



生物多样性 ›› 2010, Vol. 18 ›› Issue (3): 275-282. DOI: 10.3724/SP.J.1003.2010.275 cstr: 32101.14.SP.J.1003.2010.275
柳淑芳1, 刘进贤2,3, 庄志猛1,*( ), 高天翔3, 韩志强4, 陈大刚3
), 高天翔3, 韩志强4, 陈大刚3
收稿日期:2009-10-21
接受日期:2010-01-25
出版日期:2010-05-20
发布日期:2012-02-08
通讯作者:
庄志猛
作者简介:E-mail: zhuangzm@ysfri.ac.cn基金资助:
Shufang Liu1, Jinxian Liu2,3, Zhimeng Zhuang1,*( ), Tianxiang Gao3, Zhiqiang Han4, Dagang Chen3
), Tianxiang Gao3, Zhiqiang Han4, Dagang Chen3
Received:2009-10-21
Accepted:2010-01-25
Online:2010-05-20
Published:2012-02-08
Contact:
Zhimeng Zhuang
摘要:
舌鳎亚科鱼类因形态特征的特殊性, 其分类及系统发育关系一直存在争议。本研究测定了中国沿海14种舌鳎亚科鱼类线粒体DNA的16S rRNA和Cyt b基因的部分片段。两个基因构建的舌鳎亚科系统发育树结果显示, 中国沿海舌鳎亚科鱼类为明显的单系群, 但舌鳎亚科鱼类内部的系统发育关系与形态分类划分的亚属并不完全一致, 日本须鳎(Paraplagusis japonica)与其他舌鳎属(Cynoglossus)种类并未形成不同的分支。虽然舌鳎属内的舌鳎亚属(Cynoglossus)、拟舌鳎亚属(Cynoglossoides)和三线舌鳎亚属(Areliscus)均可以聚为独立分支, 但长吻红舌鳎(C. lighti)与短吻红舌鳎(C. joyneri)、短吻三线舌鳎(C. abbreviatus)与紫斑舌鳎(C. purpureomaculatus)、半滑舌鳎(C. semilaevis)与窄体舌鳎(C. gracilis)及褐斑三线舌鳎(C. trigrammus)这三组物种可能存在同种异名现象。这一结果提示, 基于形态学对舌鳎亚科的种属分类鉴定尚存在不足, 线粒体DNA的系统发育关系可为其分类的修订提供有意义的参考和佐证。
柳淑芳, 刘进贤, 庄志猛, 高天翔, 韩志强, 陈大刚 (2010) 舌鳎亚科鱼类单系起源和同种异名的线粒体DNA证据. 生物多样性, 18, 275-282. DOI: 10.3724/SP.J.1003.2010.275.
Shufang Liu, Jinxian Liu, Zhimeng Zhuang, Tianxiang Gao, Zhiqiang Han, Dagang Chen (2010) Monophyletic origin and synonymic phenomena in the sub-family Cynoglossinae inferred from mitochondrial DNA sequences. Biodiversity Science, 18, 275-282. DOI: 10.3724/SP.J.1003.2010.275.
| 种名 Species | 亚属名 Sub-genus | 样本量 Sample size | Cytb测定数 No. of Cytb DNA | 16S rRNA测定数 No. of 16S rRNA |
|---|---|---|---|---|
| 内群 Ingroup | ||||
| 1 双线舌鳎 Cynoglossus bilineatus | Arelia | 2 | 2 | 1 |
| 2 中华舌鳎 C. sinicus | Cynoglossus | 2 | 1 | 1 |
| 3 褐斑三线舌鳎 C. trigrammus | Areliscus | 1 | 1 | 1 |
| 4 窄体舌鳎 C. gracilis | Areliscus | 2 | 1 | 1 |
| 5 长吻红舌鳎 C. lighti | Areliscus | 2 | 2 | 1 |
| 6 短吻红舌鳎 C. joyneri | Areliscus | 2 | 2 | 1 |
| 7 黑鳃舌鳎 C. roulei | Areliscus | 1 | 1 | 1 |
| 8 紫斑舌鳎 C. purpureomaculatus | Areliscus | 2 | 1 | 1 |
| 9 短吻三线舌鳎 C. abbreviatus | Areliscus | 5 | 2 | 2 |
| 10 半滑舌鳎 C. semilaevis | Areliscus | 3 | 2 | 1 |
| 11 斑头舌鳎 C. puncticeps | Cynoglossoides | 1 | 1 | 1 |
| 12 少鳞舌鳎 C. oligolepis | Cynoglossoides | 2 | 2 | 1 |
| 13 宽体舌鳎 C. robustus | Cynoglossoides | 2 | 1 | 1 |
| 14 日本须鳎 Paraplagusis japonica | Rhinoplagusia | 1 | 1 | 1 |
| 外群 Outgroup | ||||
| 15 塞内加尔鳎 Sole senegalensis | 3 | 3 | 2 | |
| 16 人字钩嘴鳎 Heteromycteris matsubarai | 2 | 1 | 2 | |
| 17 圆斑星鲽 Verasper variegatus | 1 | 1 | 1 |
表1 本研究分析所用样品采样信息
Table 1 Information of species analyzed in the present study
| 种名 Species | 亚属名 Sub-genus | 样本量 Sample size | Cytb测定数 No. of Cytb DNA | 16S rRNA测定数 No. of 16S rRNA |
|---|---|---|---|---|
| 内群 Ingroup | ||||
| 1 双线舌鳎 Cynoglossus bilineatus | Arelia | 2 | 2 | 1 |
| 2 中华舌鳎 C. sinicus | Cynoglossus | 2 | 1 | 1 |
| 3 褐斑三线舌鳎 C. trigrammus | Areliscus | 1 | 1 | 1 |
| 4 窄体舌鳎 C. gracilis | Areliscus | 2 | 1 | 1 |
| 5 长吻红舌鳎 C. lighti | Areliscus | 2 | 2 | 1 |
| 6 短吻红舌鳎 C. joyneri | Areliscus | 2 | 2 | 1 |
| 7 黑鳃舌鳎 C. roulei | Areliscus | 1 | 1 | 1 |
| 8 紫斑舌鳎 C. purpureomaculatus | Areliscus | 2 | 1 | 1 |
| 9 短吻三线舌鳎 C. abbreviatus | Areliscus | 5 | 2 | 2 |
| 10 半滑舌鳎 C. semilaevis | Areliscus | 3 | 2 | 1 |
| 11 斑头舌鳎 C. puncticeps | Cynoglossoides | 1 | 1 | 1 |
| 12 少鳞舌鳎 C. oligolepis | Cynoglossoides | 2 | 2 | 1 |
| 13 宽体舌鳎 C. robustus | Cynoglossoides | 2 | 1 | 1 |
| 14 日本须鳎 Paraplagusis japonica | Rhinoplagusia | 1 | 1 | 1 |
| 外群 Outgroup | ||||
| 15 塞内加尔鳎 Sole senegalensis | 3 | 3 | 2 | |
| 16 人字钩嘴鳎 Heteromycteris matsubarai | 2 | 1 | 2 | |
| 17 圆斑星鲽 Verasper variegatus | 1 | 1 | 1 |
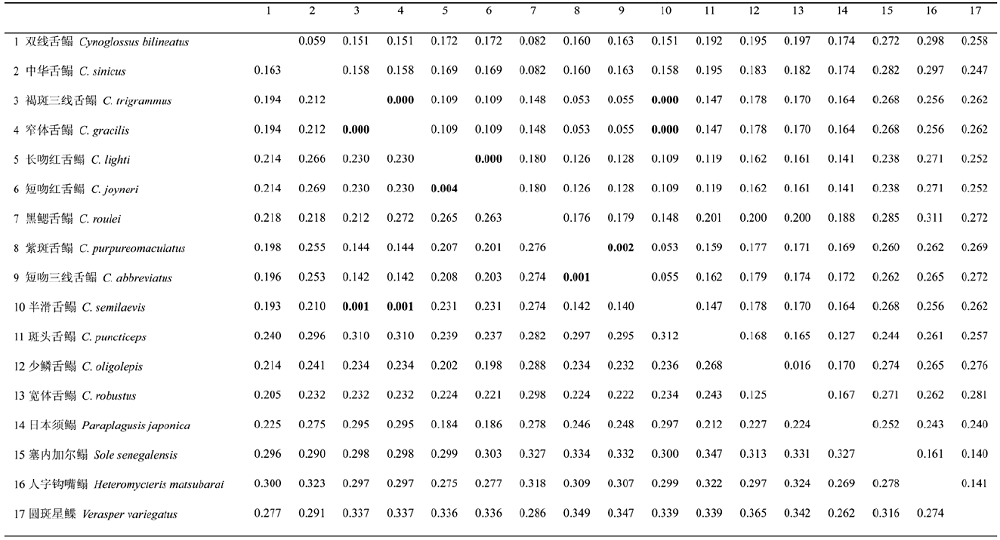 |
表2 基于Kimura双参数模型计算的物种间遗传距离(对角线下为Cyt b; 对角线上为16S rRNA)
Table 2 Pairwise distances calculated using Kimura-2-parameter model for Cyt b (below diagonal) and 16S rRNA (above diagonal)
 |
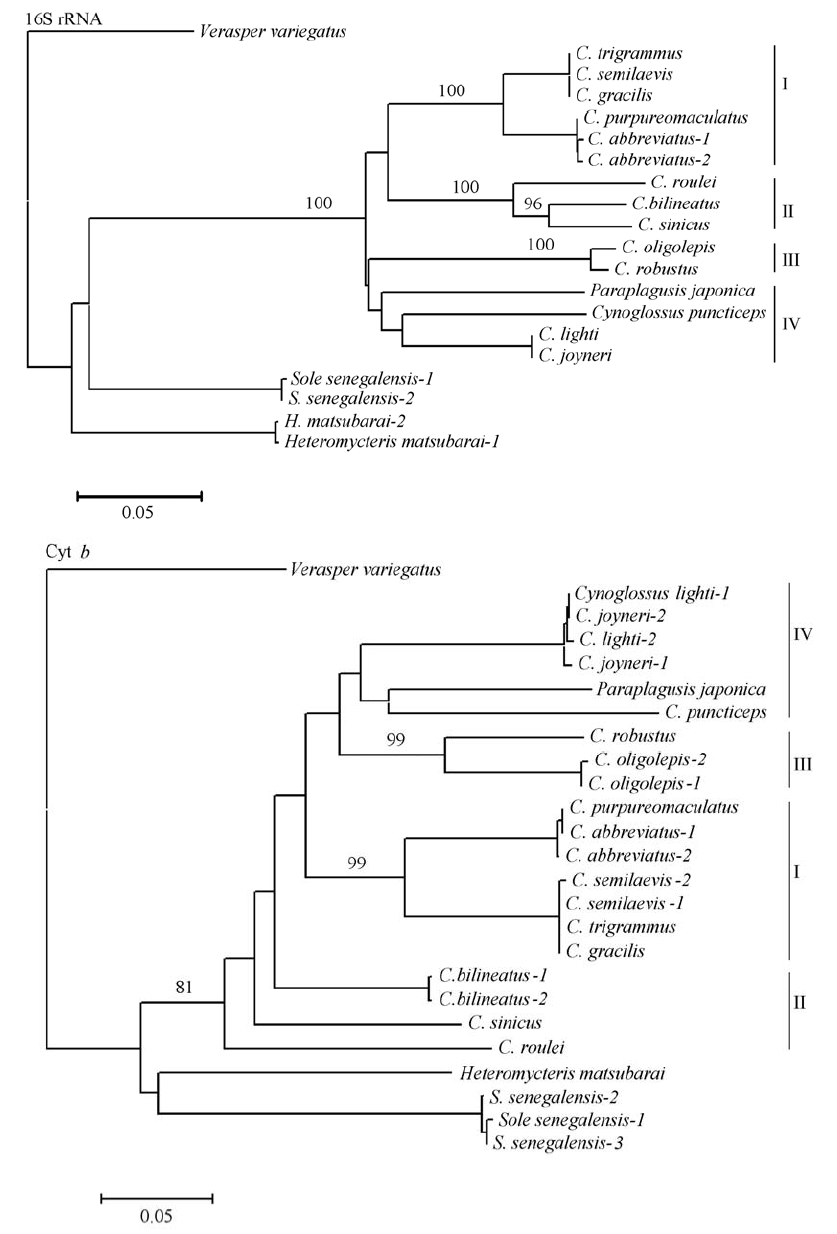
图1 以16S rRNA和Cyt b数据构建的NJ系统树。分支上的数字表示1,000次重复抽样所得的大于80%的支持率。
Fig. 1 Molecular phylogenetic tree constructed by neighbor-joining method based on 16S rRNA and Cyt b data. High bootstrap values (>80%) in 1,000 resamplings are shown at the corresponding nodes.
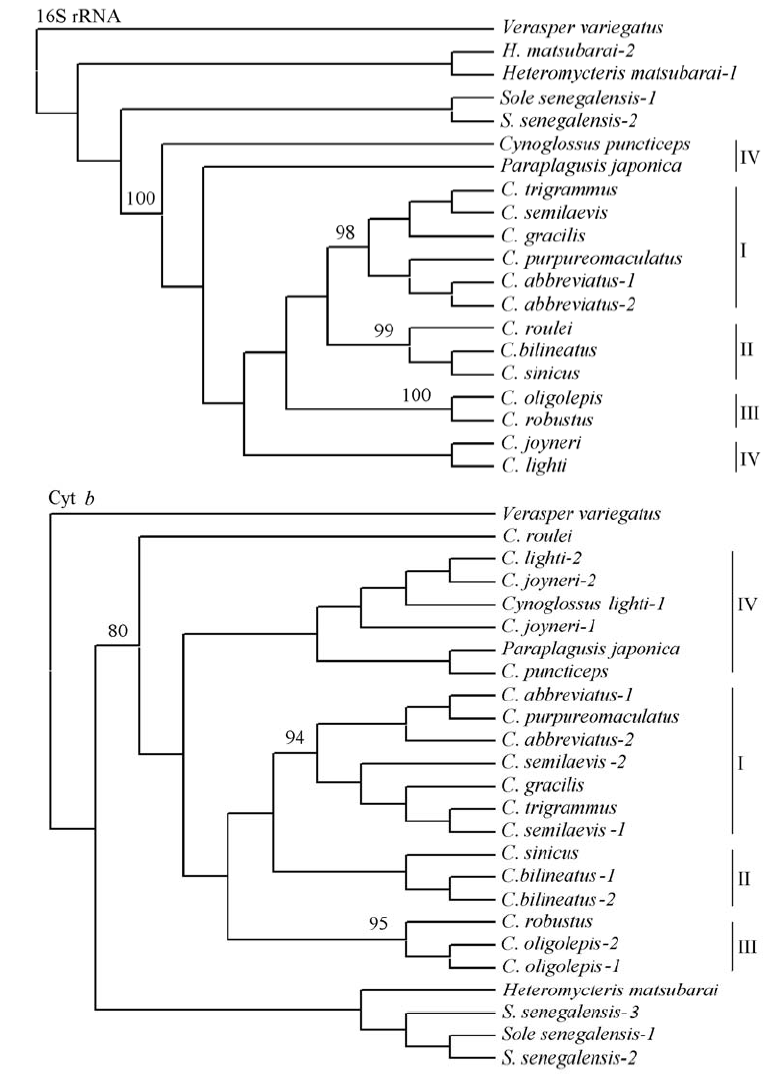
图2 以16S rRNA和Cyt b数据构建的MP系统树。分支上的数字表示1,000次重复抽样所得的大于80%的支持率。
Fig. 2 Molecular phylogenetic tree constructed by maximum parsimony based on 16S rRNA and Cyt b data. High bootstrap values (>80%) in 1,000 resamplings are shown at the corresponding nodes.
| [1] |
Briolaya J, Galtierb N, Britoc RM, Bouvetd Y (1998) Molecular phylogeny of Cyprindae inferred from cytochrome b DNA sequences. Molecular Phylogenetics and Evolution, 9, 100-108.
DOI URL PMID |
| [2] | Brown WM (1983) Evolution of animal mitochondrial DNA. In: Evolution of Genes and Proteins (eds Nei M, Koehn RK),p62. Sinauer, Sunderland, Massachusetts. |
| [3] | Chen SJ (陈姝君), He CB (赫崇波), Mu YL (木云雷), Liu WD (刘卫东), Zhou ZC (周遵春), Gao XG (高祥刚), Cong LL (丛林林) (2008) Informative efficiencies of mitochondrial genes in phylogenetic analysis of teleostean. Journal of Fishery Sciences of China (中国水产科学), 15, 12-21. (in Chinese with English abstract) |
| [4] | Cheng QT (成庆泰), Zheng BS (郑葆珊) (1987) Systematic Synopsis of Chinese Fishes (中国鱼类系统检索), pp.508-513. Science Press, Beijing. (in Chinese) |
| [5] |
Craig MT, Pondella DJ, Franek JPC, Hafner JC (2001) On the status of the serranid fish genus Epinephelus: evidence for paraphyly based upon 16S rDNA sequence. Molecular Phylogenetics and Evolution, 19, 121-130.
URL PMID |
| [6] | Ding SX (丁少雄), Wang YH (王颖汇), Wang J (王军), Zhuang X (庄轩), Su YQ (苏永全), You YZ (尤颖哲), Li QF (李祺福) (2006) Molecular phylogenetic relationships of 30 grouper species in China Seas based on 16S rDNA fragment sequences. Acta Zoologica Sinica (动物学报), 52, 504-513. (in Chinese with English abstract) |
| [7] | He SP (何舜平), Liu HZ (刘焕章), Chen YY (陈宜瑜), Kuwahara M, Nakajima T, Zhong Y (钟扬) (2004) Molecular phylogenetic relationships of Cyprindae fishes based on Cyt b DNA sequences. Science in China,Series C: Life Sciences(中国科学C辑), 34, 96-104. (in Chinese) |
| [8] |
Inoue JG, Miya M, Tsukamoto K, Nishida M (2003) Evolution of the deep-sea gulper eel mitochondrial genomes: large-scale gene rearrangements originated within the eels. Molecular Biology and Evolution, 20, 1917-1924.
DOI URL PMID |
| [9] |
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16, 111-120.
DOI URL PMID |
| [10] | Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX, Wilson AC (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proceedings of the National Academy of Sciences, USA, 86, 6196-6200. |
| [11] |
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics, 17, 1244-1245.
URL PMID |
| [12] | Li SZ (李思忠), Wang HM (王惠民) (1995) Fauna Sinica, Ostichthyes: Pleuronectiformes (中国动物志•硬骨鱼纲•鲽形目), pp.325-378. Science Press, Beijing. (in Chinese) |
| [13] | Lockhad PJ, Penny D, Meyer A (1995) Testing the phylogeny of swordtail fishes using decomposition and spectral analysis. Journal of Molecular Evolution, 41, 666-674. |
| [14] | Matsubara K (1955) Fishes Morphology and Hierarchy. Part II. Ishizaki-Shoten, Tokyo. pp.1217-1291. (in Japanese). |
| [15] | Menon AGK (1977) A Systematic Monograph of the Tongue Soles of the Genus Cynoglossus Hamilton-Buchanan (Pisces: Cynoglossidae) (Smithsonian contributions to zoology, no.238), pp.93-95. Smithsonian Institution Press, Washington. |
| [16] |
Miya M, Nishida M (2000) Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Molecular Phylogenetics and Evolution, 17, 437-455.
URL PMID |
| [17] |
Near TJ, Pesavento JJ, Cheng CH (2003) Mitochondrial DNA, morphology, and the phylogenetic relationships of Antarctic icefishes (Notothenioidei: Channichthyidae). Molecular Phylogenetics and Evolution, 28, 87-98.
DOI URL PMID |
| [18] | Ochiai A (1963) Fauna Japonica: Soleina (Pisces), pp.1-114. Biographical Society of Japan, Tokyo, Japan. |
| [19] | Ren G (任岗), Zhang Q (章群), Qian KC (钱开诚), Xu ZN (徐忠能), Lin XT (林小涛) (2007) Sequence analysis of twelve grunt fishes based on 16S ribosomal RNA gene fragments. Journal of Tropical Oceanography (热带海洋学报), 26, 48-52. (in Chinese with English abstract) |
| [20] |
Saitoh K, Miya M, Inoue JG, Ishiguro NB, Nishida M (2003) Mitochondrial genomics of ostariophysan fishes: perspectives on phylogeny and biogeography. Journal of Molecular Evolution, 56, 464-472.
URL PMID |
| [21] | Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York. |
| [22] |
Zardoya R, Meyer A (1996) Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Molecular Biology and Evolution, 13, 933-942.
DOI URL PMID |
| [1] | 陈静, 张丙昌, 刘燕晋, 武杰, 赵康, 明姣. 荒漠生物结皮细鞘丝藻类(Leptolyngbya-like)蓝藻多样性[J]. 生物多样性, 2024, 32(9): 24186-. |
| [2] | 刘君, 王宁, 崔岱宗, 卢磊, 赵敏. 大小兴安岭可培养细菌的资源多样性[J]. 生物多样性, 2019, 27(8): 903-910. |
| [3] | 张雪, 李兴安, 苏秦之, 曹棋钠, 李晨伊, 牛庆生, 郑浩. |
| [4] | 张明理. 中国西北干旱区和中亚植物区系地理研究[J]. 生物多样性, 2017, 25(2): 147-155. |
| [5] | 刘国红, 刘波, 朱育菁, 车建美, 葛慈斌, 苏明星, 唐建阳. 台湾地区芽胞杆菌物种多样性[J]. 生物多样性, 2016, 24(10): 1154-1163. |
| [6] | 葛慈斌, 郑榕, 刘波, 刘国红, 车建美, 唐建阳. 武夷山自然保护区土壤可培养芽胞杆菌 的物种多样性及分布[J]. 生物多样性, 2016, 24(10): 1164-1176. |
| [7] | 李海涛, 张保学, 高阳, 时小军, 周鹏. DNA条形码技术在海洋贝类鉴定中的实践: 以大亚湾生态监控区为例[J]. 生物多样性, 2015, 23(3): 299-305. |
| [8] | 王盼盼, 武永秀, 宋彤彤, 马春玲, 赵文, 王瑛, 孙磊. 野生及温室盆栽蕙兰可培养根内生细菌的遗传多样性[J]. 生物多样性, 2015, 23(1): 61-67. |
| [9] | 阚靖博, 李丽娜, 曲东, 王保莉. 淹水培养过程中水稻土细菌丰度与群落结构变化[J]. 生物多样性, 2014, 22(4): 508-515. |
| [10] | 曾丽萍, 张宁, 马红. 被子植物系统发育深层关系研究: 进展与挑战[J]. 生物多样性, 2014, 22(1): 21-39. |
| [11] | 何锴, 王文智, 李权, 罗培鹏, 孙悦华, 蒋学龙. DNA条形码技术在小型兽类鉴定中的探索: 以甘肃莲花山为例[J]. 生物多样性, 2013, 21(2): 197-205. |
| [12] | 库喜英, 周传江, 何舜平. 中国黄颡鱼的线粒体DNA多样性及其分子系统学[J]. 生物多样性, 2010, 18(3): 262-274. |
| [13] | 徐磊, 杨大荣. 榕树及其传粉榕小蜂的系统发育和协同进化研究现状及展望[J]. 生物多样性, 2008, 16(5): 446-453. |
| [14] | 唐文乔, 胡雪莲, 杨金权. 从线粒体控制区全序列变异看短颌鲚和湖鲚的物种有效性[J]. 生物多样性, 2007, 15(3): 224-231. |
| [15] | 许学伟, 吴敏, 迪丽拜尔·托乎提, 古丽巴哈尔·阿巴拜克利. 新疆艾比湖和伊吾湖可培养嗜盐古菌多样性[J]. 生物多样性, 2006, 14(4): 359-362. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()