



Biodiv Sci ›› 2016, Vol. 24 ›› Issue (7): 781-790. DOI: 10.17520/biods.2016038 cstr: 32101.14.biods.2016038
Special Issue: 全球气候变化下的海洋生物多样性专辑
• Original Papers • Previous Articles Next Articles
Ruoyu Liu1,2, Zhongmin Sun1,3, Jianting Yao1,4, Zimin Hu1,4,*( ), Delin Duan1,4,*(
), Delin Duan1,4,*( )
)
Received:2016-02-03
Accepted:2016-06-07
Online:2016-07-20
Published:2016-08-04
Contact:
Hu Zimin,Duan Delin
Ruoyu Liu, Zhongmin Sun, Jianting Yao, Zimin Hu, Delin Duan. Genetic diversity of the habitat-forming red alga Gracilaria vermiculophylla along Chinese coasts[J]. Biodiv Sci, 2016, 24(7): 781-790.
| 代号 Code | 地理位置 Location | 经纬度 Longitude/ Latitude | N | Np | Nh | Hn(n) | Hd | π (× 10-2) | 采样人 Collectors |
|---|---|---|---|---|---|---|---|---|---|
| 南方群体 Region South | |||||||||
| YT | 北海银滩 Yingtang beach, Beihai | 109.34 ºE, 21.45 ºN | 24 | 7 | 4 | H0(21), H2, H3, H4 | 0.239 | 0.143 | 刘若愚, 胡自民 Ruoyu Liu, Zimin Hu |
| JH | 北海金海岸 Jinhaian, Beihai | 109.11 ºE, 21.49 ºN | 33 | 1 | 2 | H0(32), H2 | 0.061 | 0.009 | 刘若愚, 胡自民 Ruoyu Liu, Zimin Hu |
| TC | 湛江特呈岛 Techeng island, Zhanshan, | 110.44 ºE, 21.15 ºN | 30 | 0 | 1 | H0(30) | 0.000 | 0.000 | 孙忠民, 姚建亭 Zhongmin Sun, Jianting Yao |
| NA | 汕头南澳岛 Nanao island, Shantou | 117.04 ºE, 23.45 ºN | 23 | 1 | 2 | H0(19), H1(4) | 0.300 | 0.047 | 孙忠民, 姚建亭 Zhongmin Sun, Jianting Yao |
| ZP | 漳州漳浦六鳌 Zhangpu liuao, Zhanzhou, | 117.77 ºE, 23.92 ºN | 35 | 0 | 1 | H0(35) | 0.000 | 0.000 | 孙忠民, 黄超华 Zhongmin Sun, Chaohua Huang |
| XL | 厦门杏林湾 Xingling bay, Xiamen | 118.08 ºE, 24.60 ºN | 5 | 1 | 2 | H0(4), H1(1) | 0.400 | 0.062 | 钟晨辉等 Chenhui Zhong et al |
| 小计 Subtotal | 150 | 8 | 5 | H0(141), H1(5), H2(2), H3, H4 | 0.116 | 0.027 | |||
| 北方群体 Region North | |||||||||
| SS | 舟山嵊泗岛 Shengsi island, Zhoushan | 122.44 ºE, 30.68 ºN | 19 | 0 | 1 | H6(19) | 0.000 | 0.000 | 胡自民 Zimin Hu |
| YY | 青岛一浴 No. 1 beach, Qingdao | 120.34 ºE, 36.05 ºN | 21 | 2 | 3 | H5(19), H7, H10 | 0.186 | 0.030 | 刘若愚 Ruoyu Liu |
| EY | 青岛二浴 No. 2 beach, Qingdao | 120.34 ºE, 36.05 ºN | 21 | 0 | 1 | H5(21) | 0.000 | 0.000 | 刘若愚 Ruoyu Liu |
| SY | 青岛三浴 No. 3 beach | 120.36 ºE, 36.05 ºN | 39 | 0 | 1 | H5(39) | 0.000 | 0.000 | 刘若愚 Ruoyu Liu |
| YH | 青岛银海国际 Yinhai, Qingdao | 120.42 ºE, 36.06 ºN | 36 | 2 | 3 | H5(26), H8(9), H13 | 0.427 | 0.069 | 刘若愚 Ruoyu Liu |
| LR | 青岛石老人 Shilaoren sea, Qingdao | 120.49 ºE, 36.09 ºN | 36 | 2 | 3 | H5(32), H9(3), H12 | 0.208 | 0.033 | 刘若愚 Ruoyu Liu |
| SD | 威海石岛 Stone island, Weihai | 122.41 ºE, 36.91 ºN | 13 | 0 | 1 | H5(13) | 0.000 | 0.000 | 李晶晶, 张杰 Jingjing Li, Jie Zhang |
| DC | 威海东楮岛 Dongchu island, Weihai | 122.56 ºE, 37.04 ºN | 32 | 1 | 2 | H5(16), H14(16) | 0.516 | 0.081 | 刘若愚, 胡自民Ruoyu Liu, Zimin Hu |
| JM | 威海鸡鸣岛 Jiming island, Weihai | 122.48 ºE, 37.45 ºN | 7 | 0 | 1 | H5(7) | 0.000 | 0.000 | 李晶晶, 张杰 Jingjing Li, Jie Zhang |
| CD | 烟台长岛 Long island, Yantai | 120.72 ºE, 37.94 ºN | 16 | 0 | 1 | H5(16) | 0.000 | 0.000 | 胡自民 Zimin Hu |
| HN | 大连黄泥川 Huangninchuan, Dalian | 121.56 ºE, 38.82 ºN | 35 | 1 | 2 | H5(34), H11 | 0.057 | 0.009 | 刘若愚, 胡自民 Ruoyu Liu, Zimin Hu |
| HS | 大连黑石礁 Heshijiao, Dalian | 121.56 ºE, 38.87 ºN | 8 | 0 | 1 | H5(8) | 0.000 | 0.000 | 刘若愚, 胡自民 Ruoyu Liu, Zimin Hu |
| ZZ | 大连獐子岛 Zhangzi island, Dalian | 122.74 ºE, 39.04 ºN | 28 | 0 | 1 | H5(28) | 0.000 | 0.000 | 姚建亭, 孙忠民 Ruoyu Liu, Zimin Hu |
| 小计 Subtotal | 311 | 9 | 10 | H5(259), H6(19), H7, H8(9), H9(3) H10, H11, H12, H13, H14(16) | 0.300 | 0.050 | |||
Table 1 Sampling information of 19 Gracilaria vermiculophylla populations, including abbreviation codes, geographic location, sample size (N), number of polymorphic sites (Np), number of haplotypes (Nh), types of haplotype (H(n)), haplotype diversity (Hd), nucleotide diversity (π), collecting date and collectors
| 代号 Code | 地理位置 Location | 经纬度 Longitude/ Latitude | N | Np | Nh | Hn(n) | Hd | π (× 10-2) | 采样人 Collectors |
|---|---|---|---|---|---|---|---|---|---|
| 南方群体 Region South | |||||||||
| YT | 北海银滩 Yingtang beach, Beihai | 109.34 ºE, 21.45 ºN | 24 | 7 | 4 | H0(21), H2, H3, H4 | 0.239 | 0.143 | 刘若愚, 胡自民 Ruoyu Liu, Zimin Hu |
| JH | 北海金海岸 Jinhaian, Beihai | 109.11 ºE, 21.49 ºN | 33 | 1 | 2 | H0(32), H2 | 0.061 | 0.009 | 刘若愚, 胡自民 Ruoyu Liu, Zimin Hu |
| TC | 湛江特呈岛 Techeng island, Zhanshan, | 110.44 ºE, 21.15 ºN | 30 | 0 | 1 | H0(30) | 0.000 | 0.000 | 孙忠民, 姚建亭 Zhongmin Sun, Jianting Yao |
| NA | 汕头南澳岛 Nanao island, Shantou | 117.04 ºE, 23.45 ºN | 23 | 1 | 2 | H0(19), H1(4) | 0.300 | 0.047 | 孙忠民, 姚建亭 Zhongmin Sun, Jianting Yao |
| ZP | 漳州漳浦六鳌 Zhangpu liuao, Zhanzhou, | 117.77 ºE, 23.92 ºN | 35 | 0 | 1 | H0(35) | 0.000 | 0.000 | 孙忠民, 黄超华 Zhongmin Sun, Chaohua Huang |
| XL | 厦门杏林湾 Xingling bay, Xiamen | 118.08 ºE, 24.60 ºN | 5 | 1 | 2 | H0(4), H1(1) | 0.400 | 0.062 | 钟晨辉等 Chenhui Zhong et al |
| 小计 Subtotal | 150 | 8 | 5 | H0(141), H1(5), H2(2), H3, H4 | 0.116 | 0.027 | |||
| 北方群体 Region North | |||||||||
| SS | 舟山嵊泗岛 Shengsi island, Zhoushan | 122.44 ºE, 30.68 ºN | 19 | 0 | 1 | H6(19) | 0.000 | 0.000 | 胡自民 Zimin Hu |
| YY | 青岛一浴 No. 1 beach, Qingdao | 120.34 ºE, 36.05 ºN | 21 | 2 | 3 | H5(19), H7, H10 | 0.186 | 0.030 | 刘若愚 Ruoyu Liu |
| EY | 青岛二浴 No. 2 beach, Qingdao | 120.34 ºE, 36.05 ºN | 21 | 0 | 1 | H5(21) | 0.000 | 0.000 | 刘若愚 Ruoyu Liu |
| SY | 青岛三浴 No. 3 beach | 120.36 ºE, 36.05 ºN | 39 | 0 | 1 | H5(39) | 0.000 | 0.000 | 刘若愚 Ruoyu Liu |
| YH | 青岛银海国际 Yinhai, Qingdao | 120.42 ºE, 36.06 ºN | 36 | 2 | 3 | H5(26), H8(9), H13 | 0.427 | 0.069 | 刘若愚 Ruoyu Liu |
| LR | 青岛石老人 Shilaoren sea, Qingdao | 120.49 ºE, 36.09 ºN | 36 | 2 | 3 | H5(32), H9(3), H12 | 0.208 | 0.033 | 刘若愚 Ruoyu Liu |
| SD | 威海石岛 Stone island, Weihai | 122.41 ºE, 36.91 ºN | 13 | 0 | 1 | H5(13) | 0.000 | 0.000 | 李晶晶, 张杰 Jingjing Li, Jie Zhang |
| DC | 威海东楮岛 Dongchu island, Weihai | 122.56 ºE, 37.04 ºN | 32 | 1 | 2 | H5(16), H14(16) | 0.516 | 0.081 | 刘若愚, 胡自民Ruoyu Liu, Zimin Hu |
| JM | 威海鸡鸣岛 Jiming island, Weihai | 122.48 ºE, 37.45 ºN | 7 | 0 | 1 | H5(7) | 0.000 | 0.000 | 李晶晶, 张杰 Jingjing Li, Jie Zhang |
| CD | 烟台长岛 Long island, Yantai | 120.72 ºE, 37.94 ºN | 16 | 0 | 1 | H5(16) | 0.000 | 0.000 | 胡自民 Zimin Hu |
| HN | 大连黄泥川 Huangninchuan, Dalian | 121.56 ºE, 38.82 ºN | 35 | 1 | 2 | H5(34), H11 | 0.057 | 0.009 | 刘若愚, 胡自民 Ruoyu Liu, Zimin Hu |
| HS | 大连黑石礁 Heshijiao, Dalian | 121.56 ºE, 38.87 ºN | 8 | 0 | 1 | H5(8) | 0.000 | 0.000 | 刘若愚, 胡自民 Ruoyu Liu, Zimin Hu |
| ZZ | 大连獐子岛 Zhangzi island, Dalian | 122.74 ºE, 39.04 ºN | 28 | 0 | 1 | H5(28) | 0.000 | 0.000 | 姚建亭, 孙忠民 Ruoyu Liu, Zimin Hu |
| 小计 Subtotal | 311 | 9 | 10 | H5(259), H6(19), H7, H8(9), H9(3) H10, H11, H12, H13, H14(16) | 0.300 | 0.050 | |||
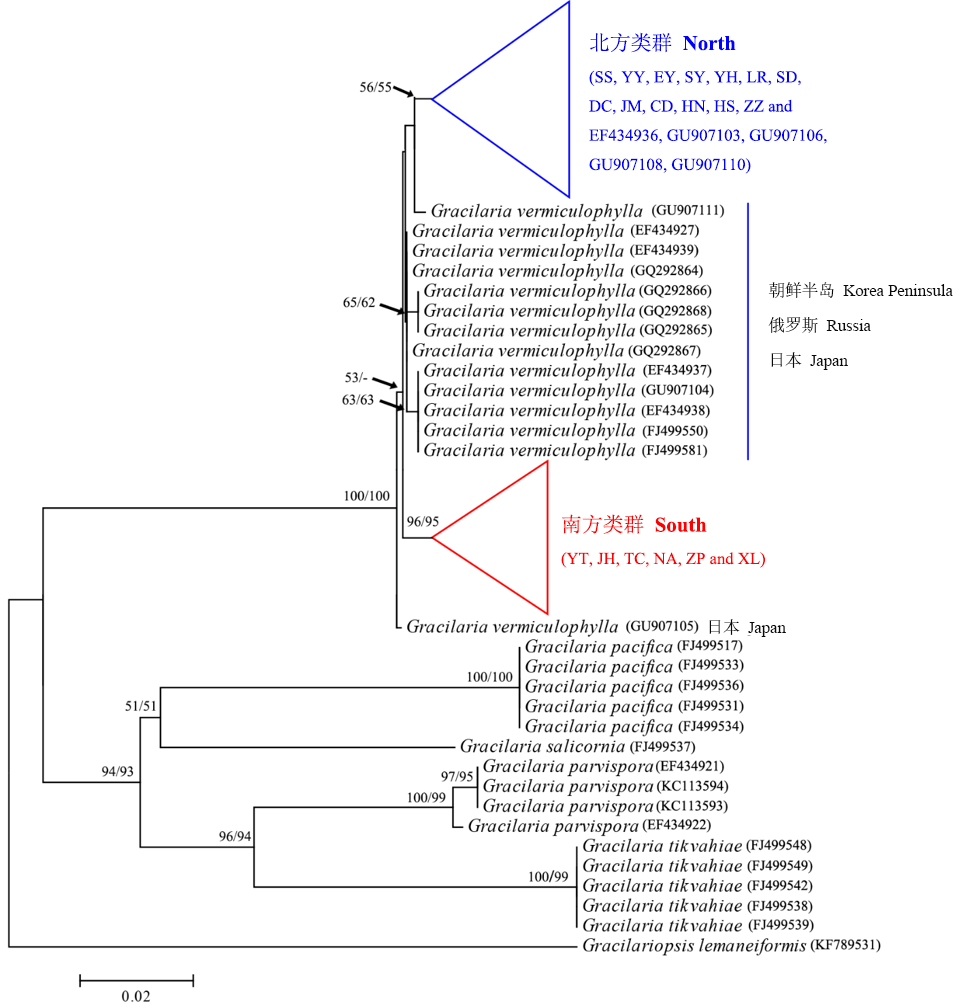
Fig. 1 Neighbor-joining (NJ) and maximum-likelihood (ML) phylogenetic trees based upon mt-DNA cox1 sequences of Gracilaria vermiculophylla and some congeneric species. Numbers indicate bootstrap values (> 50%) of NJ (left) and ML (right) inferences. Population codes are the same as in Table 1.
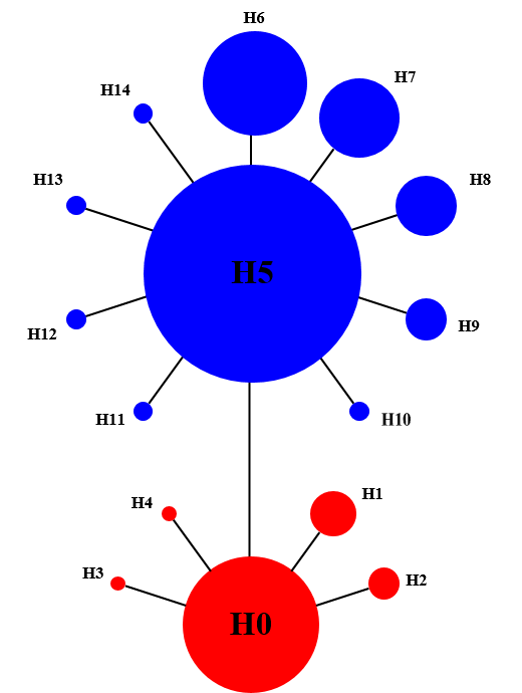
Fig. 2 Median-joining network of mitochondrial cox1 haplotypes in Gracilaria vermiculophylla populations. We only detected haplotype H0-H4 in the populations south to Xiamen, while H5-H14 only in the populations north to Shengsi Island. Population codes are the same as in Table 1.
| 单倍型 Haplotype | 单倍型数量 Number | 核苷酸多态位点 Polymorphic sites | GenBank accession number | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 25 | 52 | 64 | 65 | 112 | 151 | 250 | 346 | 352 | 370 | 406 | 433 | 460 | 517 | 558 | 562 | 593 | 604 | 605 | 634 | |||
| H0 | 141 | T | G | A | A | C | G | A | A | G | T | T | T | A | G | G | C | T | G | G | G | A | KU136384 |
| H1 | 5 | T | A | A | A | C | G | A | A | G | T | T | T | A | G | G | C | T | G | G | G | A | KF789527 |
| H2 | 2 | T | G | A | A | C | G | A | A | G | T | T | T | A | G | G | C | T | G | G | G | G | KU136385 |
| H3 | 1 | T | G | T | A | C | T | T | A | G | T | T | T | A | G | G | C | T | T | G | A | A | KU136387 |
| H4 | 1 | A | G | A | A | C | G | A | A | G | T | T | T | A | G | G | C | T | G | G | G | A | KU136386 |
| H5 | 259 | T | G | A | A | C | G | A | A | C | T | T | T | A | G | A | C | C | G | T | G | A | KU136374 |
| H6 | 19 | T | G | A | A | C | G | A | A | C | G | T | T | A | G | A | C | C | G | T | G | A | KU136375 |
| H7 | 16 | T | G | A | G | C | G | A | A | C | T | T | T | A | G | A | C | C | G | T | G | A | KU136383 |
| H8 | 9 | T | G | A | A | T | G | A | A | C | T | T | T | A | G | A | C | C | G | T | G | A | KU136377 |
| H9 | 3 | T | G | A | A | C | G | A | A | C | T | A | T | A | G | A | C | C | G | T | G | A | KU136378 |
| H10 | 1 | T | G | A | A | C | G | A | A | C | T | T | C | A | G | A | C | C | G | T | G | A | KU136379 |
| H11 | 1 | T | G | A | A | C | G | A | A | C | T | T | T | A | G | A | A | C | G | T | G | A | KU136380 |
| H12 | 1 | T | G | A | A | C | G | A | A | C | T | T | T | A | A | A | C | C | G | T | G | A | KU136381 |
| H13 | 1 | T | G | A | A | C | G | A | T | C | T | T | T | A | G | A | C | C | G | T | G | A | KU136382 |
| H14 | 1 | T | G | A | A | C | G | A | A | C | T | T | T | T | G | A | C | C | G | T | G | A | KU136376 |
Table 2 Polymorphic sites of 15 cox1 haplotypes in Gracilaria vermiculophylla. The letters with different font denote polymorphic sites.
| 单倍型 Haplotype | 单倍型数量 Number | 核苷酸多态位点 Polymorphic sites | GenBank accession number | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 25 | 52 | 64 | 65 | 112 | 151 | 250 | 346 | 352 | 370 | 406 | 433 | 460 | 517 | 558 | 562 | 593 | 604 | 605 | 634 | |||
| H0 | 141 | T | G | A | A | C | G | A | A | G | T | T | T | A | G | G | C | T | G | G | G | A | KU136384 |
| H1 | 5 | T | A | A | A | C | G | A | A | G | T | T | T | A | G | G | C | T | G | G | G | A | KF789527 |
| H2 | 2 | T | G | A | A | C | G | A | A | G | T | T | T | A | G | G | C | T | G | G | G | G | KU136385 |
| H3 | 1 | T | G | T | A | C | T | T | A | G | T | T | T | A | G | G | C | T | T | G | A | A | KU136387 |
| H4 | 1 | A | G | A | A | C | G | A | A | G | T | T | T | A | G | G | C | T | G | G | G | A | KU136386 |
| H5 | 259 | T | G | A | A | C | G | A | A | C | T | T | T | A | G | A | C | C | G | T | G | A | KU136374 |
| H6 | 19 | T | G | A | A | C | G | A | A | C | G | T | T | A | G | A | C | C | G | T | G | A | KU136375 |
| H7 | 16 | T | G | A | G | C | G | A | A | C | T | T | T | A | G | A | C | C | G | T | G | A | KU136383 |
| H8 | 9 | T | G | A | A | T | G | A | A | C | T | T | T | A | G | A | C | C | G | T | G | A | KU136377 |
| H9 | 3 | T | G | A | A | C | G | A | A | C | T | A | T | A | G | A | C | C | G | T | G | A | KU136378 |
| H10 | 1 | T | G | A | A | C | G | A | A | C | T | T | C | A | G | A | C | C | G | T | G | A | KU136379 |
| H11 | 1 | T | G | A | A | C | G | A | A | C | T | T | T | A | G | A | A | C | G | T | G | A | KU136380 |
| H12 | 1 | T | G | A | A | C | G | A | A | C | T | T | T | A | A | A | C | C | G | T | G | A | KU136381 |
| H13 | 1 | T | G | A | A | C | G | A | T | C | T | T | T | A | G | A | C | C | G | T | G | A | KU136382 |
| H14 | 1 | T | G | A | A | C | G | A | A | C | T | T | T | T | G | A | C | C | G | T | G | A | KU136376 |
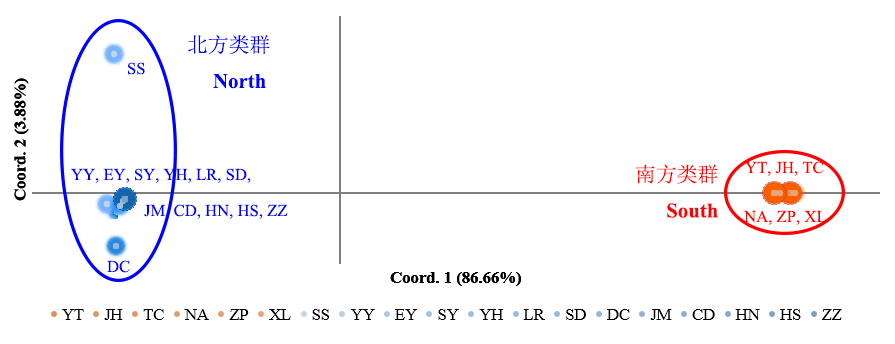
Fig. 3 Principal coordinate analysis of 461 mt-DNA cox1 sequences based on Nei’s genetic distance. The amount of variation is explained by Coord. 1 and Coord. 2, respectively. Population codes are the same as in Table 1.
| YT | JH | TC | NA | ZP | XL | SS | YY | EY | SY | YH | LR | SD | DC | JM | CD | HN | HS | ZZ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YT | 0.000 | ||||||||||||||||||
| JH | 0.000 | 0.000 | |||||||||||||||||
| TC | 0.000 | 0.000 | 0.000 | ||||||||||||||||
| NA | 0.002 | 0.001 | 0.001 | 0.000 | |||||||||||||||
| ZP | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | ||||||||||||||
| XL | 0.003 | 0.002 | 0.002 | 0.000 | 0.002 | 0.000 | |||||||||||||
| SS | 0.277 | 0.272 | 0.272 | 0.276 | 0.272 | 0.277 | 0.000 | ||||||||||||
| YY | 0.217 | 0.213 | 0.213 | 0.216 | 0.213 | 0.217 | 0.049 | 0.000 | |||||||||||
| EY | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | ||||||||||
| SY | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | |||||||||
| YH | 0.222 | 0.218 | 0.217 | 0.221 | 0.217 | 0.222 | 0.052 | 0.003 | 0.003 | 0.003 | 0.000 | ||||||||
| LR | 0.217 | 0.213 | 0.213 | 0.216 | 0.213 | 0.217 | 0.049 | 0.001 | 0.000 | 0.000 | 0.003 | 0.000 | |||||||
| SD | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | ||||||
| DC | 0.233 | 0.230 | 0.229 | 0.233 | 0.229 | 0.234 | 0.062 | 0.012 | 0.012 | 0.012 | 0.015 | 0.013 | 0.012 | 0.000 | |||||
| JM | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | ||||
| CD | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | 0.000 | |||
| HN | 0.216 | 0.212 | 0.212 | 0.215 | 0.212 | 0.216 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | 0.000 | 0.000 | ||
| HS | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | 0.000 | 0.000 | 0.000 | |
| ZZ | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Table 3 Pairwise Nei’s genetic distances among Gracilaria vermiculophylla populations. Population codes see Table 1.
| YT | JH | TC | NA | ZP | XL | SS | YY | EY | SY | YH | LR | SD | DC | JM | CD | HN | HS | ZZ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YT | 0.000 | ||||||||||||||||||
| JH | 0.000 | 0.000 | |||||||||||||||||
| TC | 0.000 | 0.000 | 0.000 | ||||||||||||||||
| NA | 0.002 | 0.001 | 0.001 | 0.000 | |||||||||||||||
| ZP | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | ||||||||||||||
| XL | 0.003 | 0.002 | 0.002 | 0.000 | 0.002 | 0.000 | |||||||||||||
| SS | 0.277 | 0.272 | 0.272 | 0.276 | 0.272 | 0.277 | 0.000 | ||||||||||||
| YY | 0.217 | 0.213 | 0.213 | 0.216 | 0.213 | 0.217 | 0.049 | 0.000 | |||||||||||
| EY | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | ||||||||||
| SY | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | |||||||||
| YH | 0.222 | 0.218 | 0.217 | 0.221 | 0.217 | 0.222 | 0.052 | 0.003 | 0.003 | 0.003 | 0.000 | ||||||||
| LR | 0.217 | 0.213 | 0.213 | 0.216 | 0.213 | 0.217 | 0.049 | 0.001 | 0.000 | 0.000 | 0.003 | 0.000 | |||||||
| SD | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | ||||||
| DC | 0.233 | 0.230 | 0.229 | 0.233 | 0.229 | 0.234 | 0.062 | 0.012 | 0.012 | 0.012 | 0.015 | 0.013 | 0.012 | 0.000 | |||||
| JM | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | ||||
| CD | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | 0.000 | |||
| HN | 0.216 | 0.212 | 0.212 | 0.215 | 0.212 | 0.216 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | 0.000 | 0.000 | ||
| HS | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | 0.000 | 0.000 | 0.000 | |
| ZZ | 0.215 | 0.212 | 0.211 | 0.215 | 0.211 | 0.215 | 0.049 | 0.000 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.012 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 变异来源 Source of variation | 自由度 df | 平方和 Sums of squares | 变异组分 Estimated variance | 变异百分比 % of variation | 分化系数 F-statistics | 基因流 Nm |
|---|---|---|---|---|---|---|
| 一个类群 One group | ||||||
| 群体间 Among populations | 18 | 434.049 | 24.114 | 93 | ||
| 群体内 Within populations | 442 | 34.103 | 0.077 | 7 | FST = 0.928* | 0.039 |
| 两个类群(南方群体, 北方群体) Two groups (Regions: South, North) | ||||||
| 类群间Among regions | 1 | 405.675 | 405.675 | 93 | FCT = 0.933* | 0.019 |
| 类群内群体间Among populations | 17 | 28.374 | 1.669 | 3 | FSC = 0.465* | |
| 群体内 Within populations | 442 | 34.103 | 0.077 | 4 | FST = 0.964* | |
Table 4 Analysis of molecular variance (AMOVA) of Gracilaria vermiculopylla populations
| 变异来源 Source of variation | 自由度 df | 平方和 Sums of squares | 变异组分 Estimated variance | 变异百分比 % of variation | 分化系数 F-statistics | 基因流 Nm |
|---|---|---|---|---|---|---|
| 一个类群 One group | ||||||
| 群体间 Among populations | 18 | 434.049 | 24.114 | 93 | ||
| 群体内 Within populations | 442 | 34.103 | 0.077 | 7 | FST = 0.928* | 0.039 |
| 两个类群(南方群体, 北方群体) Two groups (Regions: South, North) | ||||||
| 类群间Among regions | 1 | 405.675 | 405.675 | 93 | FCT = 0.933* | 0.019 |
| 类群内群体间Among populations | 17 | 28.374 | 1.669 | 3 | FSC = 0.465* | |
| 群体内 Within populations | 442 | 34.103 | 0.077 | 4 | FST = 0.964* | |
| [1] | Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48. |
| [2] | Bellorin AM, Olivera MC, Olivera EC (2004) Gracilaria vermiculophylla: a western Pacific species of Gracilariaceae (Rhodophyta) first recorded from the eastern Pacific. Phycological Research, 52, 69-79. |
| [3] | Boo GH, Kim KM, Nelson WA, Rafael RR, Yoon KJ, Boo SM (2014) Taxonomy and distribution of selected species of the agarophyte genus Gelidium (Gelidiales, Rhodophyta). Journal of Applied Phycology, 26, 1243-1251. |
| [4] | Cheang CC, Chu KH, Put AO (2010) Phylogeography of the marine macroalga Sargassum hemiphyllum (Phaeophyceae, Heterokontophyta) in northwestern Pacific. Molecular Ecology, 19, 2933-2948. |
| [5] | Chen Z, Zhang J, Fu HF, Xu ZZ, Deng KZ, Zhang JY (2012) On the validity of the species Phenacoccus solenopsis based on morphological and mitochondrial COI data, with the description of a new body color variety. Biodiversity Science, 20, 443-450. (in Chinese with English abstract) |
| [陈哲, 张姜, 傅杭飞, 许争争, 邓坤正, 张加勇 (2012) 基于形态特征和线粒体COI基因探讨扶桑绵粉蚧物种的有效性并记述一体色变异型扶桑绵粉蚧. 生物多样性, 20, 443-450.] | |
| [6] | Chu D, Liu GX, Fu HB, Xu W (2009) Phylogenetic analysis of mt-COI reveals the cryptic lineages in Phenacoccus solenopsis complex (Hemiptera: Pseudococcidae). Acta Entomologica Sinica, 52, 1261-1265. |
| [7] | Geraldino PJL, Yang EC, Kim MS, Boo SM (2009) Systematics of Hypnea asiatica sp. nov. (Hypneaceae, Rhodophyta) based on morphology and nrDNA SSU, plastid rbcL, and mitochondrial cox1. Taxon, 58, 606-616. |
| [8] | Guillemin ML, Faugeron S, Destombe C, Viard F, Correa JA, Valero M (2008) Genetic variation in wild and cultivated populations of the haploid-diploid red alga Gracilaria chilenesis: how farming practices favor asexual reproduction and heterozygosity. Evolution, 62, 1500-1519. |
| [9] | Guiry MD, Guiry GM (2016) AlgaeBase. World-wide electronic publication, National University of Ireland, Galway.. (accessed on 2016-04-12. |
| [10] | Gulbransen DJ, McGlathery KJ, Marklund M, Norris JN, Gurgel CFD (2012) Gracilaria vermiculophylla (Rhodophyta, Gracilariales) in the Virginia coastal bays, USA: cox1 analysis reveals high genetic richness of an introduced macroalga. Journal of Phycology, 48, 1278-1283. |
| [11] | Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98. |
| [12] | Hebert PD, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences, 270, 313-321. |
| [13] | Hu ZM, Zhang J, Lopez-Bautista J, Duan DL (2013) Asymmetric genetic exchange in the brown seaweed Sargassum fusiforme (Phaeophyceae) driven by oceanic currents. Marine Biology, 160, 1407-1414. |
| [14] | Hu ZM, Juan LB (2014) Adaptation mechanisms and ecological consequences of seaweed invasions: a review case of agarophyte Gracilaria vermiculophylla. Biological Invasions, 16, 967-976. |
| [15] | Kim SY, Weinberger F, Boo SM (2010) Genetic data hint at a common donor region for invasive Atlantic and Pacific populations of Gracilaria vermiculophylla (Gracilariales, Rhodophyta). Journal of Phycology, 46, 1346-1349. |
| [16] | Librado P, Roas J (2009) DNAsp v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformations, 25, 1451-1452. |
| [17] | Li JJ, Zhang J, Hu ZM, Duan DL (2013) Population genetics and demographic history of red seaweed, Palmaria palmata, from the Canada-northwest Atlantic. Biodiversity Science, 21, 306-314. (in Chinese with English abstract) |
| [李晶晶, 张杰, 胡自民, 段德麟 (2013) 加拿大西北大西洋地区掌形藻的种群遗传结构与动态变化. 生物多样性, 21, 306-314.] | |
| [18] | Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics, 28, 2537-2539. |
| [19] | Peason EA, Murray SN (1997) Patterns of reproduction, genetic diversity and genetic differentiation in California populations of the geniculate coralline alga Lithothrix aspergillum (Rhodophyta). Journal of Phycology, 33, 753-763. |
| [20] | Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics, 14, 817-818. |
| [21] | Riosmena RR, Talavera SA, Acosta VB, Gardner SC (2009) Heavy metals dynamics in seaweeds and seagrasses in Bahía Magdalena, BCS, México. Journal of Applied Phycology, 22, 283-291. |
| [22] | Robba L, Russell SJ, Barker GL, Brodie J (2006) Assessing the use of the mitochondrial Cox1 marker for use in DNA barcoding of red algae (Rhodophyta). American Journal of Botany, 93, 1101-1108. |
| [23] | Rueness J (2005) Life history and molecular sequences of Gracilaria vermiculophylla (Gracilariales, Rhodophyta), a new introduction to European waters. Phycologia, 44, 120-128. |
| [24] | Saunders GW (2005) Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences, 360, 1879-1888. |
| [25] | Saunders GW (2009) Routine DNA barcoding of Canadian Gracilariales (Rhodophyta) reveals the invasive species Gracilaria vermiculophylla in British Columbia. Molecular Ecology Resources, 9(Suppl. 1), 140-150. |
| [26] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution, 30, 2725-2729. |
| [27] | Terada R, Yamamoto H (2002) Review of Gracilaria vermiculophylla and other species in Japan and Asia. Taxonomy of Economic Seaweeds, 8, 215-224. |
| [28] | Thomsen MS, Stæhr PA, Nejrup L, Schiel DR (2013) Effects of the invasive macroalgae Gracilaria vermiculophylla on two co-occurring foundation species and associated invertebrates. Aquatic Invasions, 8, 133-145. |
| [29] | Tseng CK, Chen JF (1959) Reproductive of Gracilaria vermiculophylla and cultivation seedings inlaboratory. 90, 202-203. (in Chinese) |
| [曾呈奎, 陈椒芬 (1959) 真江蓠的繁殖习性和幼苗的室内培育. 中国科学院海洋研究所调查研究报告, 90, 202-203.] | |
| [30] | Tseng CK (2008) Seaweeds in Yellow Sea and Bohai Sea of China. Science Press, Beijing. (in Chinese) |
| [曾呈奎 (2008) 中国黄渤海海藻. 科学出版社, 北京.] | |
| [31] | Wang PX (1999) Response of western Pacific marginal seas to glacial cycles: paleoceanographic and sedimentological features. Marine Geology, 156, 5-39. |
| [32] | Xia BM, Chang CF (1999) Flora Algarum Marinarum Sinicarum Tomus II, Rhodophyta no. V Ahnfeltiales, Gigartinales, Rhodymeniales. Science Press, Beijing. (in Chinese) |
| [夏邦美, 张峻甫 (1999) 中国海藻志第二卷红藻门(第五分册): 伊谷藻目, 杉藻目, 红皮藻目. 科学出版社, 北京.] | |
| [33] | Yang EC, Kim MS, Geraldino PJL, Sahoo D, Shin JA, Boo SM (2008) Mitochondrial cox1 and plastid rbcL genes of Gracilaria vermiculophylla (Gracilariaceae, Rhodophyta). Journal of Applied Phycology, 20, 161-168. |
| [34] | Zhao XB, Pang SJ, Shan TF, Liu F (2013) Applications of three DNA barcodes in assorting intertidal red macroalgal flora in Qingdao, China. Journal of Ocean University of China, 12, 139-145. |
| [35] | Zheng WJ, Zhu SH, Shen XQ, Liu BQ, Pan ZC, Ye YF (2009) Genetic differentiation of Tegillarca granosa based on mitochondrial COI gene sequences. Zoological Research, 30, 17-23. (in Chinese with English abstract) |
| [郑文娟, 朱世华, 沈锡权, 刘必谦, 潘志崇, 叶央芳 (2009) 基于线粒体COI基因序列探讨泥蚶的遗传分化. 动物学研究, 30, 17-23.] | |
| [36] | Zhong CH, Huang RF, Lin Q, Zheng YY, Li LB, Liu B (2014) Molecular identification of a green type of Gracilaria vermiculophylla. Journal of Applied Oceanography, 33, 183-189. (in Chinese with English abstract) |
| [钟晨辉, 黄瑞芳, 林琪, 郑雅友, 李雷斌, 刘波 (2014) 一种绿色真江蓠的分子鉴定. 应用海洋学报, 33, 183-189.] |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [4] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [5] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [6] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [7] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [8] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [9] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [10] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [11] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [12] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [13] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [14] | Jing Cui, Mingfang Xu, Qun Zhang, Yao Li, Xiaoshu Zeng, Sha Li. Differences in genetic diversity of Pleuronichthys cornutus in the coastal water of China and Japan based on three mitochondrial markers [J]. Biodiv Sci, 2022, 30(5): 21485-. |
| [15] | Xinyu Cai, Xiaowei Mao, Yiqiang Zhao. Methods and research progress on the origin of animal domestication [J]. Biodiv Sci, 2022, 30(4): 21457-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()