



Biodiv Sci ›› 2008, Vol. 16 ›› Issue (2): 156-165. DOI: 10.3724/SP.J.1003.2008.07292 cstr: 32101.14.SP.J.1003.2008.07292
• Original article • Previous Articles Next Articles
Xinjun Liao1,3, Hong Chang3, Guixiang Zhang2,*( ), Donglei Wang3, Weitao Song4, Xu Han2, Zifu Zhang5
), Donglei Wang3, Weitao Song4, Xu Han2, Zifu Zhang5
Received:2007-09-17
Accepted:2007-12-24
Online:2008-03-20
Published:2008-02-20
Contact:
Guixiang Zhang
Xinjun Liao, Hong Chang, Guixiang Zhang, Donglei Wang, Weitao Song, Xu Han, Zifu Zhang. Genetic diversity of five native Chinese yak breeds based on microsatellite DNA markers[J]. Biodiv Sci, 2008, 16(2): 156-165.
| 代号 Code | 品种 Breeds | 数量 Sample size | 采集地点 Collection location |
|---|---|---|---|
| PL | 帕里牦牛 Pali yak | 62 | 西藏自治区帕里镇 Pali Town, Tibet |
| SB | 斯布牦牛 Sibu yak | 57 | 西藏自治区斯布乡斯布牦牛场 Sibu Yak Farm of Sibu Town, Tibet |
| GS | 西藏高山牦牛 Tibetan High Mountainous yak | 61 | 西藏自治区那曲地区那曲县布巴镇 Buba Town, Naqu County, Tibet |
| JL | 九龙牦牛 Jiulong yak | 60 | 四川九龙县 Jiulong County, Sichuan |
| MW | 麦洼牦牛 Maiwa yak | 59 | 四川阿坝州畜牧局牦牛选育基地 Yak Breeding Center of Aba State Livestock Bureau, Sichuan |
| GY | 大额牛 Gayal | 20 | 云南泸水县老窝乡 Laowo Town, Lushui County, Yunnan |
Table 1 Name, sample size and collecting location of the studied yak breeds
| 代号 Code | 品种 Breeds | 数量 Sample size | 采集地点 Collection location |
|---|---|---|---|
| PL | 帕里牦牛 Pali yak | 62 | 西藏自治区帕里镇 Pali Town, Tibet |
| SB | 斯布牦牛 Sibu yak | 57 | 西藏自治区斯布乡斯布牦牛场 Sibu Yak Farm of Sibu Town, Tibet |
| GS | 西藏高山牦牛 Tibetan High Mountainous yak | 61 | 西藏自治区那曲地区那曲县布巴镇 Buba Town, Naqu County, Tibet |
| JL | 九龙牦牛 Jiulong yak | 60 | 四川九龙县 Jiulong County, Sichuan |
| MW | 麦洼牦牛 Maiwa yak | 59 | 四川阿坝州畜牧局牦牛选育基地 Yak Breeding Center of Aba State Livestock Bureau, Sichuan |
| GY | 大额牛 Gayal | 20 | 云南泸水县老窝乡 Laowo Town, Lushui County, Yunnan |
| 座位 Locus | 引物序列 (5’-3’) Primer sequence | 退火温度 Annealing temp.(℃) | 片段大小 Allele Size (bp) | 观察 等位基因数 Observed allele number | 荧光标记 Fluorescence | 所在染色体 Chromosome location |
|---|---|---|---|---|---|---|
| SPS115 | F: AAAGTGACACAACAGCTTCTCCAG R: AACGAGTGTCCTAGTTTGGCTGTG | 55-60 | 234-258 | 11 | Vic | 15 |
| TGLA122 | F: CCCTCCTCCAGGTAAATCAGC R: AATCACATGGCAAATAAGTACATAC | 55-58 | 136-184 | 12 | Ned | 21 |
| TGLA53 | F: GCTTTCAGAAATAGTTTGCATTCA R: ATCTTCACATGATATTACAGCAGA | 55 | 143-191 | 13 | Pet | 16 |
| TGLA126 | F: CTAATTTAGAATGAGAGAGGCTTCT R: TTGGTCTCTATTCTCTGAATATTCC | 55-58 | 115-131 | 7 | Vic | 20 |
| TGLA227 | F: CGAATTCCAAATCTGTTAATTTGCT R: ACAGACAGAAACTCAATGAAAGCA | 55-56 | 75-105 | 10 | Pet | 18 |
| HEL13 | F: TAAGGACTTGAGATAAGGAG R: CCATCTACCTCCATCTTAAC | 51.6 | 165-191 | 11 | Pet | 11 |
| INRA005 | F: CAATCTGCATGAAGTATAAATAT R: CTTCAGGCATACCCTACACC | 55 | 120-160 | 10 | Ned | 12 |
| BM1824 | F: GAGCAAGGTGTTTTTCCAATC R: CATTCTCCAACTGCTTCCTTG | 57 | 180-200 | 7 | 6fam | 1 |
| HEL5 | F: GCAGGATCACTTGTTAGGGA R: AGACGTTAGTGTACATTAAC | 54 | 149-165 | 7 | Vic | 21 |
| BM2113 | F: GCTGCCTTCTACCAAATACCC R: CTTAGACAACAGGGGTTTGG | 55-60 | 116-146 | 13 | 6fam | 2 |
| TGLA57 | F: GCTTCCAAAACTTTACAATATGTAT R: GCTTTTTAATCCTCAGCTTGCTG | 55 | 77-99 | 9 | 6fam | 1 |
| ILSTS013 | F: ACACAAAATCAGATCAGTGG R: CTTGATCCTTATAGAACTGG | 58 | 121-135 | 11 | Vic | 9 |
| ILSTS050 | F: AAATCAGACACCCAGTTTCC R: GTTTTTCTACACGAGTTGGC | 55 | 161-183 | 13 | Pet | 21 |
| AGLA293 | F: GAAACTCAACCCAAGACAACTCAAG R: ATGACTTTATTCTCCACCTAGCAGA | 55 | 210-240 | 11 | 6fam | 5 |
| ILSTS008 | F: GAATCATGGATTTTCTGGGG R: TAGCAGTGAGTGAGGTTGGC | 58 | 175-187 | 7 | 6fam | 14 |
| TGLA73 | F: GAGAATCACCTAGAGAGAGGCA R: CTTTCTCTTTAAATTCTATATGGT | 55 | 111-143 | 14 | 6fam | 9 |
Table 2 The information about sixteen microsatellite primers
| 座位 Locus | 引物序列 (5’-3’) Primer sequence | 退火温度 Annealing temp.(℃) | 片段大小 Allele Size (bp) | 观察 等位基因数 Observed allele number | 荧光标记 Fluorescence | 所在染色体 Chromosome location |
|---|---|---|---|---|---|---|
| SPS115 | F: AAAGTGACACAACAGCTTCTCCAG R: AACGAGTGTCCTAGTTTGGCTGTG | 55-60 | 234-258 | 11 | Vic | 15 |
| TGLA122 | F: CCCTCCTCCAGGTAAATCAGC R: AATCACATGGCAAATAAGTACATAC | 55-58 | 136-184 | 12 | Ned | 21 |
| TGLA53 | F: GCTTTCAGAAATAGTTTGCATTCA R: ATCTTCACATGATATTACAGCAGA | 55 | 143-191 | 13 | Pet | 16 |
| TGLA126 | F: CTAATTTAGAATGAGAGAGGCTTCT R: TTGGTCTCTATTCTCTGAATATTCC | 55-58 | 115-131 | 7 | Vic | 20 |
| TGLA227 | F: CGAATTCCAAATCTGTTAATTTGCT R: ACAGACAGAAACTCAATGAAAGCA | 55-56 | 75-105 | 10 | Pet | 18 |
| HEL13 | F: TAAGGACTTGAGATAAGGAG R: CCATCTACCTCCATCTTAAC | 51.6 | 165-191 | 11 | Pet | 11 |
| INRA005 | F: CAATCTGCATGAAGTATAAATAT R: CTTCAGGCATACCCTACACC | 55 | 120-160 | 10 | Ned | 12 |
| BM1824 | F: GAGCAAGGTGTTTTTCCAATC R: CATTCTCCAACTGCTTCCTTG | 57 | 180-200 | 7 | 6fam | 1 |
| HEL5 | F: GCAGGATCACTTGTTAGGGA R: AGACGTTAGTGTACATTAAC | 54 | 149-165 | 7 | Vic | 21 |
| BM2113 | F: GCTGCCTTCTACCAAATACCC R: CTTAGACAACAGGGGTTTGG | 55-60 | 116-146 | 13 | 6fam | 2 |
| TGLA57 | F: GCTTCCAAAACTTTACAATATGTAT R: GCTTTTTAATCCTCAGCTTGCTG | 55 | 77-99 | 9 | 6fam | 1 |
| ILSTS013 | F: ACACAAAATCAGATCAGTGG R: CTTGATCCTTATAGAACTGG | 58 | 121-135 | 11 | Vic | 9 |
| ILSTS050 | F: AAATCAGACACCCAGTTTCC R: GTTTTTCTACACGAGTTGGC | 55 | 161-183 | 13 | Pet | 21 |
| AGLA293 | F: GAAACTCAACCCAAGACAACTCAAG R: ATGACTTTATTCTCCACCTAGCAGA | 55 | 210-240 | 11 | 6fam | 5 |
| ILSTS008 | F: GAATCATGGATTTTCTGGGG R: TAGCAGTGAGTGAGGTTGGC | 58 | 175-187 | 7 | 6fam | 14 |
| TGLA73 | F: GAGAATCACCTAGAGAGAGGCA R: CTTTCTCTTTAAATTCTATATGGT | 55 | 111-143 | 14 | 6fam | 9 |
| 座位 Locus | 帕里牦牛 Pali yak | 斯布牦牛 Sibu yak | 西藏高山牦牛 Tibetan High Mountainous yak | 九龙牦牛 Jiulong yak | 麦洼牦牛 Maiwa yak | 大额牛 Gayal | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM1824 | 192 (0.009) | ||||||||||||
| BM2113 | 116 (0.009) | 120 (0.632) | |||||||||||
| 138 (0.009) . | 132 (0.261) | ||||||||||||
| HEL5 | 149 (0.010) | 141 (0.010) | 155 (0.021) | ||||||||||
| INRA005 | 136 (0.026) | 132 (0.500) | |||||||||||
| 148 (0.009) | |||||||||||||
| TGLA126 | 127(0.009) | ||||||||||||
| TGLA53 | 161 (0.009) | 179 (0.009) | |||||||||||
| 175 (0.009) | |||||||||||||
| HEL13 | 83 (0.009) | 165 (0.009) | 191 (0.375) | ||||||||||
| 193 (0.096) | |||||||||||||
| SPS115 | 230 (0.018) | 256 (0.103) | |||||||||||
| TGLA122 | 138 (0.018) | 142 (0.008) | 166 (0.009) | ||||||||||
| 168 (0.017) | |||||||||||||
| TGLA227 | 87 (0.009) | 99 (0.010) | 101 (0.020) | 97 (0.009) | 81 (0.125) | ||||||||
| AGLA293 | 202 (0.009) | 238 (0.009) | 220 (0.017) | 234 (0.059) | |||||||||
| 242 (0.019) | |||||||||||||
| ILSTS013 | 105 (0.009) | 133(0.231) | |||||||||||
| 109 (0.009) | |||||||||||||
| ILSTS050 | 147 (0.184) 149 (0.105) 151 (0.316) | ||||||||||||
| 153 (0.316) 155 (0.079) | |||||||||||||
| TGLA57 | 31 (0.017) | 71 (0.008) | 97 (0.017) | ||||||||||
| TGLA73 | 193 (0.017) | ||||||||||||
Table 4 Unique alleles and their frequencies in the six populations studied. Allele frequency in the parentheses.
| 座位 Locus | 帕里牦牛 Pali yak | 斯布牦牛 Sibu yak | 西藏高山牦牛 Tibetan High Mountainous yak | 九龙牦牛 Jiulong yak | 麦洼牦牛 Maiwa yak | 大额牛 Gayal | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM1824 | 192 (0.009) | ||||||||||||
| BM2113 | 116 (0.009) | 120 (0.632) | |||||||||||
| 138 (0.009) . | 132 (0.261) | ||||||||||||
| HEL5 | 149 (0.010) | 141 (0.010) | 155 (0.021) | ||||||||||
| INRA005 | 136 (0.026) | 132 (0.500) | |||||||||||
| 148 (0.009) | |||||||||||||
| TGLA126 | 127(0.009) | ||||||||||||
| TGLA53 | 161 (0.009) | 179 (0.009) | |||||||||||
| 175 (0.009) | |||||||||||||
| HEL13 | 83 (0.009) | 165 (0.009) | 191 (0.375) | ||||||||||
| 193 (0.096) | |||||||||||||
| SPS115 | 230 (0.018) | 256 (0.103) | |||||||||||
| TGLA122 | 138 (0.018) | 142 (0.008) | 166 (0.009) | ||||||||||
| 168 (0.017) | |||||||||||||
| TGLA227 | 87 (0.009) | 99 (0.010) | 101 (0.020) | 97 (0.009) | 81 (0.125) | ||||||||
| AGLA293 | 202 (0.009) | 238 (0.009) | 220 (0.017) | 234 (0.059) | |||||||||
| 242 (0.019) | |||||||||||||
| ILSTS013 | 105 (0.009) | 133(0.231) | |||||||||||
| 109 (0.009) | |||||||||||||
| ILSTS050 | 147 (0.184) 149 (0.105) 151 (0.316) | ||||||||||||
| 153 (0.316) 155 (0.079) | |||||||||||||
| TGLA57 | 31 (0.017) | 71 (0.008) | 97 (0.017) | ||||||||||
| TGLA73 | 193 (0.017) | ||||||||||||
| 座位 Locus | 帕里牦牛 Pali yak | 斯布牦牛 Sibu yak | 西藏高山牦牛 Tibetan High Mountainous yak | 九龙牦牛 Jiulong yak | 麦洼牦牛 Maiwa yak | 大额牛 Gayal |
|---|---|---|---|---|---|---|
| BM1824 | 180 (0.562) | 186 (0.925) | ||||
| BM2113 | 128 (0.772) | 128 (0.583) | 128 (0.612) | 120 (0.632) | ||
| HEL5 | 137 (0.696) | 137 (0.962) | 137 (0.781) | 137 (0.894) | 137 (0.726) | |
| INRA005 | 134 (0.658) | 134 (0.543) | 132 (0.500), 134 (0.500) | |||
| TGLA126 | 117 (0.586) | 119 (0.700) | ||||
| TGLA53 | 153 (0.500) | 159 (0.500), 165 (0.500) | ||||
| HEL13 | 183 (0.644) | 183 (0.628) | 183 (0.603) | 197 (0.625) | ||
| SPS115 | 240 (0.465) | 250 (0.667) | ||||
| TGLA122 | 152 (0.388) | |||||
| TGLA227 | 75 (0.632) | 75 (0.573) | 73 (0.620) | 73 (0.561) | 73 (0.602) | 75 (0.850) |
| AGLA293 | 208 (0.860) | 208 (0.890) | 208 (0.833) | 208 (0.627) | 208 (0.737) | 226 (0.529) |
| ILSTS008 | 181 (0.553) | 181 (0.686) | 181 (0.647) | 171 (0.895) | ||
| ILSTS013 | 121 (0.593) | 121 (0.585) | 117 (0.410) | |||
| ILSTS050 | 169 (0.696) | 151 (0.316), 153 (0.316) | ||||
| TGLA57 | 73 (0.658) | 73 (0.746) | 73 (0.741) | 73 (0.636) | 73 (0.746) | 81 (0.450) |
| TGLA73 | 115 (0.246) | 125 (0.600) |
Table 5 Dominant alleles and their frequencies in the six populations. Allele frequency in the parentheses.
| 座位 Locus | 帕里牦牛 Pali yak | 斯布牦牛 Sibu yak | 西藏高山牦牛 Tibetan High Mountainous yak | 九龙牦牛 Jiulong yak | 麦洼牦牛 Maiwa yak | 大额牛 Gayal |
|---|---|---|---|---|---|---|
| BM1824 | 180 (0.562) | 186 (0.925) | ||||
| BM2113 | 128 (0.772) | 128 (0.583) | 128 (0.612) | 120 (0.632) | ||
| HEL5 | 137 (0.696) | 137 (0.962) | 137 (0.781) | 137 (0.894) | 137 (0.726) | |
| INRA005 | 134 (0.658) | 134 (0.543) | 132 (0.500), 134 (0.500) | |||
| TGLA126 | 117 (0.586) | 119 (0.700) | ||||
| TGLA53 | 153 (0.500) | 159 (0.500), 165 (0.500) | ||||
| HEL13 | 183 (0.644) | 183 (0.628) | 183 (0.603) | 197 (0.625) | ||
| SPS115 | 240 (0.465) | 250 (0.667) | ||||
| TGLA122 | 152 (0.388) | |||||
| TGLA227 | 75 (0.632) | 75 (0.573) | 73 (0.620) | 73 (0.561) | 73 (0.602) | 75 (0.850) |
| AGLA293 | 208 (0.860) | 208 (0.890) | 208 (0.833) | 208 (0.627) | 208 (0.737) | 226 (0.529) |
| ILSTS008 | 181 (0.553) | 181 (0.686) | 181 (0.647) | 171 (0.895) | ||
| ILSTS013 | 121 (0.593) | 121 (0.585) | 117 (0.410) | |||
| ILSTS050 | 169 (0.696) | 151 (0.316), 153 (0.316) | ||||
| TGLA57 | 73 (0.658) | 73 (0.746) | 73 (0.741) | 73 (0.636) | 73 (0.746) | 81 (0.450) |
| TGLA73 | 115 (0.246) | 125 (0.600) |
| 座位 Locus | 5个牦牛群体共有等位基因(bp) Common alleles in five yak populations | 6个品种共有等位基因(bp) Common alleles in six breeds | 5个牦牛群体特有等位基因数 No. of unique allele in five yak populations | 大额牛特有等位基因数 No. of unique allele in Gayal |
|---|---|---|---|---|
| BM1824 | 178, 180, 186, 190 | 186 | 1 | 0 |
| BM2113 | 118, 124, 128, 134, 144 | 124, 128 | 2 | 2 |
| HEL5 | 137, 139 | 3 | 0 | |
| INRA005 | 134, 152 | 134 | 2 | 1 |
| TGLA126 | 109, 115, 117, 119 | 119 | 1 | 0 |
| TGLA53 | 153, 157, 159, 165, 169 | 165 | 3 | 0 |
| HEL13 | 183, 189, 197, 199 | 197 | 3 | 1 |
| SPS115 | 232, 238, 240, 242, 244, 246 | 1 | 1 | |
| TGLA122 | 140, 146, 148, 150, 152, 164 | 4 | 0 | |
| TGLA227 | 73, 75 | 75 | 4 | 1 |
| AGLA293 | 208, 214, 226 | 226 | 4 | 1 |
| ILSTS008 | 179, 181 | 0 | 0 | |
| ILSTS013 | 119, 121, 123, 125, 127 | 119 | 2 | 1 |
| ILSTS050 | 167, 169, 171 | 0 | 5 | |
| TGLA57 | 73, 77, 81, 83 | 77, 81 | 2 | 1 |
| TGLA73 | 115, 117, 119, 123, 125, 127, 129, 193 | 115, 117, 123, 125 | 1 | 0 |
Table 6 The common alleles and the number of unique alleles in the six populations
| 座位 Locus | 5个牦牛群体共有等位基因(bp) Common alleles in five yak populations | 6个品种共有等位基因(bp) Common alleles in six breeds | 5个牦牛群体特有等位基因数 No. of unique allele in five yak populations | 大额牛特有等位基因数 No. of unique allele in Gayal |
|---|---|---|---|---|
| BM1824 | 178, 180, 186, 190 | 186 | 1 | 0 |
| BM2113 | 118, 124, 128, 134, 144 | 124, 128 | 2 | 2 |
| HEL5 | 137, 139 | 3 | 0 | |
| INRA005 | 134, 152 | 134 | 2 | 1 |
| TGLA126 | 109, 115, 117, 119 | 119 | 1 | 0 |
| TGLA53 | 153, 157, 159, 165, 169 | 165 | 3 | 0 |
| HEL13 | 183, 189, 197, 199 | 197 | 3 | 1 |
| SPS115 | 232, 238, 240, 242, 244, 246 | 1 | 1 | |
| TGLA122 | 140, 146, 148, 150, 152, 164 | 4 | 0 | |
| TGLA227 | 73, 75 | 75 | 4 | 1 |
| AGLA293 | 208, 214, 226 | 226 | 4 | 1 |
| ILSTS008 | 179, 181 | 0 | 0 | |
| ILSTS013 | 119, 121, 123, 125, 127 | 119 | 2 | 1 |
| ILSTS050 | 167, 169, 171 | 0 | 5 | |
| TGLA57 | 73, 77, 81, 83 | 77, 81 | 2 | 1 |
| TGLA73 | 115, 117, 119, 123, 125, 127, 129, 193 | 115, 117, 123, 125 | 1 | 0 |
| 品种 Breeds | 帕里牦牛 PL | 斯布牦牛 SB | 西藏高山牦牛 GS | 九龙牦牛 JL | 麦洼牦牛 MW | 大额牛 GY |
|---|---|---|---|---|---|---|
| 帕里牦牛 PL | 0.0861 | 0.0707 | 0.1266 | 0.0944 | 0.7136 | |
| 斯布牦牛 SB | 0.0793 | 0.0715 | 0.1071 | 0.0615 | 0.7052 | |
| 西藏高山牦牛 GS | 0.0827 | 0.0649 | 0.0970 | 0.0723 | 0.6955 | |
| 九龙牦牛 JL | 0.1632 | 0.1659 | 0.1168 | 0.0987 | 0.6515 | |
| 麦洼牦牛 MW | 0.0838 | 0.0738 | 0.0937 | 0.1239 | 0.6836 | |
| 大额牛 GY | 1.1508 | 1.4356 | 1.6049 | 1.2457 | 1.4628 |
Table 7 Nei’s genetic distance (DA) (above diagonal) and Nei’s standard genetic distance (DS) (below diagonal) among the six studied populations. Breed codes see Table 1.
| 品种 Breeds | 帕里牦牛 PL | 斯布牦牛 SB | 西藏高山牦牛 GS | 九龙牦牛 JL | 麦洼牦牛 MW | 大额牛 GY |
|---|---|---|---|---|---|---|
| 帕里牦牛 PL | 0.0861 | 0.0707 | 0.1266 | 0.0944 | 0.7136 | |
| 斯布牦牛 SB | 0.0793 | 0.0715 | 0.1071 | 0.0615 | 0.7052 | |
| 西藏高山牦牛 GS | 0.0827 | 0.0649 | 0.0970 | 0.0723 | 0.6955 | |
| 九龙牦牛 JL | 0.1632 | 0.1659 | 0.1168 | 0.0987 | 0.6515 | |
| 麦洼牦牛 MW | 0.0838 | 0.0738 | 0.0937 | 0.1239 | 0.6836 | |
| 大额牛 GY | 1.1508 | 1.4356 | 1.6049 | 1.2457 | 1.4628 |
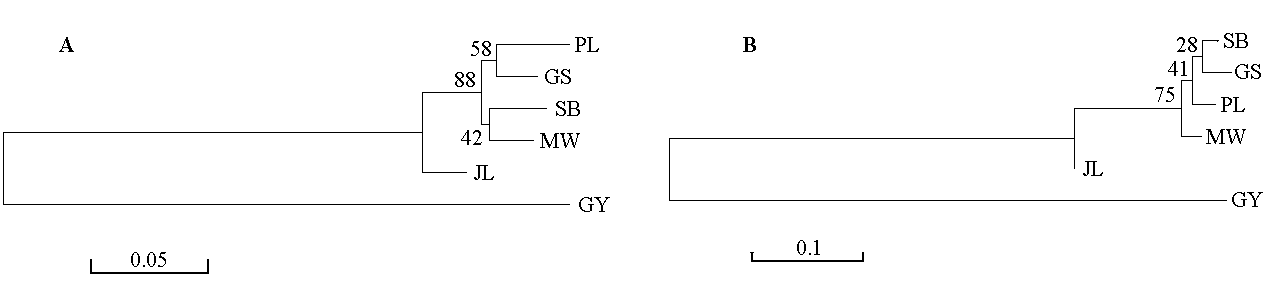
Fig. 1 Dendrograms of six populations based on Nei’s genetic distance (DA) (A) and Nei’s standard genetic distance (DS) (B) using neighbor-joining method. Breed codes see Table 1.
| 品种 Breeds | 帕里牦牛 PL | 斯布牦牛 SB | 西藏高山牦牛 GS | 九龙牦牛 JL | 麦洼牦牛 MW | 大额牛 GY |
|---|---|---|---|---|---|---|
| 帕里牦牛 PL | 1.0000 | 0.9530 | 0.9530 | 0.9340 | 0.9530 | 0.3450 |
| 斯布牦牛 SB | 0.9530 | 1.0000 | 0.9600 | 0.9340 | 0.9560 | 0.3450 |
| 西藏高山牦牛 GS | 0.9510 | 0.9600 | 1.0000 | 0.9340 | 0.9560 | 0.3450 |
| 九龙牦牛 JL | 0.9110 | 0.9100 | 0.9340 | 1.0000 | 0.9340 | 0.3450 |
| 麦洼牦牛 MW | 0.9510 | 0.9560 | 0.9460 | 0.9310 | 1.0000 | 0.3450 |
| 大额牛 GY | 0.2420 | 0.2500 | 0.1650 | 0.3450 | 0.2370 | 1.0000 |
Table 8 Fuzzy similarity relation matrix (above diagonal) and fuzzy containing relation matrix (below diagonal) among the six populations. Breed codes see Table 1.
| 品种 Breeds | 帕里牦牛 PL | 斯布牦牛 SB | 西藏高山牦牛 GS | 九龙牦牛 JL | 麦洼牦牛 MW | 大额牛 GY |
|---|---|---|---|---|---|---|
| 帕里牦牛 PL | 1.0000 | 0.9530 | 0.9530 | 0.9340 | 0.9530 | 0.3450 |
| 斯布牦牛 SB | 0.9530 | 1.0000 | 0.9600 | 0.9340 | 0.9560 | 0.3450 |
| 西藏高山牦牛 GS | 0.9510 | 0.9600 | 1.0000 | 0.9340 | 0.9560 | 0.3450 |
| 九龙牦牛 JL | 0.9110 | 0.9100 | 0.9340 | 1.0000 | 0.9340 | 0.3450 |
| 麦洼牦牛 MW | 0.9510 | 0.9560 | 0.9460 | 0.9310 | 1.0000 | 0.3450 |
| 大额牛 GY | 0.2420 | 0.2500 | 0.1650 | 0.3450 | 0.2370 | 1.0000 |
| [1] |
Botstein D, White RL, Skolnick M, David RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32,314-331.
URL PMID |
| [2] | Cai L(蔡立) (1992) Chinese Yak (中国牦牛). China Agricultural Press, Beijing. (in Chinese) |
| [3] | Chang GB(常国斌) (2004) Study on the Level of Evolutionary Divergence Between Two Wild and Domestic Quail Species (两种野生鹌鹑与家鹑进化趋异水平的研究). PhD dissertation, Yangzhou University, Yangzhou. (in Chinese with English summary). |
| [4] | Chang H(常洪) (1995) Essentials of Livestock Genetic Resources (家畜遗传资源学纲要). China Agricultural Press, Beijing. (in Chinese) |
| [5] | Editorial Committee of Chinese Yak (中国牦牛学编委会) (1989) Chinese Yak (中国牦牛学). Sichuan Science and Technology Press, Chengdu. (in Chinese) |
| [6] |
Guo SC, Savolainen P, Su JP, Zhang Q, Qi DL, Zhou J, Zhong Y, Zhao XQ, Liu JQ (2006) Origin of mitochondrial DNA diversity of domestic yaks. BMC Evolutionary Biology, 6,73.
DOI URL PMID |
| [7] |
Hanotte O, Okomo M, Verjee Y, Regee E, Teale A (1997) A polymorphic Y chromosome microsatellite locus in cattle. Animal Genetics, 28,318-319.
URL PMID |
| [8] | Lai SJ(赖松家), Wang L(王玲), Liu YP(刘益平), Li XW(李学伟) (2005) Study on mitochondrial DNA genetic polymorphism of some yak breeds in China. Acta Genetica Sinica (遗传学报), 32,463-470. (in Chinese with English abstract) |
| [9] |
Lai SJ, Chen SY, Liu YP, Yao YG (2007) Mitochondrial DNA sequence diversity and origin of Chinese domestic yak. Animal Genetics, 38,77-80.
URL PMID |
| [10] | Li QF(李齐发), Li YX(李隐侠), Zhao XB(赵兴波), Liu ZS(刘振山), Zhang QB(张庆波), Song DW(宋大伟), Qu XG(屈旭光), Li N(李宁), Xie Z(谢庄) (2006) Sequencing cytochrome b gene of mitochondrial DNA in yak and researching its origin and taxonomic status . Acta Veterinaria et Zootechnica Sinica (畜牧兽医学报), 37,1118-1123. (in Chinese with English abstract) |
| [11] | Lu SD(卢圣栋) (1993) Experimental Technique of Modern Molecular Biology (现代分子生物学实验技术). Higher Education Press, Beijing. (in Chinese) |
| [12] | Luo YF(罗永发), Wang ZG(王志刚), Li JQ(李加琪), Zhang GX(张桂香), Chen YS(陈瑶生), Liang Y(梁勇), Yu FQ(于福清), Song WT(宋卫涛), Zhang ZF(张自富) (2006) Genetic variation and genetic relationship among 13 Chinese and introduced cattle breeds using microsatellite DNA markers. Biodiversity Science (生物多样性), 14,498-507. (in Chinese with English abstract) |
| [13] | Ma YH(马月辉), Chen YC(陈幼春), Feng WQ(冯维祺), Wang DY(王端云) (2002) Germplasm of Chinese domestic animals and their conservation. Review of China Agricultural Science and Technology (中国农业科技导报), 3,37-42. (in Chinese with English abstract) |
| [14] | Nei M (1972) Genetic distance between populations. The American Naturalist, 106,283-292. |
| [15] |
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution, 19,153-170.
URL PMID |
| [16] |
Nguyen TT, Genini S, Ménétrey F, Malek M, Vögeli P, Goe MR, Stranzinger G (2005) Application of bovine microsatellite markers for genetic diversity analysis of Swiss yak (Poephagus grunniens). Animal Genetics, 36,484-489.
DOI URL PMID |
| [17] | Ota T (1993) DISPAN: Genetic Distance and Phylogenetic Analysis. Institute of Molecular Evolutionary Genetics, The Pennsylvania State University. |
| [18] | Qi XB (2004) Genetic Diversity, Differentiation, and Relationship of Domestic Yak Populations: A Microsatellite and Mitochondrial DNA Study. PhD dissertation, Lanzhou University, Lanzhou. |
| [19] |
Qi XB, Han JL, Lkhagva B, Chekarova I, Badamdorj D, Rege JEO, Hanotte O (2005) Genetic diversity and differentiation of Mongolian and Russian yak populations. Journal of Animal Breeding and Genetics, 122,117-126.
DOI URL PMID |
| [20] | Qiu H(邱怀), Qin ZR(秦志锐), Chen YC(陈幼春), Wang DA(王东安), Wei WY(韦文雅), Li KL(李孔亮), Li RY(李忍益), Zou XQ(邹霞青), Guo JX(郭经恂), Tong BQ(童碧泉), Ji YL(冀一伦), Cai L(蔡立) (1986) Bovine Breeds in China (中国牛品种志). Shanghai Science and Technology Press, Shanghai. (in Chinese) |
| [21] | Raymond M, Rousset F (1995) Population genetics software for exact tests and ecumenicism. Journal of Heredity, 86,248-249. |
| [22] | Tu ZC(涂正超), Zhang YP(张亚平), Qiu H(邱怀) (1998) Mitochondrial DNA polymorphism and genetic diversity in Chinese yaks. Acta Genetica Sinica (遗传学报), 25,205-212. (in Chinese with English abstract) |
| [23] |
Tu ZC, Zhang YP, Qiu H (1997) Genetic diversity and divergence in Chinese yak ( Bos grunniens) populations inferred from blood protein electrophoresis . Biochemical Genetics, 35,13-16.
URL PMID |
| [24] | Wang DL(王冬蕾), Chang H(常洪), Yang JX(杨军香), Zhang GX(张桂香), Wang ZG(王志刚), Yu B(于波), Liao XJ(廖信军), Song WT(宋卫涛), Han X(韩旭) (2007) Analysis on the genetic structure of 8 Asia buffalo populations. Hereditas (Beijing) (遗传), 29,1103-1109. (in Chinese with English abstract) |
| [25] | Zhong JC(钟金城), Chen ZH(陈智华), Zi XD(字向东), Wen YL(文勇立), Ma L(马力) (2001) Cluster analysis of yak breeds. Journal of Southwest Nationalities College (National Science Edition) (西南民族学院学报(自然科学版)), 27,92-94. ( in Chinese with English abstract ). |
| [26] | Zhong JC(钟金城), Zhao SJ(赵素君), Chen ZH(陈智华), Ma ZJ(马志杰) (2006) Study on genetic diversity and classification of the yak. Scientia Agricultura Sinica (中国农业科学), 39,389-397. (in Chinese with English abstract) |
| [1] | Wei Yi, Yi Ai, Meng Wu, Liming Tian, Tserang Donko Mipam. Soil archaeal community responses to different grazing intensities in the alpine meadows of the Qinghai-Tibetan Plateau [J]. Biodiv Sci, 2025, 33(1): 24339-. |
| [2] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [3] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [4] | Qingqing Liu, Zhijun Dong. Population genetic structure of Gonionemus vertens based on the mitochondrial COI sequence [J]. Biodiv Sci, 2018, 26(11): 1204-1211. |
| [5] | Jing Zhang, Yuan Li, Na Song, Longshan Lin, Tianxiang Gao. Species identification and phylogenetic relationship of Thryssa species in the coastal waters of China [J]. Biodiv Sci, 2016, 24(8): 888-895. |
| [6] | Ruoyu Liu, Zhongmin Sun, Jianting Yao, Zimin Hu, Delin Duan. Genetic diversity of the habitat-forming red alga Gracilaria vermiculophylla along Chinese coasts [J]. Biodiv Sci, 2016, 24(7): 781-790. |
| [7] | Xiao Luo, Feng Li, Jing Chen, Zhigang Jiang. The taxonomic status of badgers in the Qinghai Lake area and evolutionary history of Meles [J]. Biodiv Sci, 2016, 24(6): 694-700. |
| [8] | Yani Hu, Zongwen Zhang, Bin Wu, Jia Gao, Yanqin Li. Genetic relationships of buckwheat species based on the sequence analysis of ITS and ndhF-rpl32 [J]. Biodiv Sci, 2016, 24(3): 296-303. |
| [9] | Rong Zhou, Jiaqi Li, You Li, Naifa Liu, Fengjie Fang, Limin Shi, Ying Wang. Genetic variation in rusty-necklaced partridge (Alectoris magna) detected by mitochondrial DNA [J]. Biodiv Sci, 2012, 20(4): 451-459. |
| [10] | Xiaohong He, Xiuli Han, Weijun Guan, Kechuan Tian, Wenbin Zhang, Yuehui Ma. Genetic variability and relationship of 10 Bactrian camel populations revealed by microsatellite markers [J]. Biodiv Sci, 2012, 20(2): 199-206. |
| [11] | Biyun Chen, Qiong Hu, Christina Dixelius, Guoqing Li, Xiaoming Wu. Genetic diversity in Sclerotinia sclerotiorum assessed with SRAP markers [J]. Biodiv Sci, 2010, 18(5): 509-515. |
| [12] | Junhong Zhang, Huahong Huang, Zaikang Tong, Longjun Cheng, Yuelong Liang, Yiliang Chen. Genetic diversity in six natural populations of Betula luminifera from southern China [J]. Biodiv Sci, 2010, 18(3): 233-240. |
| [13] | Gen Zhang, Yilong Xi, Yinghao Xue, Xin Hu, Xianling Xiang, Xinli Wen. Effects of coal ash pollution on the genetic diversity of Brachionus calyciflorus based on rDNA ITS sequences [J]. Biodiv Sci, 2010, 18(3): 241-250. |
| [14] | Hongtuo Fu, Hui Qiao, Jianhua Yao, Yongsheng Gong, Yan Wu, Sufei Jiang, Yiwei Xiong. Genetic diversity in five Macrobrachium hainanense populations using SRAP markers [J]. Biodiv Sci, 2010, 18(2): 145-149. |
| [15] | Yinghui Ling, Yuejiao Cheng, Yanping Wang, Weijun Guan, Jianlin Han, Baoling Fu, Qianjun Zhao, Xiaohong He, Yabin Pu, Yuehui Ma. Genetic diversity of 23 Chinese indigenous horse breeds revealed by microsatellite markers [J]. Biodiv Sci, 2009, 17(3): 240-247. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()