



生物多样性 ›› 2013, Vol. 21 ›› Issue (3): 306-314. DOI: 10.3724/SP.J.1003.2013.09029 cstr: 32101.14.SP.J.1003.2013.09029
李晶晶1,2, 张杰1,2, 胡自民1,*( ), 段德麟1,*(
), 段德麟1,*( )
)
收稿日期:2013-01-30
接受日期:2013-03-28
出版日期:2013-05-20
发布日期:2013-06-05
通讯作者:
胡自民,段德麟
基金资助:
Jingjing Li1,2, Jie Zhang1,2, Zimin Hu1,*( ), Delin Duan1,*(
), Delin Duan1,*( )
)
Received:2013-01-30
Accepted:2013-03-28
Online:2013-05-20
Published:2013-06-05
Contact:
Hu Zimin,Duan Delin
摘要:
探讨古气候波动(如更新世末期冰期)对典型生物的时空分布和有效种群大小变动的影响是生物地理学和进化遗传学的重要研究课题。本文利用线粒体cox2-3序列和RAPD两种分子标记, 对分布于加拿大-西北大西洋地区8个地点(共138个个体)的掌形藻(Palmaria palmata)进行谱系地理学研究, 试图阐明当更新世冰期来临时掌形藻如何衍生出适应性的进化机制, 并形成当前的地理分布格局。结果表明, 线粒体cox2-3间区序列共检测出11个单倍型, 其中1个单倍型(C3)在所有种群中都有分布, 并位于星状基因谱系的中心位置, 可认为是祖先单倍型。St. Lawrence湾内北部的两个种群多样性最高, 与其他地理种群分化最明显, 这与基于RAPD数据的STRUCTURE聚类分析结果相一致。根据掌形藻遗传多样性及其单倍型谱系结构特征, 推测掌形藻在加拿大-西北大西洋沿岸存在多个冰期避难所。分子多态性分析(AMOVA)显示掌形藻的遗传变异主要来自种群内, 而St. Lawrence湾和Fundy湾群组间的遗传变异较小。cox2-3序列的Bayesian skyline plots分析结果反映出掌形藻种群在加拿大-西北大西洋沿岸经历了轻微的种群扩张, 时间大概在0.18-0.13百万年前。St. Lawrence湾和Fundy湾群组间的K2P遗传距离为0.2%, 相应的分化时间大约在0.36百万年前。由此推测, 更新世末期的冰期及间冰期是影响掌形藻种群结构及变动的重要古气候环境因子。
李晶晶, 张杰, 胡自民, 段德麟 (2013) 加拿大-西北∇大西洋地区掌形藻的种群遗传结构与动态变化. 生物多样性, 21, 306-314. DOI: 10.3724/SP.J.1003.2013.09029.
Jingjing Li,Jie Zhang,Zimin Hu,Delin Duan (2013) Population genetics and demographic history of red seaweed, Palmaria palmata, from the Canada-northwest Atlantic. Biodiversity Science, 21, 306-314. DOI: 10.3724/SP.J.1003.2013.09029.
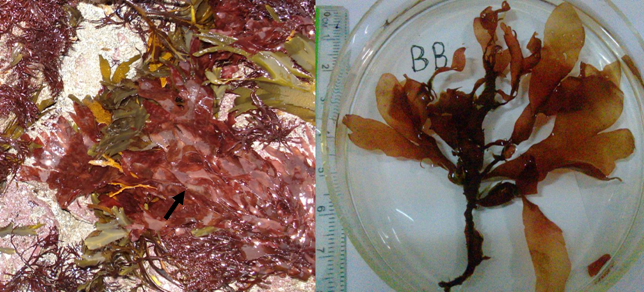
图1 掌形藻的外部形态图片。(a)掌形藻的典型栖息地环境, 常见附生于皱波角叉菜(Chondrus crispus)和齿缘墨角藻(Fucus serratus)的藻体上。于2008年拍摄自爱尔兰的Clare郡潮间带; (b)紫红色的掌形藻外部形态, 采自加拿大的Bonne Bay。
Fig. 1 Morphological characters of Palmaria palmata. (a) Living intertidal habitat of P. palmata from County Clare, Ireland in 2008. It is frequently found living on thallus of red marcroalga Chondrus crispus and brown macroalga Fucus serratus; (b) Purple morphological features of P. palmata from Bonne Bay, Canada.
| 简称 Code | 种群 Population | 经纬度 Locality | N | Nh | H | h (SD) | π (SD) |
|---|---|---|---|---|---|---|---|
| Gulf of St. Lawrence | |||||||
| LA | L’Anse Amour, Canada | 52.32°N, 56.60°W | 25 | 3 | C1, C2, C3 | 0.640 (0.052) | 0.0024 (0.0020) |
| BB | Bonne Bay, Canada | 49.51°N, 57.92°W | 31 | 3 | C1, C3, C4 | 0.540 (0.043) | 0.0018 (0.0017) |
| GP | Gaspe, Canada | 48.88°N, 64.50°W | 11 | 1 | C3 | 0.000 (0.000) | 0.0000 (0.0000) |
| RS | Rimouski, Canada | 48.47°N, 68.51°W | 24 | 2 | C3, C5 | 0.083 (0.075) | 0.0002 (0.0005) |
| Bay of Fundy | |||||||
| MB | Maces Bay, Canada | 45.11°N, 66.75°W | 8 | 2 | C3, C6 | 0.250 (0.180) | 0.0008 (0.0011) |
| VB | Victoria Beach, Canada | 44.69°N, 65.75°W | 9 | 3 | C3, C7, C8 | 0.417 (0.191) | 0.0014 (0.0016) |
| LT | Letete, Canada | 45.06°N, 66.89°W | 12 | 2 | C3, C9 | 0.303 (0.148) | 0.0009 (0.0012) |
| WH | White Head, Canada | 44.63°N, 66.72°W | 18 | 3 | C3, C10, C11 | 0.451 (0.117) | 0.0015 (0.0015) |
表1 掌形藻8个种群的简称、位置分布、样本量(N)、单倍型个数(Nh)、单倍型种类(H)、单倍型多样性(h)及核苷酸多样性(π)
Table 1 Sampling details of eight Palmaria palmata populations, including abbreviation codes, geographic location, sample size (N), number of haplotypes (Nh), types of haplotype (H), haplotype diversity (h), and nucleotide diversity (π)
| 简称 Code | 种群 Population | 经纬度 Locality | N | Nh | H | h (SD) | π (SD) |
|---|---|---|---|---|---|---|---|
| Gulf of St. Lawrence | |||||||
| LA | L’Anse Amour, Canada | 52.32°N, 56.60°W | 25 | 3 | C1, C2, C3 | 0.640 (0.052) | 0.0024 (0.0020) |
| BB | Bonne Bay, Canada | 49.51°N, 57.92°W | 31 | 3 | C1, C3, C4 | 0.540 (0.043) | 0.0018 (0.0017) |
| GP | Gaspe, Canada | 48.88°N, 64.50°W | 11 | 1 | C3 | 0.000 (0.000) | 0.0000 (0.0000) |
| RS | Rimouski, Canada | 48.47°N, 68.51°W | 24 | 2 | C3, C5 | 0.083 (0.075) | 0.0002 (0.0005) |
| Bay of Fundy | |||||||
| MB | Maces Bay, Canada | 45.11°N, 66.75°W | 8 | 2 | C3, C6 | 0.250 (0.180) | 0.0008 (0.0011) |
| VB | Victoria Beach, Canada | 44.69°N, 65.75°W | 9 | 3 | C3, C7, C8 | 0.417 (0.191) | 0.0014 (0.0016) |
| LT | Letete, Canada | 45.06°N, 66.89°W | 12 | 2 | C3, C9 | 0.303 (0.148) | 0.0009 (0.0012) |
| WH | White Head, Canada | 44.63°N, 66.72°W | 18 | 3 | C3, C10, C11 | 0.451 (0.117) | 0.0015 (0.0015) |
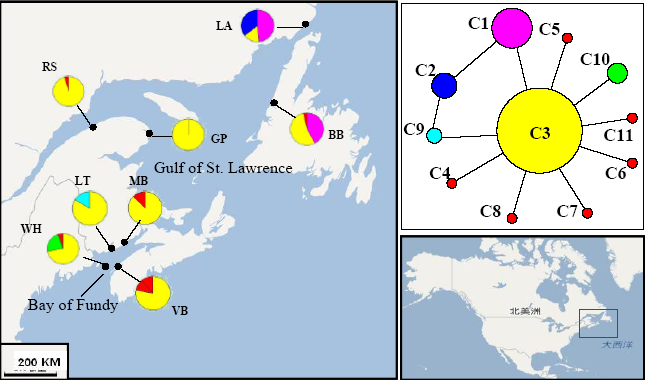
图2 掌形藻验证的线粒体cox2-3单倍型在各种群中的分布及单倍型谱系图(种群代号同表1)
Fig. 2 Distribution of mitochondrial cox2-3 haplotypes of Palmaria palmata populations and median-joining haplotype network. Population codes correspond to those in Table 1.
| 变异来源 Source of variations | 自由度 d.f. | 变异组成 Variance components | 变异百分比 % of variation | 固定指数 Fixation indices |
|---|---|---|---|---|
| 群组间 Among groups | 1 | 0.0409 | 10.12 | FCT = 0.10118NS |
| 种群间 Among populations within groups | 6 | 0.1246 | 34.09 | Fsc = 0.37928*** |
| 种群内 Within populations | 130 | 0.2075 | 55.79 | FST = 0.44509*** |
表2 掌形藻种群的分子多态性分析(AMOVA)。掌形藻的8个种群被分为St. Lawrence湾和Fundy湾2个群组。
Table 2 Molecular variance (AMOVA) of Palmaria palmata populations. The eight populations were clustered into two groups: the Gulf of St. Lawrence and the Bay of Fundy.
| 变异来源 Source of variations | 自由度 d.f. | 变异组成 Variance components | 变异百分比 % of variation | 固定指数 Fixation indices |
|---|---|---|---|---|
| 群组间 Among groups | 1 | 0.0409 | 10.12 | FCT = 0.10118NS |
| 种群间 Among populations within groups | 6 | 0.1246 | 34.09 | Fsc = 0.37928*** |
| 种群内 Within populations | 130 | 0.2075 | 55.79 | FST = 0.44509*** |
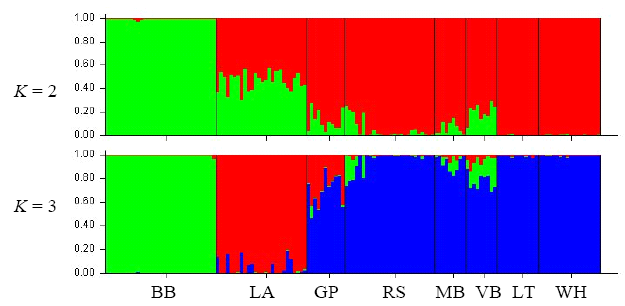
图3 基于RAPD数据的STRUCTURE种群遗传结构分析。每个竖条代表1个个体多位点基因型的组合, 不同颜色代表模拟后的聚类群。种群简称同表1。
Fig. 3 Population structuring analysis based on RAPD data with STRUCTURE. Each vertical bar indicates the multi-locus genotype of one individual, and colors represent the K virtual clusters. Population codes are the same as Table 1.
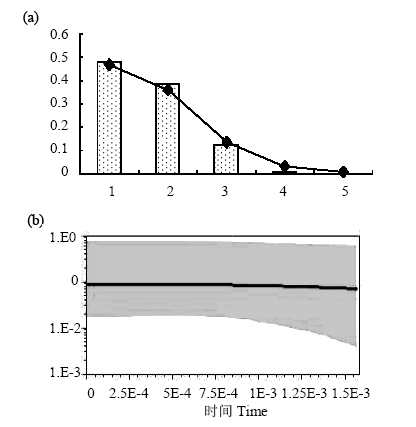
图4 掌形藻种群规模的历史变动。(a)歧点分布图(mismatch distribution), 横坐标表示序列间差异碱基数, 纵坐标表示概率, 柱状图为观测值, 曲线为种群扩张模型下的预期分布; (b) Bayesian skyline plots分析, 表示有效种群大小随时间的变动。灰色区域的上下限表示95%的HPD分析置信区间。
Fig. 4 Demographic history of Palmaria palmata. (a) Mismatch distributions of Palmaria palmata populations. The abscissa indicates the number of pairwise differences between compared sequences. The ordinate is the frequency for each value. Bars represent the observed distribution of pairwise differences, while the solid line shows the expected distribution; (b) Bayesian skyline plots show effective population size as a function of time. The upper and lower limits of grey trend represent the 95% confidence intervals of higher probability density (HPD) analysis.
| 22 | Krebes L, Blank M, Bastrop R (2011) Phylogeography, historical demography and postglacial colonization routes of two amphi-Atlantic distributed amphipods. Systematics and Biodiversity, 9, 259-273. |
| 23 | Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452. |
| 24 | Lindstrom SC, Olsen JL, Stam WT (1996) Recent radiation of the Palmariaceae (rhodophyta). Journal of Phycology, 32, 457-468. |
| 25 | Lindstrom SC, Olsen JL, Stam WT (1997) Postglacial recolonization and the biogeography of Palmaria mollis (Rhodophyta) along the Northeast Pacific coast. Canadian Journal of Botany, 75, 1887-1896. |
| 26 | Lister A, Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society B: Biological Sciences, 359, 183-195. |
| 27 | Maggs CA, Castilho R, Foltz D, Henzler C, Jolly MT, Kelly J, Olsen J, Perez KE, Stam W, Väinölä R, Viard F, Wares J (2008) Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology, 89, S108-S122. |
| 28 | Olsen JL, Zechman FW, Hoarau G, Coyer JA, Stam WT, Valero M, Åberg P (2010) The phylogeographic architecture of the fucoid seaweed Ascophyllum nodosum: an intertidal ‘marine tree’ and survivor of more than one glacial- interglacial cycle. Journal of Biogeography, 37, 842-856. |
| 29 | Panova M, Blakeslee AMH, Miller AW, Mäkinen T, Ruiz GM, Johannesson K, André C (2011) Glacial history of the North Atlantic marine snail, Littorina saxatilis, inferred from distribution of mitochondrial DNA lineages. PLoS ONE, 6, e17511. |
| 30 | Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics, 14, 817-818. |
| 31 | Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959. |
| 32 | Provan J, Bennett KD (2008) Phylogeographic insights into cryptic glacial refugia. Trends in Ecology and Evolution, 23, 564-571. |
| 33 | Provan J, Wattier RA, Maggs CA (2005) Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Molecular Ecology, 14, 793-803. |
| 34 | Ray N, Currat M, Excoffier L (2003) Intra-deme molecular diversity in spatially expanding populations. Molecular Biology and Evolution, 20, 76-86. |
| 35 | Riggs SR, Snyder SW, Hine AC, Mearns DL (1996) Hardbottom morphology and relationship to the geologic framework: mid-Atlantic continental shelf. Journal of Sedimentary Research, 66, 830-846. |
| 36 | Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution, 9, 552-569. |
| 37 | Saunders GW (2004) A chronicle of the convoluted systematics of the red algal orders Palmariales and Rhodymeniales (Florideophyceae, Rhodophyta). CEMAR Occasional Notes in Phycology, 1, 1-16. |
| 38 | Strimmer K, Pybus OG (2001) Exploring the demographic history of DNA sequences using the generalized skyline plot. Molecular Biology and Evolution, 18, 2298-2305. |
| 39 | Svendsen JI, Alexanderson H, Astakhov VI, Demidov I, Dowdeswell JA, Funder S, Gataullin V, Henriksen M, Hjort C, Houmark-Nielsen M, Hubberten HW, Ingólfsson O, Jakobsson M, Kjaer KH, Larsen E, Lokrantz H, Lunkka JP, Lyså A, Mangerud J, Matiouchkov A, Murray A, Möller P, Niessen F, Nikolskaya O, Polyak L, Saarnisto M, Siegert C, Siegert MJ, Spielhagen RF, Stein R (2004) Late quaternary ice sheet history of northern Eurasia. Quaternary Science Reviews, 23, 1229-1271. |
| 40 | Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585-595. |
| 41 | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731-2739. |
| 42 | Teasdale BW, Klein AS (2010) Genetic variation and biogeographical boundaries within the red alga Porphyra umbilicalis (Bangiales, Rhodophyta). Botanica Marina, 53, 417-431. |
| 43 | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. |
| 44 | Vermeij GJ (1991) Anatomy of an invasion: the trans-Arctic interchange. Paleobiology, 17, 281-307. |
| 45 | Victoria M, Miagostovich MP, Ferreira MSR, Vieira CB, Fioretti JM, Leite JPG, Colina R, Cristina J (2009) Bayesian coalescent inference reveals high evolutionary rates and expansion of Norovirus populations. Infection, Genetics and Evolution, 9, 927-932. |
| 46 | Wares JP (2002) Community genetics in the Northwestern Atlantic intertidal. Molecular Ecology, 11, 1131-1144. |
| 47 | Wares JP, Cunningham CW (2001) Phylogeography and historical ecology of the North Atlantic intertidal. Evolution, 55, 2455-2469. |
| 48 | Zuccarello GC, Burger G, West JA, King RJ (1999) A mitochondrial marker for red algal intraspecific relationships. Molecular Ecology, 8, 1443-1447. |
| 49 | Zuccarello GC, West JA (2002) Phylogeography of the Bostrychia calliptera-B. pinnata complex (Rhodomelaceae, Rhodophyta) and divergence rates based on nuclear, mitochondrial and plastid DNA markers. Phycologia, 41, 49-60. |
| 1 | Addison JA, Hart MW (2005) Colonization, dispersal, and hybridization influence phylogeography of North Atlantic sea urchins (Strongylocentrotus droebachiensis). Evolution, 59, 532-543. |
| 2 | Albaina N, Olsen JL, Couceiro L, Ruiz JM, Barreiro R (2012) Recent history of the European Nassarius nitidus (Gastropoda): phylogeographic evidence of glacial refugia and colonization pathways. Marine Biology, 159, 1871-1884. |
| 3 | Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48. |
| 4 | Bigg GR, Cunningham CW, Ottersen G, Pogson GH, Wadley MR, Williamson P (2008) Ice-age survival of Atlantic cod: agreement between palaeoecology models and genetics. Proceedings of the Royal Society B: Biological Sciences, 275, 163-172. |
| 5 | Campo D, Molares J, Garcia L, Fernandez-Rueda P, Garcia- Gonzalez C, Garcia-Vazquez E (2010) Phylogeography of the European stalked barnacle (Pollicipes pollicipes): identification of glacial refugia. Marine Biology, 157, 147-156. |
| 6 | Charbit S, Ritz C, Philippon G, Peyaud V, Kageyama M (2007) Numerical reconstructions of the Northern Hemisphere ice sheets through the last glacial-interglacial cycle. Climate of the Past, 3, 15-37. |
| 7 | Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution, 22, 1185-1192. |
| 8 | Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biology, 4, 699-710. |
| 9 | Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969-1973. |
| 10 | Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611-2620. |
| 11 | Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Molecular Ecology, 13, 853-864. |
| 12 | Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567. |
| 13 | Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479-491. |
| 14 | Flint RF (1940) Pleistocene features of the Atlantic Coastal Plain. American Journal of Science, 238, 757-787. |
| 15 | Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147, 915-925. |
| 16 | Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society, 58, 247-276. |
| 17 | Hoarau G, Coyer JA, Veldsink JH, Stam WT, Olsen JL (2007) Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Molecular Ecology, 16, 3606-3616. |
| 18 | Hu ZM, Guiry MD, Critchley AT, Duan DL (2010) Phylogeographic patterns indicate transatlantic migration from Europe to North America in the red seaweed Chondrus Crispus (Gigartinales, Rhodophyta). Journal of Phycology, 46, 889-900. |
| 19 | Hu ZM, Li W, Li JJ, Duan DL (2011) Post-Pleistocene demographic history of the North Atlantic endemic Irish moss Chondrus crispus: glacial survival, spatial expansion and gene flow. Journal of Evolutionary Biology, 24, 505-517. |
| 20 | Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16, 111-120. |
| 21 | Kingman JFC (1982) On the genealogy of large populations. Journal of Applied Probability, 19, 27-43. |
| [1] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [2] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [3] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [4] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [5] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [6] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [7] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [8] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [9] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [10] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [11] | 崔静, 徐明芳, 章群, 李瑶, 曾晓舒, 李莎. 基于3种线粒体标记探讨中日沿海角木叶鲽遗传多样性差异[J]. 生物多样性, 2022, 30(5): 21485-. |
| [12] | 孙军, 宋煜尧, 施义锋, 翟键, 燕文卓. 近十年中国海洋生物多样性研究进展[J]. 生物多样性, 2022, 30(10): 22526-. |
| [13] | 张丹, 马松梅, 魏博, 王春成, 张林, 闫涵. 中国梭梭属植物历史分布格局及其驱动机制[J]. 生物多样性, 2022, 30(1): 21192-. |
| [14] | 栗冬梅, 杨卫红, 李庆多, 韩茜, 宋秀平, 潘虹, 冯云. 巴尔通体在滇西南蝙蝠中高度流行并具有丰富的遗传变异特征[J]. 生物多样性, 2021, 29(9): 1245-1255. |
| [15] | 姚志, 郭军, 金晨钟, 刘勇波. 中国纳入一级保护的极小种群野生植物濒危机制[J]. 生物多样性, 2021, 29(3): 394-408. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn