期:2012-07-03
接受日期:2012-12-18
出版日期:2013-01-20
发布日期:2013-02-04
通讯作者:
徐刚标
基金资助:
Xueqin Wu, Gangbiao Xu*( ), Yan Liang, Xiangbao Shen
), Yan Liang, Xiangbao Shen
Received:2012-07-03
Accepted:2012-12-18
Online:2013-01-20
Published:2013-02-04
Contact:
Xu Gangbiao
摘要:
迁地保护是珍稀濒危植物保护的重要措施。观光木(Tsoongiodendron odorum)是古老的孑遗树种, 被列为国家二级重点保护植物, 营建迁地保护林是对其进行保护的重要手段, 但已经营建的迁地保护林每个种群目前只保存下来几十株个体。为了评价这些迁地保护林的遗传多样性现状, 作者采用ISSR分子标记对南岭地区观光木3个人工迁地保护种群和4个自然种群的遗传多样性进行了比较。结果显示: 16条ISSR引物共扩增出362个条带, 其中301个为多态条带, 多态条带百分比(P)为83.2%, 各种群P值为37.9-62.2%, 平均为53.1%, 表明观光木在物种和种群水平都具有较高的遗传多样性。自然种群总体P值和Shannon信息指数(I)(80.9%, 0.3629)均高于人工迁地保护种群(66.6%, 0.2990), 说明人工迁地保护种群遗传多样性较低。种群结构分析表明3个人工迁地保护种群均有可能来源于源口自然种群。4个自然种群间遗传分化系数(GST)为0.2495, 表明自然种群间存在显著遗传隔离, 基因流受阻和生态环境差异是造成种群遗传分化的主要原因。在今后的迁地保护工作中, 我们建议从不同生态地理区收集种质材料, 并在不同生态类型区开展迁地保护工作, 同时开展观光木种群生态生殖生物学研究。
吴雪琴, 徐刚标, 梁艳, 申响保 (2013) 南岭地区观光木自然和人工迁地保护种群的遗传多样性. 生物多样性, 21, 71-79. DOI: 10.3724/SP.J.1003.2013.09138.
Xueqin Wu,Gangbiao Xu,Yan Liang,Xiangbao Shen (2013) Genetic diversity of natural and planted populations of Tsoongiodendron odorum from the Nanling Mountains. Biodiversity Science, 21, 71-79. DOI: 10.3724/SP.J.1003.2013.09138.
| 种群 Population codes | 地点 Location | 经度 Longitude | 纬度 Latitude | 海拔 Altitude (m) | 样本大小 Sample size |
|---|---|---|---|---|---|
| 自然种群 Natural populations | |||||
| 弄相山 NXS | 贵州弄相山省级自然保护区 Nongxiangshan Provincial Nature Reserve, Guizhou | 109°12′ E | 25°46′ N | 230 | 21 |
| 源口 YK | 湖南源口省级自然保护区 Yuankou Provincial Nature Reserve, Hunan | 110°59′ E | 24°58′ N | 610 | 26 |
| 连山 LS | 广东连山省级自然保护区 Lianshan Provincial Nature Reserve, Guangdong | 112°01′ E | 24°25′ N | 580 | 28 |
| 南昆山 NKS | 广东南昆山省级自然保护区 Nankunshan Provincial Nature Reserve, Guangdong | 113°52′ E | 23°38′ N | 457 | 32 |
| 人工种群 Planted populations | |||||
| 车八岭 CBL | 广东车八岭国家级自然保护区 Chebaling National Nature Reserve, Guangdong | 114°15′ E | 24°43′ N | 350 | 18 |
| 鼎湖山 DHS | 广东鼎湖山国家级自然保护区 Dinghushan National Nature Reserve, Guangdong | 112°32′ E | 23°10′ N | 245 | 10 |
| 大顶山 DDS | 广东南岭国家级自然保护区大顶山管理处 Dadingshan Management Office, Nanling Nature Reserve, Guangdong | 113°03′ E | 24°43′ N | 516 | 28 |
表1 观光木种群采集信息及样本大小
Table 1 Collection location and number of individuals tested of Tsoongiodendron odorum
| 种群 Population codes | 地点 Location | 经度 Longitude | 纬度 Latitude | 海拔 Altitude (m) | 样本大小 Sample size |
|---|---|---|---|---|---|
| 自然种群 Natural populations | |||||
| 弄相山 NXS | 贵州弄相山省级自然保护区 Nongxiangshan Provincial Nature Reserve, Guizhou | 109°12′ E | 25°46′ N | 230 | 21 |
| 源口 YK | 湖南源口省级自然保护区 Yuankou Provincial Nature Reserve, Hunan | 110°59′ E | 24°58′ N | 610 | 26 |
| 连山 LS | 广东连山省级自然保护区 Lianshan Provincial Nature Reserve, Guangdong | 112°01′ E | 24°25′ N | 580 | 28 |
| 南昆山 NKS | 广东南昆山省级自然保护区 Nankunshan Provincial Nature Reserve, Guangdong | 113°52′ E | 23°38′ N | 457 | 32 |
| 人工种群 Planted populations | |||||
| 车八岭 CBL | 广东车八岭国家级自然保护区 Chebaling National Nature Reserve, Guangdong | 114°15′ E | 24°43′ N | 350 | 18 |
| 鼎湖山 DHS | 广东鼎湖山国家级自然保护区 Dinghushan National Nature Reserve, Guangdong | 112°32′ E | 23°10′ N | 245 | 10 |
| 大顶山 DDS | 广东南岭国家级自然保护区大顶山管理处 Dadingshan Management Office, Nanling Nature Reserve, Guangdong | 113°03′ E | 24°43′ N | 516 | 28 |
| 引物 Primer | 序列(5′ to 3′) Sequence(5′ to 3′) | 最适温度 Optimum temperature (℃) | 总条带数 No. of bands | 多态条带数 No. of polymorphic bands | 多态条带百分比 % of polymorphic bands |
|---|---|---|---|---|---|
| UBC814 | (CT)8A | 51 | 28 | 25 | 89.3 |
| UBC815 | (CT)8G | 53 | 21 | 20 | 95.2 |
| UBC818 | (CA)8G | 54 | 22 | 19 | 86.4 |
| UBC822 | (TC)8A | 53 | 25 | 19 | 76.0 |
| UBC824 | (TC)8G | 51 | 25 | 20 | 80.0 |
| UBC835 | (AG)8YC | 55 | 23 | 17 | 73.9 |
| UBC840 | (GA)8YT | 57 | 25 | 20 | 80.0 |
| UBC841 | (GA)8YC | 57 | 26 | 20 | 76.9 |
| UBC844 | (CT)8RC | 57 | 21 | 16 | 76.2 |
| UBC845 | (CT)8RG | 54 | 18 | 14 | 77.8 |
| UBC853 | (TC)8RT | 54 | 18 | 16 | 88.9 |
| UBC854 | (TC)8RG | 54 | 23 | 21 | 91.3 |
| UBC866 | (CTC)6 | 60 | 23 | 18 | 78.3 |
| UBC874 | (CCCT)4 | 55 | 23 | 21 | 91.3 |
| UBC879 | (CTTCA)3 | 41 | 22 | 20 | 90.9 |
| UBC881 | (GGGTG)3 | 54 | 19 | 15 | 79.0 |
| 平均 Mean | 22.63 | 18.82 | 83.2 |
表2 ISSR引物序列及扩增结果
Table 2 ISSR primers and their amplification results with 163 Tsoongiodendron odorum individuals
| 引物 Primer | 序列(5′ to 3′) Sequence(5′ to 3′) | 最适温度 Optimum temperature (℃) | 总条带数 No. of bands | 多态条带数 No. of polymorphic bands | 多态条带百分比 % of polymorphic bands |
|---|---|---|---|---|---|
| UBC814 | (CT)8A | 51 | 28 | 25 | 89.3 |
| UBC815 | (CT)8G | 53 | 21 | 20 | 95.2 |
| UBC818 | (CA)8G | 54 | 22 | 19 | 86.4 |
| UBC822 | (TC)8A | 53 | 25 | 19 | 76.0 |
| UBC824 | (TC)8G | 51 | 25 | 20 | 80.0 |
| UBC835 | (AG)8YC | 55 | 23 | 17 | 73.9 |
| UBC840 | (GA)8YT | 57 | 25 | 20 | 80.0 |
| UBC841 | (GA)8YC | 57 | 26 | 20 | 76.9 |
| UBC844 | (CT)8RC | 57 | 21 | 16 | 76.2 |
| UBC845 | (CT)8RG | 54 | 18 | 14 | 77.8 |
| UBC853 | (TC)8RT | 54 | 18 | 16 | 88.9 |
| UBC854 | (TC)8RG | 54 | 23 | 21 | 91.3 |
| UBC866 | (CTC)6 | 60 | 23 | 18 | 78.3 |
| UBC874 | (CCCT)4 | 55 | 23 | 21 | 91.3 |
| UBC879 | (CTTCA)3 | 41 | 22 | 20 | 90.9 |
| UBC881 | (GGGTG)3 | 54 | 19 | 15 | 79.0 |
| 平均 Mean | 22.63 | 18.82 | 83.2 |
| 种群 Population | 多态条带百分比 % of polymorphic bands | Shannon信息指数 Shannon information index | |
|---|---|---|---|
| 自然种群 Natural populations | |||
| 弄相山 NXS | 59.4 | 0.2848 | |
| 源口 YK | 55.0 | 0.2618 | |
| 连山 LS | 61.9 | 0.2995 | |
| 南昆山 NKS | 62.2 | 0.2926 | |
| 总体 Total | 80.9 | 0.3629 | |
| 人工种群 Planted populations | |||
| 车八岭 CBL | 47.2 | 0.2388 | |
| 鼎湖山 DHS | 37.9 | 0.2137 | |
| 大顶山 DDS | 48.3 | 0.2473 | |
| 总体 Total | 66.6 | 0.2990 | |
| 平均 Mean level | 53.1 | 0.2626 | |
| 物种 Species level | 83.2 | 0.3621 | |
表3 7个观光木种群遗传多样性分析
Table 3 Genetic diversity among the seven populations of Tsoongiodendron odorum based on ISSR markers
| 种群 Population | 多态条带百分比 % of polymorphic bands | Shannon信息指数 Shannon information index | |
|---|---|---|---|
| 自然种群 Natural populations | |||
| 弄相山 NXS | 59.4 | 0.2848 | |
| 源口 YK | 55.0 | 0.2618 | |
| 连山 LS | 61.9 | 0.2995 | |
| 南昆山 NKS | 62.2 | 0.2926 | |
| 总体 Total | 80.9 | 0.3629 | |
| 人工种群 Planted populations | |||
| 车八岭 CBL | 47.2 | 0.2388 | |
| 鼎湖山 DHS | 37.9 | 0.2137 | |
| 大顶山 DDS | 48.3 | 0.2473 | |
| 总体 Total | 66.6 | 0.2990 | |
| 平均 Mean level | 53.1 | 0.2626 | |
| 物种 Species level | 83.2 | 0.3621 | |
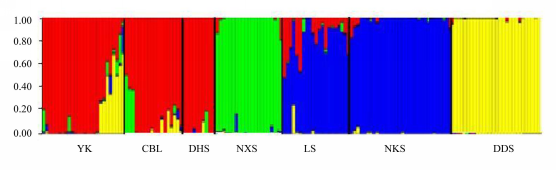
图2 观光木163株个体的遗传结构。纵坐标显示个体所属各类群的比例。种群代号见表1。
Fig. 2 The genetic structure of 163 Tsoongiodendron odorum individuals. The vertical axis shows the proportion of individuals in different clusters. Population codes see Table 1.
| 种群 Population | 弄相山 NXS | 源口 YK | 连山 LS | 南昆山 NKS | 车八岭 CBL | 鼎湖山 DHS | 大顶山 DDS |
|---|---|---|---|---|---|---|---|
| 弄相山 NXS | **** | ||||||
| 源口 YK | 0.2895* | **** | |||||
| 连山 LS | 0.2739* | 0.1858* | **** | ||||
| 南昆山 NKS | 0.3332* | 0.2699* | 0.1581* | **** | |||
| 车八岭 CBL | 0.2657* | 0.2231* | 0.2461* | 0.2982* | **** | ||
| 鼎湖山 DHS | 0.2951* | 0.2041* | 0.2338* | 0.3067* | 0.1544* | **** | |
| 大顶山 DDS | 0.3174* | 0.1784* | 0.2266* | 0.2788* | 0.1867* | 0.2212* | **** |
表4 观光木种群间遗传分化系数(ΦST)估算值
Table 4 Pairwise estimated ΦST values among Tsoongiodendron odorum populations
| 种群 Population | 弄相山 NXS | 源口 YK | 连山 LS | 南昆山 NKS | 车八岭 CBL | 鼎湖山 DHS | 大顶山 DDS |
|---|---|---|---|---|---|---|---|
| 弄相山 NXS | **** | ||||||
| 源口 YK | 0.2895* | **** | |||||
| 连山 LS | 0.2739* | 0.1858* | **** | ||||
| 南昆山 NKS | 0.3332* | 0.2699* | 0.1581* | **** | |||
| 车八岭 CBL | 0.2657* | 0.2231* | 0.2461* | 0.2982* | **** | ||
| 鼎湖山 DHS | 0.2951* | 0.2041* | 0.2338* | 0.3067* | 0.1544* | **** | |
| 大顶山 DDS | 0.3174* | 0.1784* | 0.2266* | 0.2788* | 0.1867* | 0.2212* | **** |
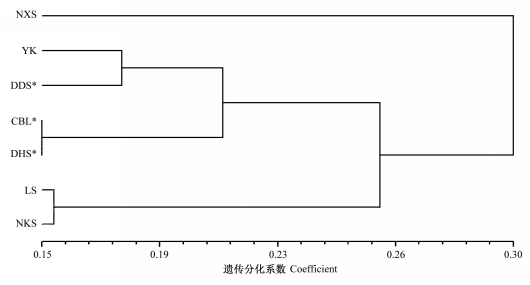
图3 观光木种群间ΦST的UPGMA聚类图。种群代号见表1。*示人工种群。
Fig. 3 Dendrogram of UPGMA based on ΦST of different Tsoongiodendron odorum populations. Population codes see Table 1. * indicates planted populations.
| 1 | Chen T (陈涛), Chang HT (张宏达) (1994) The Floristic geography of Nanling Mountain Range, China. I. Floristic composition and characteristics.Journal of Tropical and Subtropical Botany(热带亚热带植物学报), 2(1), 10-23. (in Chinese with English abstract) |
| 2 | Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication.Cell, 127, 1309-1321. |
| 3 | Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611-2620. |
| 4 | Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.1, an Integrated Software for Population Genetics Data Analysis. Zoological Institute, University of Berne, Berne. |
| 5 | Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479-491. |
| 6 | Falush D, Stephensb M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies.Genetics, 164, 1567-1587. |
| 7 | Ferreira DK, Nazareno AG, Mantovani A, Bittencourt R, Sebbenn AM, dos Reis MS (2012) Genetic analysis of 50-year old Brazilian pine (Araucaria angustifolia) plantations: implications for conservation planning. Conservation Genetics, 13, 435-442. |
| 8 | Finkeldey R, Hattemer HH (2007) Tropical Forest Genetics. Springer, Berlin. |
| 9 | Frankham R, Ballou JD, Mcinnes KH (2002) Introduction to Conservation Genetics, pp. 266-276. Cambridge University Press, Cambridge, UK. |
| 10 | Fu LK (傅立国), Jin JM (金鉴明) (1992) China Plant Red Data Book——Rare and Endangered Plants, Vol. 1 (中国红皮书——稀有濒危植物, 第一册), pp. 454-455. Science Press, Beijing. (in Chinese) |
| 11 | Gauli A, Gailing O, Stefenon VM, Finkeldey R (2009) Genetic similarity of natural populations and plantations of Pinus roxburghii Sarg. in Nepal.Annals of Forest Science, 66, 703. |
| 12 | Hamrick JL, Godt MJ (1990) Allozyme diversity in plant species. In: Plant Population Genetics, Breeding and Genetic Resources (eds Brown AHD, Clegg MT, Kahler AL, Weir BS), pp. 43-63. Sinauer, Sunderland. |
| 13 | Huang HW (黄宏文), Zhang Z (张征) (2012) Current status and prospects of ex situ cultivation and conservation of plants in China.Biodiversity Science(生物多样性), 20, 559-571. (in Chinese with English abstract) |
| 14 | Huang JX (黄久香), Zhuang XY (庄雪影) (2002a) Comparison of genetic diversity of Tsoongiodendron odorum in southern China by RAPD markers. Journal of South China Agricultural University (Natural Science Edition) (华南农业大学学报(自然科学版)), 23(2), 54-57. (in Chinese with English abstract) |
| 15 | Huang JX (黄久香), Zhuang XY (庄雪影) (2002b) Genetic diversity of the populations of Tsoongiodendron odorum.Acta Phytoecologica Sinica(植物生态学报), 26, 413-419. (in Chinese with English abstract) |
| 16 | Jiang JH, Wang J, Kang M, Sun W, Huang HW (2011) Isolation and characterization of microsatellite loci in Tsoongiodendron odorum (Magnoliaceae).American Journal of Botany, 98, 284-286. |
| 17 | Li NW (李乃伟), He SA (贺善安), Shu XC (束晓春), Wang Q (汪庆), Xia B (夏冰), Peng F (彭峰) (2011) Genetic diversity and structure analyses of wild and ex-situ conservation populations of Taxus chinensis var. mairei based on ISSR marker.Journal of Plant Resources and Environment(植物资源与环境学报), 20(1), 25-30. (in Chinese with English abstract) |
| 18 | Li SH (李松海), Xie AD (谢安德), Bi LY (贲丽云), Wang LH (王凌晖) (2011) Research status of precious tree species Tsoongiodendron odorum and its prospect.Journal of Southern Agriculture(南方农业学报), 42, 968-971. (in Chinese with English abstract) |
| 19 | Liu YH (刘玉壶) (1984) A preliminary study on the taxonomy of the family Magnoliaceae.Acta Phytotaxonomica Sinica(植物分类学报), 22, 89-109. (in Chinese with English abstract) |
| 20 | Navascués M, Emerson BC (2007) Natural recovery of genetic diversity by gene flow in reforested areas of the endemic Canary Island pine, Pinus canariensis.Forest Ecology and Management, 244, 122-128. |
| 21 | Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants.Molecular Ecology, 13, 1143-1156. |
| 22 | Pandey M, Gailing O, Leinemann L, Finkeldey R (2004) Molecular markers provide evidence for long-distance planting material transfer during plantation establishment of Dalbergia sissoo Roxb. in Nepal. Annals of Forest Science, 61, 603-606. |
| 23 | Primack PB, Ma KP (马克平) (2009) A Primer of Conservation Biology (保护生物学简明教程), pp. 121-132. Higher Education Press, Beijing. (in Chinese) |
| 24 | Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data.Genetics, 155, 945-959. |
| 25 | Rohlf FJ (2000) NTSYS-pc version 2.1, Numerical Taxonomy and Multivariate Analysis System. Exeter Software, Setauket, New York. |
| 26 | Salas-Leiva DE, Mayor-Durán VM, Toro-Perea N (2009) Genetic diversity of black mangrove (Avicennia germinans) in natural and reforested areas of Salamanca Island Parkway, Colombian Caribbean.Hydrobiologia, 620, 17-24. |
| 27 | Sharma K, Degen B, von Wuehlisch G, Singh NB (2002) Allozyme variation in eight natural populations of Pinus roxburghii Sarg. in India.Silvae Genetica, 51, 246-253. |
| 28 | Shi QF (史全芬), Yang J (杨佳), Li XD (李晓东), Li XW (李新伟), Li JQ (李建强) (2005) Genetic diversity of cultivated populations of Metasequoia glyptostroboides.Acta Botanica Yunnanica(云南植物研究), 27, 403-412. (in Chinese with English abstract) |
| 29 | Sozen E, Ozaydin B (2010) A study of genetic variation in endemic plant Centaurea wiedemanniana by using RAPD markers. Ekoloji, 19, 1-8. |
| 30 | Stefenon VM, Gailing O, Finkeldey R (2008) Genetic structure of plantations and the conservation of genetic resources of Brazilian pine (Araucaria angustifolia).Forest Ecology and Management, 255, 2718-2725. |
| 31 | The Office of Forestry Division of China Forestry Department, PRC (中华人民共和国林业部林业区划办公室) (1987) China Division of Forest (中国林业区划), pp. 220-225. China Forestry Publishing House, Beijing. (in Chinese) |
| 32 | Wang X (王霞), Wang J (王静), Jiang JH (蒋敬虎), Kang M (康明) (2012) Genetic diversity and the mating system in a fragmented population of Tsoongiodendron odorum. Biodiversity Science(生物多样性), 20, 676-684. (in Chinese with English abstract) |
| 33 | Wang ZS, Liu H, Xu WX, Wei N, An SQ ( 2010) Genetic diversity in young and mature cohorts of cultivated and wild populations of Picea asperata Mast (Pinaceae), a spruce endemic in western China.European Journal of Forest Research, 129, 719-728. |
| 34 | Willi Y, van Buskirk J, Hoffmann AA (2006) Limits to the adaptive potential of small populations.Annual Review of Ecology, Evolution, and Systematics, 37, 433-458. |
| 35 | Wu XQ (吴雪琴), Xu GB (徐刚标), Liang Y (梁艳), Wang JQ (王家庆) (2012) Establishment and optimization of Tsoongiodendron odorum ISSR-PCR reaction system. Journal of Central South University of Forestry &Technology(中南林业科技大学学报), 32(7), 76-79. (in Chinese with English abstract) |
| 36 | Wu ZY (吴则焰), Liu JF (刘金福), Hong W (洪伟), Pan DM (潘东明), Zheng SQ (郑世群) (2011) Genetic diversity of natural and planted Glyptostrobus pensilis populations: a comparative study.Chinese Journal of Applied Ecology(应用生态学报), 22, 873-879. (in Chinese with English abstract) |
| 37 | Xing FW (邢福武), Chen HF (陈红锋), Wang FG (王发国), Chen ZM (陈振明), Zeng QW (曾庆文) (2011) Inventory of Plant Species Diversity in Nanling Mountains (南岭植物物种多样性编目), pp. 1-2. Huazhong University of Science and Technology Press, Wuhan. (in Chinese) |
| 38 | Yang YS, Chen GS, Guo JF, Lin P (2004) Decomposition dynamic of fine roots in a mixed forest of Cunninghamia lanceolata and Tsoongiodendron odorum in mid-subtropics.Annals of Forest Science, 61, 65-72 |
| 39 | Yeh FC, Yang RC, Boyle T (1999)POPGENE version 1.31, Software Microsoft Window-based Freeware for Population Genetic Analysis. University of Alberta and Centre for International Forestry Research, Alberta. |
| 40 | Zeng QW (曾庆文), Gao ZZ (高泽正), Xing FW (邢福武), Zhang DX (张奠湘), Gong X (龚洵) (2004) Pollination biology of an endangered species: Tsoongiodendron odorum (Magnoliaceae). In: Proceedings of the Fifth National Symposium on the Conservation and Sustainable Use of Biodiversity in China (第5届生物多样性保护和持续利用研讨会论文集) (ed. Biodiversity Committee of the Chinese Academy of Sciences (中国科学院生物多样性委员会)), pp. 199-204. China Meteorological Press, Beijing. (in Chinese with English abstract) |
| 41 | Zhu TB, Meng TZ, Zhang JB, Yin YF, Cai ZC, Yang WY, Zhong WH (2012) Nitrogen mineralization, immobilization turnover, heterotrophic nitrification, and microbial groups in acid forest soils of subtropical China.Biology and Fertility of Soil, doi: 10.1007/s00374-012-0725-y. |
| [1] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [2] | 陈进, 杨玺. 关于植物迁地保护若干问题的讨论[J]. 生物多样性, 2024, 32(2): 24064-. |
| [3] | 刘夙, 杜诚, 胡永红. 迁地保护应当具有明确的灵活性[J]. 生物多样性, 2024, 32(2): 24037-. |
| [4] | 任海, 何拓, 文世峰, 董晖. 中国特色、世界一流国家植物园的主要特征[J]. 生物多样性, 2023, 31(9): 23192-. |
| [5] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [6] | 文世峰, 周志华, 何拓, 董晖, 袁良琛, 卢泽洋, 王泳腾, 郭琳, 舒江平, 李开凡. 《国家植物园体系布局方案》编制背景、程序、思路和重点考虑[J]. 生物多样性, 2023, 31(9): 23193-. |
| [7] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [8] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [9] | 李仕裕, 张奕奇, 邹璞, 宁祖林, 廖景平. 广东省植物园植物多样性迁地保护现状及发展建议[J]. 生物多样性, 2023, 31(6): 22647-. |
| [10] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [11] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [12] | 王利松, 湛青青, 廖景平, 黄宏文. 我国迁地保护的国家重点保护、受威胁和特有维管植物多样性[J]. 生物多样性, 2023, 31(2): 22495-. |
| [13] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [14] | 许再富. 对国家植物园体系建设“统筹原则”的一些见解[J]. 生物多样性, 2023, 31(1): 22470-. |
| [15] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn