



生物多样性 ›› 2014, Vol. 22 ›› Issue (1): 21-39. DOI: 10.3724/SP.J.1003.2014.13189 cstr: 32101.14.SP.J.1003.2014.13189
收稿日期:2013-08-16
接受日期:2013-12-07
出版日期:2014-01-20
发布日期:2014-02-10
通讯作者:
马红
基金资助:
Liping Zeng, Ning Zhang, Hong Ma*( )
)
Received:2013-08-16
Accepted:2013-12-07
Online:2014-01-20
Published:2014-02-10
Contact:
Ma Hong
摘要:
被子植物系统发育学是研究被子植物及其各类群间亲缘关系与进化历史的学科。从20世纪90年代起, 核苷酸和氨基酸序列等分子数据开始被广泛运用于被子植物系统发育研究, 经过20多年的发展, 从使用单个或联合少数几个细胞器基因, 到近期应用整个叶绿体基因组来重建被子植物的系统发育关系, 目、科水平上的被子植物系统发育框架已被广泛接受。在这个框架中, 基部类群、主要的5个分支(即真双子叶植物、单子叶植物、木兰类、金粟兰目和金鱼藻目)、每个分支所包含的目以及几个大分支包括的核心类群等都具有高度支持。与此同时, 细胞器基因还存在一些固有的问题, 例如单亲遗传、系统发育信息量有限等, 因此近年来双亲遗传的核基因在被子植物系统发育研究中的重要性逐渐得到关注, 并在不同分类阶元的研究中都取得了一定进展。但是, 被子植物系统发育中仍然存在一些难以确定的关系, 例如被子植物5个分支之间的关系、真双子叶植物内部某些类群的位置等。本文简述了20多年来被子植物系统发育深层关系的主要研究进展, 讨论了被子植物系统发育学常用的细胞器基因和核基因的选用, 已经确定和尚未确定系统发育位置的主要类群, 以及研究中尚存在的问题和可能的解决方法。
曾丽萍, 张宁, 马红 (2014) 被子植物系统发育深层关系研究: 进展与挑战. 生物多样性, 22, 21-39. DOI: 10.3724/SP.J.1003.2014.13189.
Liping Zeng,Ning Zhang,Hong Ma (2014) Advances and challenges in resolving the angiosperm phylogeny. Biodiversity Science, 22, 21-39. DOI: 10.3724/SP.J.1003.2014.13189.
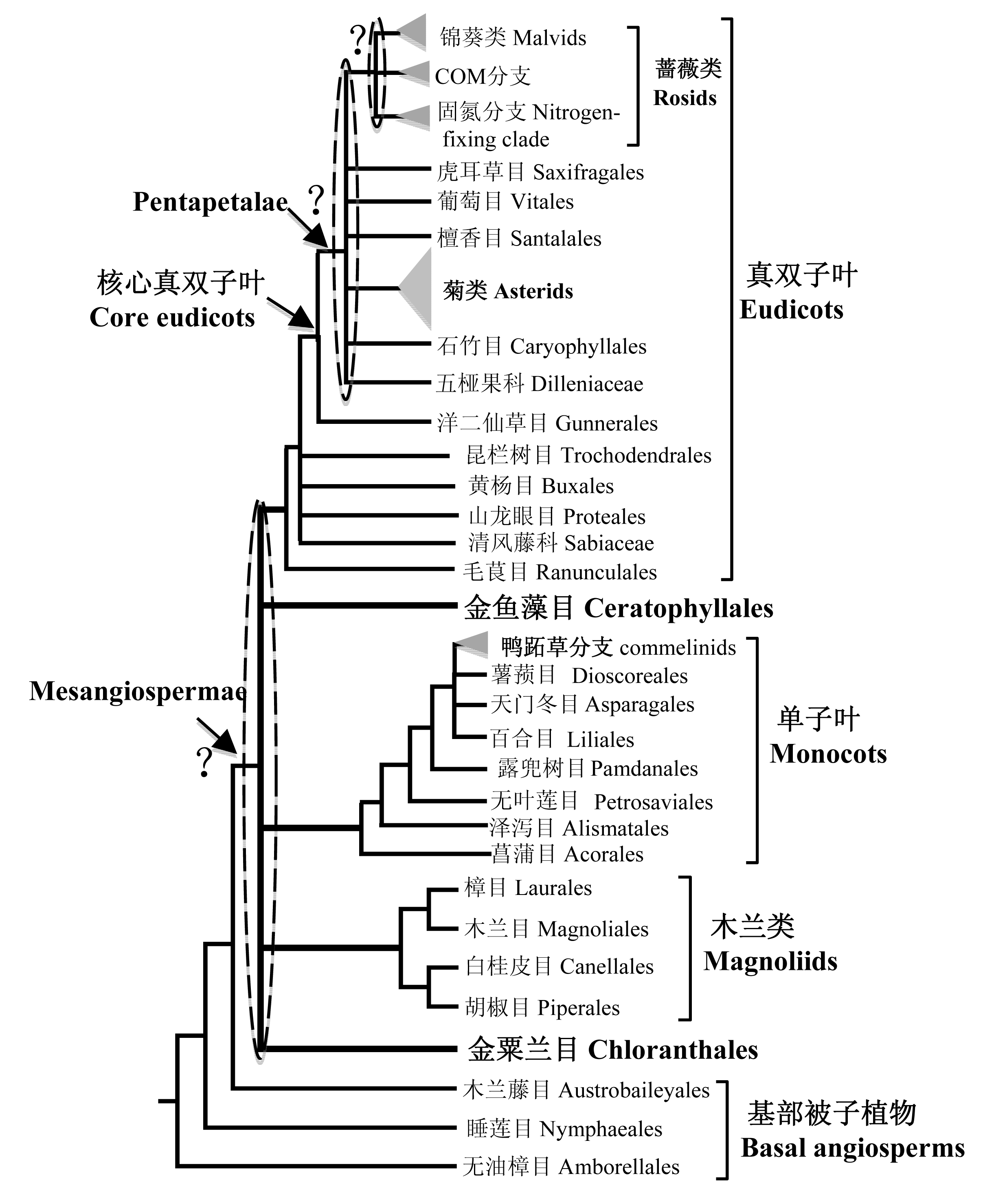
图1 被子植物各大类群关系图。除了位于最基部的3个目之外, 其余所有被子植物形成单系群。这个单系群由5个类群组成: 金粟兰目、木兰类、单子叶植物、金鱼藻目和真双子叶植物, 但是这5个分支的关系没有被确定, 因此五者的关系在树上以梳子结构来表示。单子叶植物中, 菖蒲目被认为是最基部的类群; 鸭跖草分支则是其核心类群, 包含4个目, 在树上以三角形表示。木兰类4个目的关系已经确定。毛茛目被认为位于真双子叶植物的最基部; 而在核心真双子叶植物中, 洋二仙草目被认为位于最基部, 其他的类群形成一个被称为Pentaperalae的单系群, 蔷薇类和菊类是其核心类群, 但是Pentaperalae内部各个类群的关系并没有被最终确定。蔷薇类中, COM分支与固氮分支还是锦葵类形成姊妹群仍然有争议。
Fig. 1 The relationships of major lineages of angiosperms. Except for three basal orders, all angiosperms formed a monophyletic group composed of five major lineages: Chloranthaceae, magnoliids, monocots, Ceratophyllaceae and eudicots. However, their relationships are not clear. Among monocots, Acorales is the basal order and commelinids (shown as triangle in this figure) with four orders are the core groups. The relationships among four magnoliid orders have been clarified. In eudicots, Ranunculales is sister to all other eudicots and Gunnerales is sister to all other core eudicots, which formed a monophyletic group known as Pentapetalae. Rosids and asterids are core groups of Pentapetalae, but there are many uncertain relationships in Pentapetalae. In rosids, it is still uncertain whether the sister group of the COM clade is the nitrogen-fixing clade or malvids.
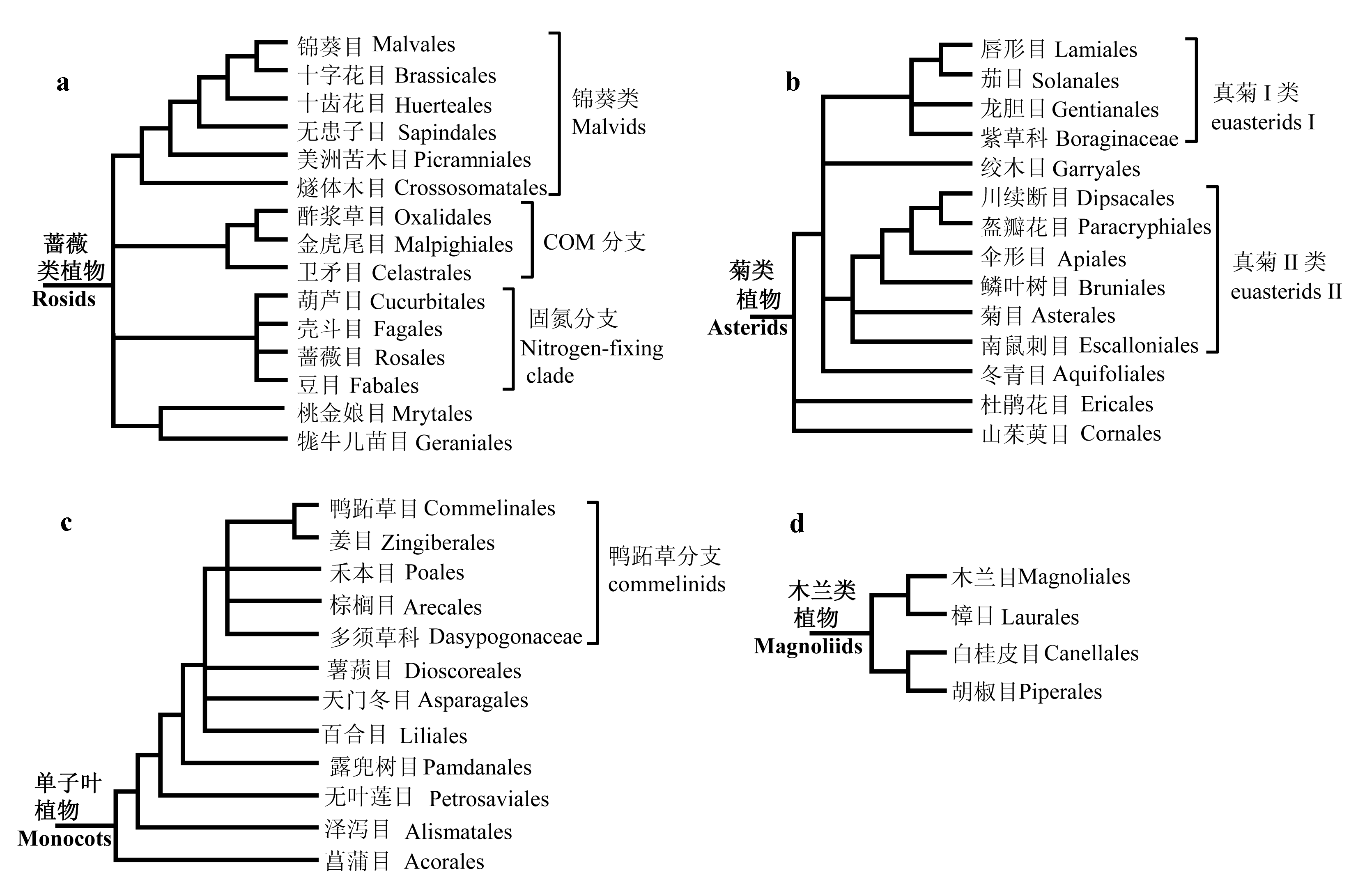
图2 蔷薇类植物、菊类植物、单子叶植物和木兰类植物内部关系图。(a)蔷薇类主要包含3个高支持率的分支, 即锦葵类植物、COM分支和固氮分支, 但是COM分支的姊妹群是固氮分支还是锦葵类植物仍然有争议, 因此在图中以梳齿结构显示。(b)菊类植物主要分为真菊I类和II类, 杜鹃花目和山茱萸目位于其基部; 真菊I类内部的关系尚不清楚; 绞木目和冬青目曾被认为分别位于真菊I类和II类的基部, 但最近来自核基因的研究结果表明二者以姊妹群形式位于真菊I类的基部。(c)单子叶类植物的核心类群是鸭跖草分支, 但该分支内部几个目的关系并不是很清楚; 菖蒲目是单子叶植物的最基部类群。(d)木兰类植物中, 木兰目与樟目、白桂皮目与胡椒目互为姊妹群。
Fig. 2 The relationships within rosids, asterids, monocots and magnoliids, respectively. (a) Rosids contain three clades: malvids, COM and the nitrogen-fixing clade, but whether the sister group of the COM clade is the nitrogen-fixing clade or malvids remains unclear. (b) Asterids are composed of euasterid I, euasterid II and two basal orders: Ericales and Cornales. The relationships among euasterids I are not clear. Garryales and Aquifoliales were respectively regarded as the basal group of euasterids I and II; however, new evidence based on nuclear genes suggested that these two together formed a sister group of other euasterids I. (c) Commelinids is the core group of monocots, and Acorales is the basal order. The relationships among commelinids are not clear. (d) In magnollids, Magnoliales and Laurales are sister groups, and Canellales and Piperales are sister groups.
| [1] | Álvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference.Molecular Phylogenetics and Evolution, 29, 417-434. |
| [2] | Álvarez I, Cronn R, Wendel JF (2005) Phylogeny of the New World diploid cottons (Gossypium L., Malvaceae) based on sequences of three low-copy nuclear genes.Plant Systematics and Evolution, 252, 199-214. |
| [3] | Álvarez I, Costa A, Feliner GN (2008) Selecting single-copy nuclear genes for plant phylogenetics: a preliminary analysis for the Senecioneae (Asteraceae).Journal of Molecular Evolution, 66, 276-291. |
| [4] | Anderberg AA, Baldwin BG, Bayer RG, Breitwieser J, Jeffrey C, Dillon MO, Eldenas P, Funk V, Garcia-Jacas N, Hind DJN (2007) Compositae. In: The Families and Genera of Vascular Plants (ed. Kubitzki K), pp. 61-588, Springer. |
| [5] | Aoki S, Uehara K, Imafuku M, Hasebe M, Ito M (2004) Phylogeny and divergence of basal angiosperms inferred from APETALA-3 and PISTILLATA-like MADS-box genes.Journal of Plant Research, 117, 229-244. |
| [6] | APG (1998) An ordinal classification for the families of flowering plants.Annals of the Missouri Botanical Garden, 85, 531-553. |
| [7] | APG (2003) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II.Botanical Journal of the Linnean Society, 141, 399-436. |
| [8] | Bailey CD, Doyle JJ (1999) Potential phylogenetic utility of the low-copy nuclear gene pistillata in dicotyledonous plants: comparison to nrDNA ITS and trnL intron in Sphaerocardamum and other Brassicaceae.Molecular Phylogenetics and Evolution, 13, 20-30. |
| [9] | Bailey CD, Koch MA, Mayer M, Mummenhoff K, O'Kane SL, Warwick SI, Windham MD, Al-Shehbaz IA (2006) Toward a global phylogeny of the Brassicaceae.Molecular Biology and Evolution, 23, 2142-2160. |
| [10] | Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny.Annals of the Missouri Botanical Garden, 82, 247-277. |
| [11] | Barkman TJ, Chenery G, McNeal JR, Lyons-Weiler J, Ellisens WJ, Moore G, Wolfe AD, dePamphilis CW (2000) Independent and combined analyses of sequences from all three genomic compartments converge on the root of flowering plant phylogeny. Proceedings of the National Academy of Sciences,USA, 97, 13166-13171. |
| [12] | Bergthorsson U, Adams KL, Thomason B, Palmer JD (2003) Widespread horizontal transfer of mitochondrial genes in flowering plants.Nature, 424, 197-201. |
| [13] | Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, vos Rutger A, Gittleman JL, Purvis A (2007) The delayed rise of present-day mammals.Nature, 446, 507-512. |
| [14] | Bremer B, Bremer K, Chase M, Fay M, Reveal J, Soltis D, Soltis P, Stevens P (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III.Botanical Journal of the Linnean Society, 161, 105-121. |
| [15] | Bremer K (2000) Early Cretaceous lineages of monocot flowering plants. Proceedings of the National Academy of Sciences,USA, 97, 4707-4711. |
| [16] | Burleigh JG, Hilu KW, Soltis DE (2009) Inferring phylogenies with incomplete data sets: a 5-gene, 567-taxon analysis of angiosperms.BMC Evolutionary Biology, 9, 61. |
| [17] | Cantino PD, Doyle JA, Graham SW, Judd WS, Olmstead RG, Soltis DE, Soltis PS, Donoghue MJ (2007) Towards a phylogenetic nomenclature of Tracheophyta.Taxon, 56, 1E-44E. |
| [18] | Carpenter KJ (2006) Specialized structures in the leaf epidermis of basal angiosperms: morphology, distribution, and homology.American Journal of Botany, 93, 665-681. |
| [19] | Chapman MA, Chang JC, Weisman D, Kesseli RV, Burke JM (2007) Universal markers for comparative mapping and phylogenetic analysis in the Asteraceae (Compositae).Theoretical and Applied Genetics, 115, 747-755. |
| [20] | Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD, Duvall MR, Price RA, Hills HG, Qiu YL (1993) Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL.Annals of the Missouri Botanical Garden, 80, 528-580. |
| [21] | Choi HK, Luckow MA, Doyle J, Cook DR (2006) Development of nuclear gene-derived molecular markers linked to legume genetic maps.Molecular Genetics and Genomics, 276, 56-70. |
| [22] | Collins TM, Fedrigo O, Naylor GJP (2005) Choosing the best genes for the job: the case for stationary genes in genome-scale phylogenetics.Systematic Biology, 54, 493-500. |
| [23] | Corriveau JL, Coleman AW (1988) Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species.American Journal of Botany, 75, 1443-1458. |
| [24] | Couvreur TL, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K (2010) Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae).Molecular Biology and Evolution, 27, 55-71. |
| [25] | Crane PR, Lidgard S (1989) Angiosperm diversification and paleolatitudinal gradients in Cretaceous floristic diversity.Science, 246, 675-678. |
| [26] | Crane PR, Herendeen P, Friis EM (2004) Fossils and plant phylogeny.American Journal of Botany, 91, 1683-1699. |
| [27] | Crawley S, Hilu K (2012) Impact of missing data, gene choice, and taxon sampling on phylogenetic reconstruction: the Caryophyllales (angiosperms).Plant Systematics and Evolution, 298, 297-312. |
| [28] | Cronquist A (1968) The Evolution and Classification of Flowering Plants. Houghton Mifflin, Boston. |
| [29] | Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW (2002) Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences.American Journal of Botany, 89, 132-144. |
| [30] | Curto MA, Puppo P, Ferreira D, Nogueira M, Meimberg H (2012) Development of phylogenetic markers from single-copy nuclear genes for multi locus, species level analyses in the mint family (Lamiaceae).Molecular Phylogenetics and Evolution, 63, 758-767. |
| [31] | Darwin F, Seward AC (1903) More Letters of Charles Darwin. Cambridge University Press, London, UK. |
| [32] | Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V (2004) Darwin's abominable mystery: insights from a supertree of the angiosperms. Proceedings of the National Academy of Sciences,USA, 101, 1904-1909. |
| [33] | Davis CC, Wurdack KJ (2004) Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales.Science, 305, 676-678. |
| [34] | Davis JI, Stevenson DW, Petersen G, Seberg O, Campbell LM, Freudenstein JV, Goldman DH, Hardy CR, Michelangeli FA, Simmons MP, Mark P, Specht CD, Vergara-Silva F, Gandolfo M (2004) A phylogeny of the monocots, as inferred from rbcL and atpA sequence variation, and a comparison of methods for calculating jackknife and bootstrap values.Systematic Botany, 29, 467-510. |
| [35] | Day A, Ellis T (1984) Chloroplast DNA deletions associated with wheat plants regenerated from pollen: possible basis for maternal inheritance of chloroplasts.Cell, 39, 359-368. |
| [36] | Delsuc F, Brinkmann H, Philippe H (2005) Phylogenomics and the reconstruction of the tree of life.Nature Reviews Genetics, 6, 361-375. |
| [37] | Denton AL, McConaughy BL, Hall BD (1998) Usefulness of RNA polymerase II coding sequences for estimation of green plant phylogeny.Molecular Biology and Evolution, 15, 1082-1085. |
| [38] | Dilcher DL (1989) The occurrence of fruits with affinities to Ceratophyllaceae in lower and mid-Cretaceous sediments.American Journal of Botany, 76, 162. |
| [39] | Doyle JA, Hotton CL (1991) Diversification of early angiosperm pollen in a cladistic context. In: Pollen and Spores: Pattern of Diversification (eds Blackmore S, Barnes SH), pp. 169-195. Clarendon Press, Oxford , England. |
| [40] | Drinnan AN, Crane PR, Friis EM, Pedersen KR (1991) Angiosperm flowers and tricolpate pollen of buxaceous affinity from the Potomac Group (mid-Cretaceous) of eastern North America.American Journal of Botany, 78, 153-176. |
| [41] | Duarte JM, Wall PK, Edger PP, Landherr LL, Ma H, Pires JC, Leebens-Mack J, dePamphilis CW (2010) Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels.BMC Evolutionary Biology, 10, 61. |
| [42] | Eaton DA, Ree RH (2013) Inferring phylogeny and introgression using RADseq Data: an example from flowering plants (Pedicularis: Orobanchaceae).Systematic Biology, 62, 689-706. |
| [43] | Ebersberger I, Strauss S, von Haeseler A (2009) HaMStR: Profile hidden markov model based search for orthologs in ESTs.BMC Evolutionary Biology, 9, 157. |
| [44] | Eklund H, Doyle JA, Herendeen PS (2004) Morphological phylogenetic analysis of living and fossil Chloranthaceae.International Journal of Plant Sciences, 165, 107-151. |
| [45] | Emshwiller E, Doyle JJ (1999) Chloroplast-expressed glutamine synthetase (ncpGS): potential utility for phylogenetic studies with an example from Oxalis (Oxalidaceae).Molecular Phylogenetics and Evolution, 12, 310-319. |
| [46] | Endress PK, Doyle JA (2009) Reconstructing the ancestral angiosperm flower and its initial specializations.American Journal of Botany, 96, 22-66. |
| [47] | Evans RC, Alice LA, Campbell CS, Kellogg EA, Dickinson TA (2000) The granule-bound starch synthase (GBSSI) gene in the Rosaceae: multiple loci and phylogenetic utility.Molecular Phylogenetics and Evolution, 17, 388-400. |
| [48] | Farrell BD (1998) “Inordinate fondness” explained: why are there so many beetles?Science, 281, 555-559. |
| [49] | Fauré S, Noyer JL, Carreel F, Horry JP, Bakry F, Lanaud C (1994) Maternal inheritance of chloroplast genome and paternal inheritance of mitochondrial genome in bananas (Musa acuminata).Current Genetics, 25, 265-269. |
| [50] | Finet C, Timme RE, Delwiche CF, Marlétaz F (2010) Multigene phylogeny of the green lineage reveals the origin and diversification of land plants.Current Biology, 20, 2217-2222. |
| [51] | Fishbein M, Hibsch-Jetter C, Soltis DE, Hufford L (2001) Phylogeny of Saxifragales (angiosperms, eudicots): analysis of a rapid, ancient radiation.Systematic Biology, 50, 817-847. |
| [52] | Fortune P, Schierenbeck KA, Ainouche A, Jacquemin J, Wendel JF, Ainouche M (2007) Evolutionary dynamics of Waxy and the origin of hexaploid Spartina species (Poaceae).Molecular Phylogenetics and Evolution, 43, 1040-1055. |
| [53] | Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains.The EMBO Journal, 18, 4679-4688. |
| [54] | Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K (2011) Cabbage family affairs: the evolutionary history of Brassicaceae.Trends in Plant Science, 16, 108-116. |
| [55] | Friedman WE (2009) The meaning of Darwin’s “abominable mystery”.American Journal of Botany, 96, 5-21. |
| [56] | Friis E, Crane P, Pedersen K (1986) Floral evidence for Cretaceous chloranthoid angiosperms.Nature, 320, 163-164. |
| [57] | Friis EM, Crane PR, Pedersen KR, Bengtson S, Donoghue PC, Grimm GW, Stampanoni M (2007) Phase-contrast X-ray microtomography links Cretaceous seeds with Gnetales and Bennettitales.Nature, 450, 549-552. |
| [58] | Fulton TM, van der Hoeven R, Eannetta NT, Tanksley SD (2002) Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants.The Plant Cell, 14, 1457-1467. |
| [59] | Funk VA, Randall BA, Sterling-Keeley ER, Chan R, Watson L, Gemeinholzer B, Schilling E, Panero JL, Baldwin BG, Nuria GJ (2005) Everywhere but Antarcita: using a supertree to understand the diversity and distribution of the Compositae.Biologiske Skrifter, 55, 343-374. |
| [60] | Galloway GL, Malmberg RL, Price RA (1998) Phylogenetic utility of the nuclear gene arginine decarboxylase: an example from Brassicaceae.Molecular Biology and Evolution, 15, 1312-1320. |
| [61] | Goremykin VV, Hirsch-Ernst KI, Wölfl S, Hellwig FH (2003) Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm.Molecular Biology and Evolution, 20, 1499-1505. |
| [62] | Goremykin VV, Viola R, Hellwig FH (2009) Removal of noisy characters from chloroplast genome-scale data suggests revision of phylogenetic placements of Amborella and Ceratophyllum.Journal of Molecular Evolution, 68, 197-204. |
| [63] | Goremykin VV, Nikiforova SV, Bininda-Emonds OR (2010) Automated removal of noisy data in phylogenomic analyses.Journal of Molecular Evolution, 71, 319-331. |
| [64] | Goremykin VV, Nikiforova SV, Biggs PJ, Zhong B, Delange P, Martin W, Woetzel S, Atherton RA, Mclenachan PA, Lockhart PJ (2013) The evolutionary root of flowering plants.Systematic Biology, 62, 50-61. |
| [65] | Gouy M, Li W (1989) Molecular phylogeny of the kingdoms Animalia, Plantae, and Fungi.Molecular Biology and Evolution, 6, 109-122. |
| [66] | Graham SW, Zgurski JM, McPherson MA, Cherniawsky DM, Saarela JM, Horne EF, Smith SY, Wong WA, O’Brien HE, Biron VL (2006) Robust inference of monocot deep phylogeny using an expanded multigene plastid data set.Aliso, 22, 3-20. |
| [67] | Graybeal A (1998) Is it better to add taxa or characters to a difficult phylogenetic problem?Systematic Biology, 47, 9-17. |
| [68] | Gu X, Fu YX, Li WH (1995) Maximum likelihood estimation of the heterogeneity of substitution rate among nucleotide sites.Molecular Biology and Evolution, 12, 546-557. |
| [69] | Hamby RK, Zimmer EA (1988) Ribosomal RNA sequences for inferring phylogeny within the grass family (Poaceae).Plant Systematics and Evolution, 160, 29-37 |
| [70] | Herendeen PS, Magallon-Puebla S, Lupia R, Crane PR, Kobylinska J (1999) A preliminary conspectus of the Allon flora from the late Cretaceous (late Santonian) of central Georgia, USA.Annals of the Missouri Botanical Garden, 86, 407-471. |
| [71] | Hillis DM, Pollock DD, McGuire JA, Zwickl DJ (2003) Is sparse taxon sampling a problem for phylogenetic inference? Systematic Biology, 52, 124. |
| [72] | Hilu KW, Liang HP (1997) The matK gene: sequence variation and application in plant systematics.American Journal of Botany, 84, 830-830. |
| [73] | Hilu KW, Borsch T, Müller K, Soltis DE, Soltis PS, Savolainen V, Chase MW, Powell MP, Alice LA, Evans R (2003) Angiosperm phylogeny based on matK sequence information.American Journal of Botany, 90, 1758-1776. |
| [74] | Huang SX, Sirikhachornkit A, Faris JD, Su XJ, Gill BS, Haselkorn R, Gornicki P (2002) Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses.Plant Molecular Biology, 48, 805-820. |
| [75] | Jansen RK, Cai ZQ, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences,USA, 104, 19369-19374. |
| [76] | Jansen RK, Saski C, Lee SB, Hansen AK, Daniell H (2011) Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus.Molecular Biology and Evolution, 28, 835-847. |
| [77] | Jeffroy O, Brinkmann H, Delsuc F, Philippe H (2006) Phylogenomics: the beginning of incongruence?Trends in Genetics, 22, 225-231. |
| [78] | Jian SG, Soltis PS, Gitzendanner MA, Moore MJ, Li RQ, Hendry TA, Qiu YL, Dhingra A, Bell CD, Soltis DE (2008) Resolving an ancient, rapid radiation in Saxifragales.Systematic Biology, 57, 38-57. |
| [79] | Jiao YN, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang HY, Soltis PS (2011) Ancestral polyploidy in seed plants and angiosperms.Nature, 473, 97-100. |
| [80] | Johnson BR, Borowiec ML, Chiu JC, Lee EK, Atallah J, Ward PS (2013) Phylogenomics resolves evolutionary relationships among ants, bees, and wasps.Current Biology, 23, 2058-2062. |
| [81] | Judd WS, Olmstead RG (2004) A survey of tricolpate (eudicot) phylogenetic relationships.American Journal of Botany, 91, 1627-1644. |
| [82] | Kim K, Jansen RK (1995)ndhF sequence evolution and the major clades in the sunflower family. Proceedings of the National Academy of Sciences,USA, 92, 10379-10383. |
| [83] | Kim ST, Sultan SE, Donoghue MJ (2008) Allopolyploid speciation in Persicaria (Polygonaceae): insights from a low-copy nuclear region. Proceedings of the National Academy of Sciences,USA, 105, 12370-12375. |
| [84] | Kingman JF (1982) On the genealogy of large populations.Journal of Applied Probability, 19, 27-43. |
| [85] | Kingman JF (2000) Origins of the coalescent: 1974-1982.Genetics, 156, 1461-1463. |
| [86] | Kocot KM, Cannon JT, Todt C, Citarella MR, Kohn AB, Meyer A, Santos SR, Schander C, Moroz LL, Lieb B, Halanych KM (2011) Phylogenomics reveals deep molluscan relationships.Nature, 477, 452-456. |
| [87] | Kong HZ (2001) Comparative morphology of leaf epidermis in the Chloranthaceae.Botanical Journal of the Linnean Society, 136, 279-294. |
| [88] | Krak K, Álvarez I, Caklová P, Costa A, Chrtek J, Fehrer J (2012) Development of novel low-copy nuclear markers for Hieraciinae (Asteraceae) and their perspective for other tribes.American Journal of Botany, 99, e74-e77. |
| [89] | Kubatko LS, Carstens BC, Knowles LL (2009) STEM: species tree estimation using maximum likelihood for gene trees under coalescence.Bioinformatics, 25, 971-973. |
| [90] | Lee EK, Cibrian-Jaramillo A, Kolokotronis SO, Katari MS, Stamatakis A, Ott M, Chiu JC, Little DP, Stevenson DW, McCombie WR (2011) A functional phylogenomic view of the seed plants.PLoS Genetics, 7, e1002411. |
| [91] | Li DZ, Gao LM, Li H, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, Yang JB (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences,USA, 108, 19641-19646. |
| [92] | Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes.Genome Research, 13, 2178-2189. |
| [93] | Lin ZG, Kong HZ, Nei M, Ma H (2006) Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proceedings of the National Academy of Sciences,USA, 103, 10328-10333. |
| [94] | Liu L (2008) BEST: Bayesian estimation of species trees under the coalescent model.Bioinformatics, 24, 2542-2543. |
| [95] | Liu L, Yu LL, Pearl DK, Edwards SV (2009) Estimating species phylogenies using coalescence times among sequences.Systematic Biology, 58, 468-477. |
| [96] | Liu L, Yu LL, Pearl DK (2010) Maximum tree: a consistent estimator of the species tree.Journal of Mathematical Biology, 60, 95-106. |
| [97] | Löhne C, Borsch T (2005) Molecular evolution and phylogenetic utility of the petD group II intron: a case study in basal angiosperms.Molecular Biology and Evolution, 22, 317-332. |
| [98] | Lonsdale D, Brears T, Hodge T, Melville SE, Rottmann W (1988) The plant mitochondrial genome: homologous recombination as a mechanism for generating heterogeneity.Philosophical Transactions of the Royal Society of London B: Biological Sciences, 319, 149-163. |
| [99] | Lopez P, Casane D, Philippe H (2002) Heterotachy, an important process of protein evolution.Molecular Biology and Evolution, 19, 1-7. |
| [100] | Magallón S, Castillo A (2009) Angiosperm diversification through time.American Journal of Botany, 96, 349-365. |
| [101] | Magallón S, Crane PR, Herendeen PS (1999) Phylogenetic pattern, diversity, and diversification of eudicots.Annals of the Missouri Botanical Garden, 96, 297-372. |
| [102] | Ma H, Yanofsky MF, Meyerowitz EM (1990) Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proceedings of the National Academy of Sciences,USA, 87, 3821-3825. |
| [103] | Mason-Gamer RJ, Weil CF, Kellogg EA (1998) Granule-bound starch synthase: structure, function, and phylogenetic utility.Molecular Biology and Evolution, 15, 1658-1673. |
| [104] | Mathews S, Donoghue MJ (1999) The root of angiosperm phylogeny inferred from duplicate phytochrome genes.Science, 286, 947-950. |
| [105] | Meng SW, Chen ZD, Li DZ, Liang HX (2002) Phylogeny of Saururaceae based on mitochondrial matR gene sequence data.Journal of Plant Research, 115, 0071-0076. |
| [106] | Mitsui Y, Chen ST, Zhou ZK, Peng CI, Deng YF, Setoguchi H (2008) Phylogeny and biogeography of the genus Ainsliaea (Asteraceae) in the Sino-Japanese region based on nuclear rDNA and plastid DNA sequence data.Annals of Botany, 101, 111-124. |
| [107] | Moore MJ, Bell CD, Soltis PS, Soltis DE (2007) Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms.Proceedings of the National Academy of Sciences, USA, 104, 19363-19368. |
| [108] | Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE (2010) Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots.Proceedings of the National Academy of Sciences, USA, 107, 4623-4628. |
| [109] | Moore MJ, Hassan N, Gitzendanner MA, Bruenn RA, Croley M, Vandeventer A, Horn JW, Dhingra A, Brockington SF, Latvis M (2011) Phylogenetic analysis of the plastid inverted repeat for 244 species: insights into deeper-level angiosperm relationships from a long, slowly evolving sequence region.International Journal of Plant Sciences, 172, 541-558. |
| [110] | Moreau CS, Bell CD, Vila R, Archibald SB, Pierce NE (2006) Phylogeny of the ants: diversification in the age of angiosperms.Science, 312, 101-104. |
| [111] | Mort ME, Crawford DJ (2004) The continuing search: low-copy nuclear sequences for lower-level plant molecular phylogenetic studies.Taxon, 53, 257-261. |
| [112] | Ness RW, Graham SW, Barrett SC (2011) Reconciling gene and genome duplication events: using multiple nuclear gene families to infer the phylogeny of the aquatic plant family Pontederiaceae.Molecular Biology and Evolution, 28, 3009-3018. |
| [113] | Ng M, Yanofsky MF (2001) Function and evolution of the plant MADS-box gene family.Nature Reviews Genetics, 2, 186-195. |
| [114] | Nomura N, Takaso T, Peng CI, Kono Y, Oginuma K, Mitsui Y, Setoguchi H (2010) Molecular phylogeny and habitat diversification of the genus Farfugium (Asteraceae) based on nuclear rDNA and plastid DNA.Annals of Botany, 106, 467-482. |
| [115] | O'Brien KP, Remm M, Sonnhammer EL (2005) Inparanoid: a comprehensive database of eukaryotic orthologs.Nucleic Acids Research, 33, D476-D480. |
| [116] | Olmstead RG, Palmer JD (1994) Chloroplast DNA systematics: a review of methods and data analysis.American Journal of Botany, 81, 1205-1224. |
| [117] | Palmer JD, Herbon LA (1988) Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence.Journal of Molecular Evolution, 28, 87-97. |
| [118] | Palmer JD (1990) Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant.Nature, 348, 337-339. |
| [119] | Panero JL, Funk VA (2008) The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed.Molecular Phylogenetics and Evolution, 47, 757-782. |
| [120] | Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, Baurain D (2011) Resolving difficult phylogenetic questions: why more sequences are not enough.PLoS Biology, 9, e1000602. |
| [121] | Phillips MJ, Delsuc F, Penny D (2004) Genome-scale phylogeny and the detection of systematic biases.Molecular Biology and Evolution, 21, 1455-1458. |
| [122] | Piep MB, Gunn BF, Roberts C (2007) Flora of North America. Oxford University Press, New York. |
| [123] | Qiu YL, Lee JH, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen ZD, Savolainen V, Chase MW (1999) The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes.Nature, 402, 404-407. |
| [124] | Qiu YL, Dombrovska O, Lee JH, Li LB, Whitlock BA, Bernasconi-Quadroni F, Rest JS, Davis CC, Borsch T, Hilu KW (2005) Phylogenetic analyses of basal angiosperms based on nine plastid, mitochondrial, and nuclear genes.International Journal of Plant Sciences, 166, 815-842. |
| [125] | Qiu YL, Li L, Wang B, Xue JY, Hendry TA, Li RQ, Brown JW, Liu Y, Hudson GT, Chen ZD (2010) Angiosperm phylogeny inferred from sequences of four mitochondrial genes.Journal of Systematics and Evolution, 48, 391-425. |
| [126] | Roelants K, Gower DJ, Wilkinson M, Loader SP, Biju S, Guillaume K, Moriau L, Bossuyt F (2007) Global patterns of diversification in the history of modern amphibians.Proceedings of the National Academy of Sciences, USA, 104, 887-892. |
| [127] | Rokas A, Williams BL, King N, Carroll SB (2003) Genome-scale approaches to resolving incongruence in molecular phylogenies.Nature, 425, 798-804. |
| [128] | Rokas A, Carroll SB (2006) Bushes in the tree of life.PLoS Biology, 4, e352. |
| [129] | Salichos L, Rokas A (2013) Inferring ancient divergences requires genes with strong phylogenetic signals.Nature, 497, 327-331. |
| [130] | Sang T (2002) Utility of low-copy nuclear gene sequences in plant phylogenetics.Critical Reviews in Biochemistry and Molecular Biology, 37, 121-147. |
| [131] | Savolainen V, Chase MW, Hoot SB, Morton CM, Soltis DE, Bayer C, Fay MF, De Bruijn AY, Sullivan S, Qiu YL (2000) Phylogenetics of flowering plants based on combined analysis of plastid atpB and rbcL gene sequences.Systematic Biology, 49, 306-362. |
| [132] | Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallón S, Lupia R (2004) Ferns diversified in the shadow of angiosperms.Nature, 428, 553-557. |
| [133] | Shaw, Ruan TZ, Glenn T, Liu L (2013) STRAW: Species TRee Analysis Web server.Nucleic Acids Research, 41, 238-241. |
| [134] | Small RL, Wendel JF (2000) Phylogeny, duplication, and intraspecific variation of Adh sequences in new world diploid cottons (Gossypium L., Malvaceae).Molecular Phylogenetics and Evolution, 16, 73-84. |
| [135] | Smith SA, Beaulieu JM, Donoghue MJ (2010) An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants.Proceedings of the National Academy of Sciences, USA, 107, 5897-5902. |
| [136] | Smith SA, Beaulieu JM, Stamatakis A, Donoghue MJ (2011) Understanding angiosperm diversification using small and large phylogenetic trees.American Journal of Botany, 98, 404-414. |
| [137] | Smith SA, Wilson NG, Goetz FE, Feehery C, Andrade SC, Rouse GW, Giribet G, Dunn CW (2012) Corrigendum: resolving the evolutionary relationships of molluscs with phylogenomic tools.Nature, 493, 708-708. |
| [138] | Soltis DE, Albert VA, Savolainen V, Hilu K, Qiu YL, Chase MW, Farris JS, Stefanović S, Rice DW, Palmer JD (2004) Genome-scale data, angiosperm relationships, and ‘ending incongruence’: a cautionary tale in phylogenetics.Trends in Plant Science, 9, 477-483. |
| [139] | Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, Wall PK, Soltis PS (2009) Polyploidy and angiosperm diversification.American Journal of Botany, 96, 336-348. |
| [140] | Soltis DE, Smith SA, Cellinese N, Wurdack KJ, Tank DC, Brockington SF, Refulio-Rodriguez NF, Walker JB, Moore MJ, Carlsward BS, Bell CD, Latvis M, Crawley S, Black C, Diouf D, Xi Z, Rushworth CA, Gitzendanner MA, Sytsma KJ, Qiu YL, Hilu KW, Davis CC, Sanderson MJ, Beaman RS, Olmstead RG, Judd WS, Donoghue MJ, Soltis PS (2011) Angiosperm phylogeny: 17 genes, 640 taxa.American Journal of Botany, 98, 704-730. |
| [141] | Soltis DE, Soltis PS, Endress PK, Chase MW (2005) Phylogeny and evolution of angiosperms. Sunderland, Massachusetts. |
| [142] | Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF (2000) Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences.Botanical Journal of the Linnean Society, 133, 381-461. |
| [143] | Soltis DE, Soltis PS, Nickrent DL, Johnson LA, Hahn WJ, Hoot SB, Sweere JA, Kuzoff RK, Kron KA, Chase MW (1997) Angiosperm phylogeny inferred from 18S ribosomal DNA sequences.Annals of the Missouri Botanical Garden, 84, 1-49. |
| [144] | Soltis DE, Soltis PS, Zanis MJ (2002) Phylogeny of seed plants based on evidence from eight genes.American Journal of Botany, 89, 1670-1681. |
| [145] | Soltis DE, Soltis PS (2004) Amborella not a “basal angiosperm”? Not so fast.American Journal of Botany, 91, 997-1001. |
| [146] | Soltis PS, Soltis DE (2013) Angiosperm phylogeny: a framework for studies of genome evolution.Plant Genome Diversity, 2, 1-11. |
| [147] | Soltis PS, Soltis DE, Chase MW (1999) Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology.Nature, 402, 402-404. |
| [148] | Steele PR, Guisinger-Bellian M, Linder CR, Jansen RK (2008) Phylogenetic utility of 141 low-copy nuclear regions in taxa at different taxonomic levels in two distantly related families of rosids.Molecular Phylogenetics and Evolution, 48, 1013-1026. |
| [149] | Stefanović S, Rice DW, Palmer JD (2004) Long branch attraction, taxon sampling, and the earliest angiosperms: Amborella or monocots?BMC Evolutionary Biology, 4, 35. |
| [150] | Stevens P (2001) Angiosperm phylogeny website.. |
| [151] | Strand A, Leebens-Mack J, Milligan B (1997) Nuclear DNA-based markers for plant evolutionary biology.Molecular Ecology, 6, 113-118. |
| [152] | Sun G, Ji Q, Dilcher DL, Zheng SL, Nixon KC, Wang XF (2002) Archaefructaceae, a new basal angiosperm family.Science, 296, 899-904. |
| [153] | Sun G, Dilcher DL, Wang HS, Chen ZD (2011) A eudicot from the Early Cretaceous of China.Nature, 471, 625-628. |
| [154] | Sun YX, Moore MJ, Meng AP, Soltis PS, Soltis DE, Li JQ, Wang HC (2013) Complete plastid genome sequencing of trochodendraceae reveals a significant expansion of the inverted repeat and suggests a paleogene divergence between the two extant species.PLoS ONE, 8, e60429. |
| [155] | Swamy B (1953) The morphology and relationships of the Chloranthaceae.Journal of the Arnold Arboretum, 34, 375-408. |
| [156] | Tang L, Zou XH, Achoundong G, Potgieter C, Second G, Zhang DY, Ge S (2010) Phylogeny and biogeography of the rice tribe (Oryzeae): evidence from combined analysis of 20 chloroplast fragments.Molecular Phylogenetics and Evolution, 54, 266-277. |
| [157] | Tank DC, Sang T (2001) Phylogenetic utility of the Glycerol-3-Phosphate acyltransferase gene: evolution and implications in Paeonia (Paeoniaceae).Molecular Phylogenetics and Evolution, 19, 421-429. |
| [158] | Thorne RF (1992) Classification and geography of the flowering plants.The Botanical Review, 58, 225-327. |
| [159] | Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S (2002) Agricultural sustainability and intensive production practices.Nature, 418, 671-677. |
| [160] | Townsend JP, Su Z, Tekle YI (2012) Phylogenetic signal and noise: predicting the power of a data set to resolve phylogeny.Systematic Biology, 61, 835-849. |
| [161] | Wang HX, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE (2009) Rosid radiation and the rapid rise of angiosperm-dominated forests.Proceedings of the National Academy of Sciences, USA, 106, 3853-3858. |
| [162] | Wang X, Zhao L, Eaton D, Li D, Guo Z (2013) Identification of SNP markers for inferring phylogeny in temperate bamboos (Poaceae: Bambusoideae) using RAD sequencing.Molecular Ecology Resources, 13, 938-945. |
| [163] | Weiss CA, Garnaat CW, Mukai K, Hu Y, Ma H (1994) Isolation of cDNAs encoding guanine nucleotide-binding protein beta-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1).Proceedings of the National Academy of Sciences, USA, 91, 9554-9558. |
| [164] | Wen J, Nie ZL, Soejima A, Meng Y (2007) Phylogeny of Vitaceae based on the nuclear GAI1 gene sequences.Canadian Journal of Botany, 85, 731-745. |
| [165] | Wen J, Xiong Z, Nie ZL, Mao L, Zhu Y, Kan XZ, Ickert-Bond SM, Gerrath J, Zimmer EA, Fang XD (2013) Transcriptome sequences resolve deep relationships of the grape family.PLoS ONE, 8, e74394. |
| [166] | Westwood JH, Yoder JI, Timko MP, dePamphilis CW (2010) The evolution of parasitism in plants.Trends in Plant Science, 15, 227-235. |
| [167] | Weng ML, Ruhlman TA, Gibby M, Jansen RK (2012) Phylogeny, rate variation, and genome size evolution of Pelargonium (Geraniaceae).Molecular Phylogenetics and Evolution, 64, 654-670. |
| [168] | Wilf P, Labandeira CC, Kress WJ, Staines CL, Windsor DM, Allen AL, Johnson KR (2000) Timing the radiations of leaf beetles: Hispines on gingers from latest Cretaceous to recent.Science, 289, 291-294. |
| [169] | Won H, Renner SS (2003) Horizontal gene transfer from flowering plants to Gnetum.Proceedings of the National Academy of Sciences, USA, 100, 10824-10829. |
| [170] | Worberg A, Quandt D, Barniske AM, Löhne C, Hilu KW, Borsch T (2007) Phylogeny of basal eudicots: insights from non-coding and rapidly evolving DNA.Organisms Diversity and Evolution, 7, 55-77. |
| [171] | Wortley AH, Rudall PJ, Harris DJ, Scotland RW (2005) How much data are needed to resolve a difficult phylogeny? Case study in Lamiales.Systematic Biology, 54, 697-709. |
| [172] | Wu FN, Mueller LA, Crouzillat D, Pétiard V, Tanksley SD (2006) Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade.Genetics, 174, 1407-1420. |
| [173] | Wu ZQ, Ge S (2012) The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts.Molecular Phylogenetics and Evolution, 62, 573-578. |
| [174] | Xi ZX, Ruhfel BR, Schaefer H, Amorim AM, Sugumaran M, Wurdack KJ, Endress PK, Matthews ML, Stevens PF, Mathews S (2012) Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales.Proceedings of the National Academy of Sciences, USA, 109, 17519-17524. |
| [175] | Xu GX, Ma H, Nei M, Kong HZ (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification.Proceedings of the National Academy of Sciences, USA, 106, 835-840. |
| [176] | Yang ZH (1994) Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods.Journal of Molecular Evolution, 39, 306-314. |
| [177] | Yuan YW, Liu C, Marx HE, Olmstead RG (2009) The pentatricopeptide repeat (PPR) gene family, a tremendous resource for plant phylogenetic studies.New Phytologist, 182, 272-283. |
| [178] | Yuan YW, Liu C, Marx HE, Olmstead RG (2010) An empirical demonstration of using pentatricopeptide repeat (PPR) genes as plant phylogenetic tools: phylogeny of Verbenaceae and the Verbena complex.Molecular Phylogenetics and Evolution, 54, 23-35. |
| [179] | Zanis MJ, Soltis DE, Soltis PS, Mathews S, Donoghue MJ (2002) The root of the angiosperms revisited.Proceedings of the National Academy of Sciences, USA, 99, 6848-6853. |
| [180] | Zhang N, Zeng LP, Shan HY, Ma H (2012) Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms.New Phytologist, 195, 923-937. |
| [181] | Zhang YJ, Ma PF, Li DZ (2011) High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae).PLoS ONE, 6, e20596. |
| [182] | Zhu X, Chase MW, Qiu YL, Kong HZ, Dilcher DL, Li JH, Chen ZD (2007) Mitochondrial matR sequences help to resolve deep phylogenetic relationships in rosids.BMC Evolutionary Biology, 7, 217. |
| [183] | Zimmer EA, Wen J (2013) Reprint of: using nuclear gene data for plant phylogenetics: progress and prospects.Molecular Phylogenetics and Evolution, 66, 539-550. |
| [1] | 朱瑞良, 马晓英, 曹畅, 曹子寅. 中国苔藓植物多样性研究进展[J]. 生物多样性, 2022, 30(7): 22378-. |
| [2] | 李潮,金锦锦,罗锦桢,王春晖,王俊杰,赵俊. 唐鱼养殖种群与广州附近4个野生种群的遗传关系[J]. 生物多样性, 2020, 28(4): 474-484. |
| [3] | 王伟, 刘阳. 植物生命之树重建的现状、问题和对策建议[J]. 生物多样性, 2020, 28(2): 176-188. |
| [4] | 杜新宇, 卢金梅, 李德铢. 石松类和蕨类植物质体基因组结构演化研究进展[J]. 生物多样性, 2019, 27(11): 1172-1183. |
| [5] | 杨计平, 李策, 陈蔚涛, 李跃飞, 李新辉. 西江中下游鳤的遗传多样性与种群动态历史[J]. 生物多样性, 2018, 26(12): 1289-1295. |
| [6] | 周秋杰, 蔡亚城, 黄伟伦, 吴伟, 代色平, 王峰, 周仁超. 野牡丹属两个海南特有种与同属广布种自然杂交的分子证据[J]. 生物多样性, 2017, 25(6): 638-646. |
| [7] | 舒江平, 刘莉, 沈慧, 戴锡玲, 王全喜, 严岳鸿. 基于系统基因组学分析揭示早期陆生植物的复杂网状进化关系[J]. 生物多样性, 2017, 25(6): 675-682. |
| [8] | 张明理. 中国西北干旱区和中亚植物区系地理研究[J]. 生物多样性, 2017, 25(2): 147-155. |
| [9] | 柳淑芳, 刘进贤, 庄志猛, 高天翔, 韩志强, 陈大刚. 舌鳎亚科鱼类单系起源和同种异名的线粒体DNA证据[J]. 生物多样性, 2010, 18(3): 275-282. |
| [10] | 徐磊, 杨大荣. 榕树及其传粉榕小蜂的系统发育和协同进化研究现状及展望[J]. 生物多样性, 2008, 16(5): 446-453. |
| [11] | 唐文乔, 胡雪莲, 杨金权. 从线粒体控制区全序列变异看短颌鲚和湖鲚的物种有效性[J]. 生物多样性, 2007, 15(3): 224-231. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn