



Biodiv Sci ›› 2022, Vol. 30 ›› Issue (9): 22103. DOI: 10.17520/biods.2022103 cstr: 32101.14.biods.2022103
Previous Articles Next Articles
Fan Xia1, Jing Yang1, Jian Li2, Yang Shi3, Lixin Gai4, Wenhua Huang5, Jingwei Zhang4, Nan Yang6, Fuli Gao1, Yingying Han1, Weidong Bao1,*( )
)
Received:2022-03-09
Accepted:2022-07-11
Online:2022-09-20
Published:2022-09-09
Contact:
Weidong Bao
About author:First author contact:# Co-first author
Fan Xia, Jing Yang, Jian Li, Yang Shi, Lixin Gai, Wenhua Huang, Jingwei Zhang, Nan Yang, Fuli Gao, Yingying Han, Weidong Bao. Gut bacterial composition of four leopard cat subpopulations in Beijing[J]. Biodiv Sci, 2022, 30(9): 22103.
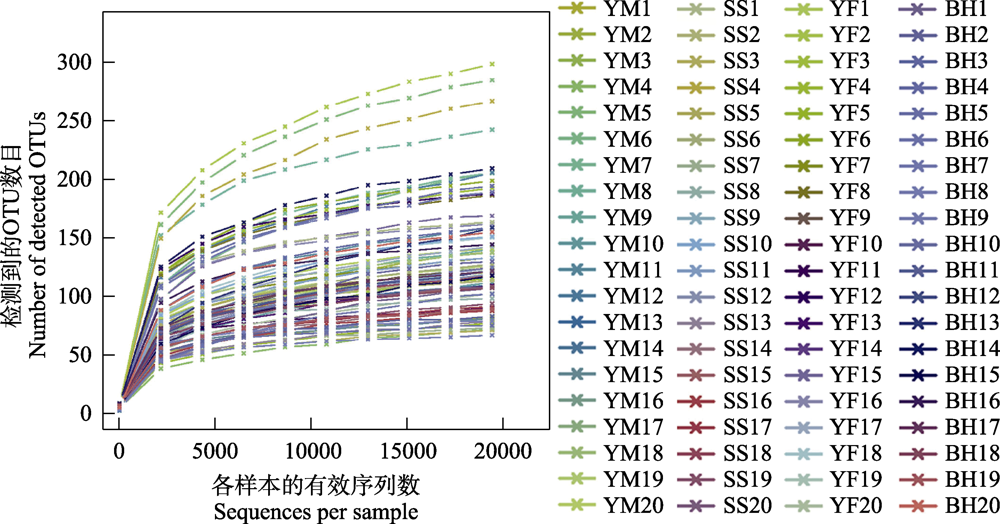
Fig. 2 Rarefaction curves of all samples for analysis of gut bacteria of leopard cat in Beijing. Each curve represents a sample and is marked with different color.
| 亚种群 Subpopulation | Q20碱基百分比 Q20 (%) | Q30碱基百分比 Q30 (%) | GC碱基百分比 GC (%) | 测序深度 Good’s coverage |
|---|---|---|---|---|
| 松山 SS | 94.32 | 91.52 | 53.749 | 0.999 |
| 云蒙山 YM | 92.58 | 89.07 | 54.014 | 0.999 |
| 云峰山 YF | 92.91 | 89.63 | 52.819 | 0.999 |
| 百花山 BH | 91.69 | 88.12 | 53.125 | 0.999 |
Table 1 Sequencing quality parameters in gut bacterial analysis of leopard cat in Beijing (average value). SS, Songshan; YM, Yunmengshan; YF, Yunfengshan; BH, Baihuashan.
| 亚种群 Subpopulation | Q20碱基百分比 Q20 (%) | Q30碱基百分比 Q30 (%) | GC碱基百分比 GC (%) | 测序深度 Good’s coverage |
|---|---|---|---|---|
| 松山 SS | 94.32 | 91.52 | 53.749 | 0.999 |
| 云蒙山 YM | 92.58 | 89.07 | 54.014 | 0.999 |
| 云峰山 YF | 92.91 | 89.63 | 52.819 | 0.999 |
| 百花山 BH | 91.69 | 88.12 | 53.125 | 0.999 |
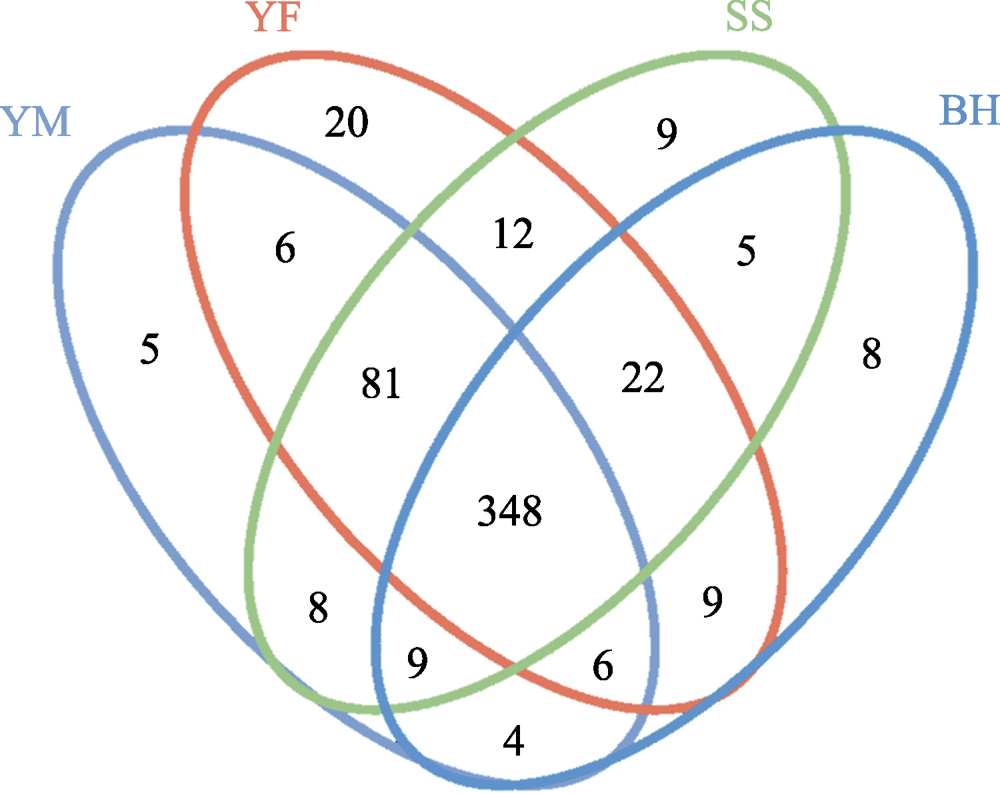
Fig. 3 Venn diagram of the OTU numbers of gut bacteria of leopard cat at four sampling areas in Beijing. The ellipses of four colors in the figure represent four sampling areas. YM, Yunmengshan; YF, Yunfengshan; SS, Songshan; BH, Baihuashan. The numbers represent the unique or shared number of OTUs of the four sampling areas.
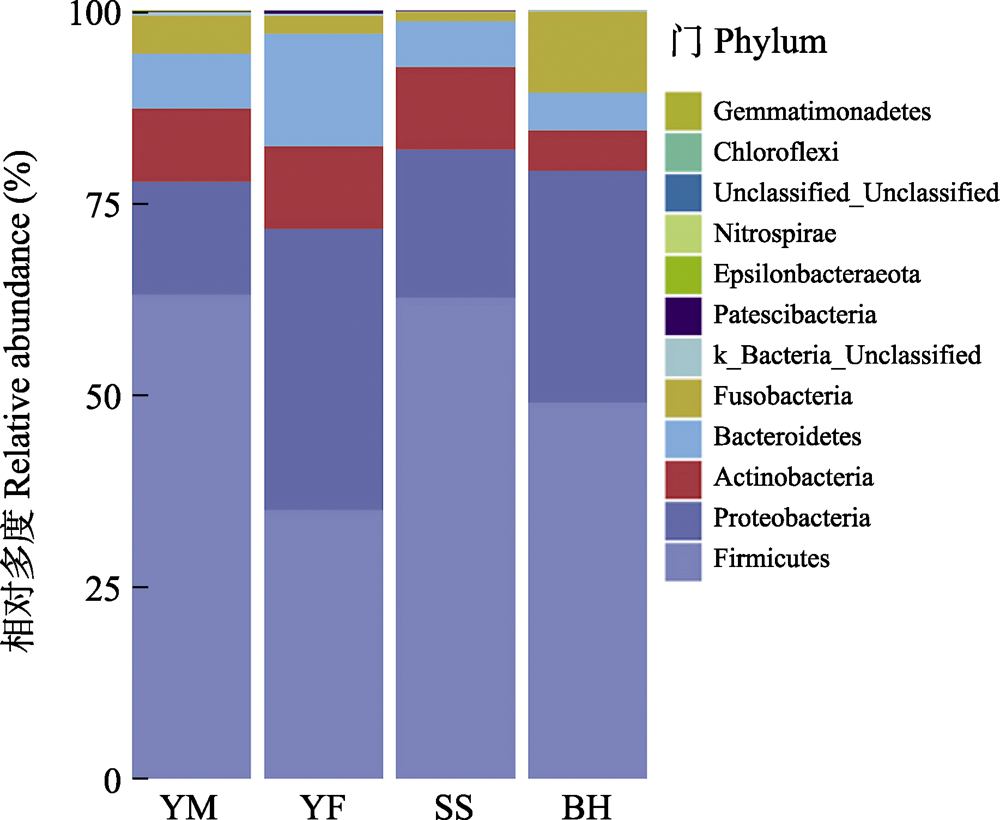
Fig. 4 The relative abundance of gut bacteria of leopard cat at four sampling areas in Beijing at the phylum level. The abscissa is the four sampling areas, and different colors represent the classification of bacteria at the phylum level. Full names of the four sampling areas see Fig. 3.
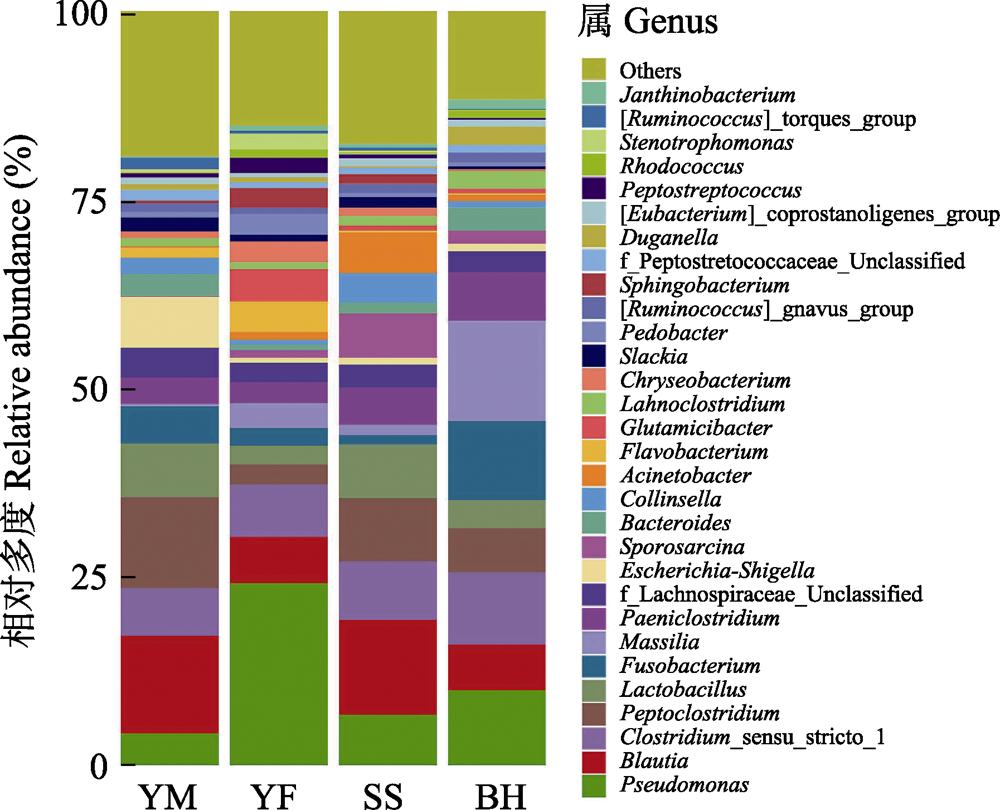
Fig. 5 The relative abundance of gut bacteria of leopard cat at four sampling areas in Beijing at genus level. Different colors represent the classification of bacteria at the genus level. Full names of the four sampling areas see Fig. 3.
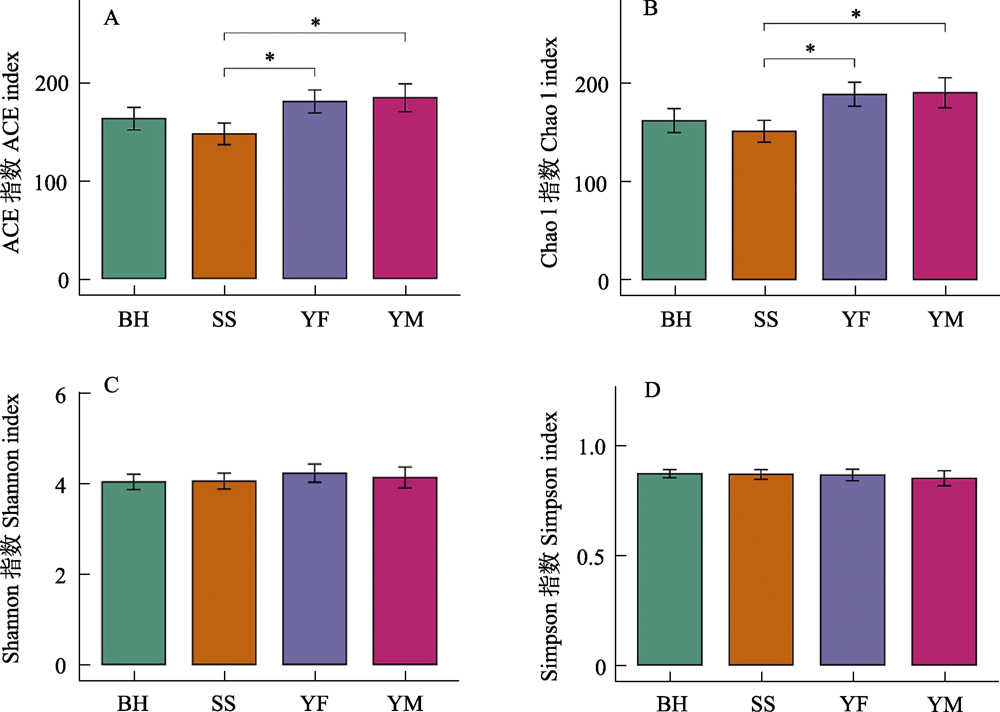
Fig. 6 Differences in α-diversity of gut bacteria of leopard cat at four sampling areas in Beijing. * P < 0.05. Full names of the four sampling areas see Fig. 3.
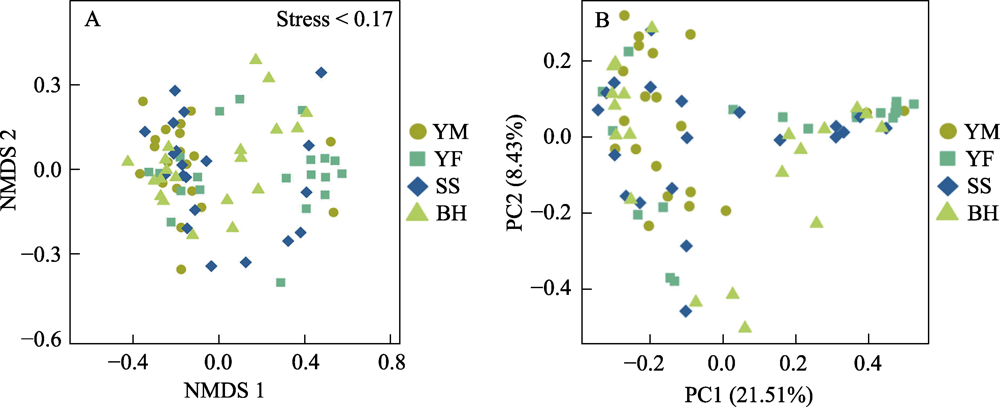
Fig. 7 Analysis of gut bacteria distance based on non-metric multidimensional scaling (NMDS) and principal coordinates analysis (PCoA) of leopard cat at four sampling areas in Beijing. In NMDS analysis, each point represents one sample, and the distance between points represents difference degree. In PCoA analysis, the closer the distance between the sample points is, the higher the similarity is. Full names of the four sampling areas see Fig. 3.
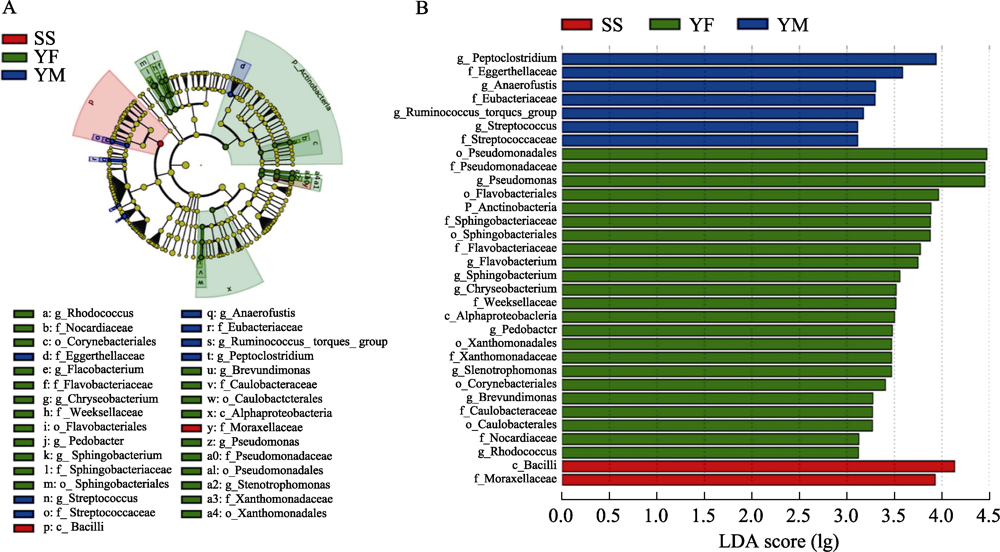
Fig. 8 LEfSe analysis of gut bacterial composition among three leopard cat subpopulations in Beijing. Figure A is the cluster evolutionary tree of gut bacteria of three leopard cat subpopulations, and figure B shows the LDA scores of various bacteria under different classification levels. Different colors indicate sampling areas with red for Songshan, green for Yunfengshan, and blue for Yunmengshan.
| [1] |
Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M, Gillis M, Wilson BA, Nelson KE, White BA, Leigh SR (2013) Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. The ISME Journal, 7, 1344-1353.
DOI URL |
| [2] |
An C, Okamoto Y, Xu SY, Eo KY, Kimura J, Yamamoto N (2017) Comparison of fecal microbiota of three captive carnivore species inhabiting Korea. The Journal of Veterinary Medical Science, 79, 542-546.
DOI URL |
| [3] |
Arboleya S, Watkins C, Stanton C, Ross RP (2016) Gut bifidobacteria populations in human health and aging. Frontiers in Microbiology, 7, 1204.
DOI PMID |
| [4] | Bao WD, Li XJ, Shi Y (2005) Comparative analysis of food habits in carnivores from three areas of Beijing. Zoological Research, 26, 118-122. (in Chinese with English abstract) |
| [鲍伟东, 李晓京, 史阳 (2005) 北京市三个区域食肉类动物食性的比较分析. 动物学研究, 26, 118-122.] | |
| [5] | Chen W, Gao W, Fu BQ (2002) Mammal Records of Beijing. Beijing Press, Beijing. (in Chinese) |
| [陈卫, 高武, 傅必谦 (2002) 北京兽类志. 北京出版社, 北京.] | |
| [6] | Clarke K, Gorley RN (2006) Primer v6: User Manual/Tutorial. Plymouth Marine Laboratory, Plymouth. |
| [7] |
Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, Gougis S, Rizkalla S, Batto JM, Renault P, Doré J, Zucker JD, Clément K, Ehrlich SD (2013) Dietary intervention impact on gut microbial gene richness. Nature, 500, 585-588.
DOI URL |
| [8] |
Doré J, Blottière H (2015) The influence of diet on the gut microbiota and its consequences for health. Current Opinion in Biotechnology, 32, 195-199.
DOI PMID |
| [9] | Du LH, Wang XP, Chen JQ, Liu GL, Wu JG, Zhang ZX (2012) Comprehensive Scientific Investigation Report of Beijing Songshan Nature Reserve. China Forestry Publishing House, Beijing. (in Chinese) |
| [杜连海, 王小平, 陈峻崎, 刘桂林, 吴记贵, 张志翔 (2012) 北京松山自然保护区综合科学考察报告. 中国林业出版社, 北京.] | |
| [10] | Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science, 308, 1635-1638. |
| [11] |
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27, 2194-2200.
DOI PMID |
| [12] | Eo SH, Ko BJ, Lee BJ, Seomun H, Kim S, Kim MJ, Kim JH, An J (2016) A set of microsatellite markers for population genetics of leopard cat (Prionailurus bengalensis) and cross-species amplification in other felids. Biochemical Systematics & Ecology, 66, 196-200. |
| [13] | Fan YQ, Yang J, Zhang HL, Jiang J, Jiang WJ, Tang SP, Bao WD (2020) Food composition of medium sized carnivores in Beijing Songshan Nature Reserve. Journal of Biology, 37(1), 59-62. (in Chinese with English abstract) |
| [范雅倩, 杨婧, 张洪亮, 蒋健, 蒋万杰, 唐书培, 鲍伟东 (2020) 北京松山自然保护区中型捕食动物的食物构成分析. 生物学杂志, 37(1), 59-62.] | |
| [14] |
Godoy-Vitorino F, Goldfarb KC, Karaoz U, Leal S, Garcia-Amado MA, Hugenholtz P, Tringe SG, Brodie EL, Dominguez-Bello MG (2012) Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. The ISME Journal, 6, 531-541.
DOI URL |
| [15] | Han SY, Guan Y, Dou HL, Yang HT, Yao M, Ge JP, Feng LM (2019) Comparison of the fecal microbiota of two free-ranging Chinese subspecies of the leopard (Panthera pardus) using high-throughput sequencing. PeerJ, 7, e6684. |
| [16] |
Hooper LV, Littman DR, MacPherson AJ (2012) Interactions between the microbiota and the immune system. Science, 336, 1268-1273.
DOI PMID |
| [17] |
Kaoutari AE, Armougom F, Gordon JI, Raoult D, Henrissat B (2013) The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nature Reviews Microbiology, 11, 497-504.
DOI PMID |
| [18] | Ko BJ, An J, Seomun H, Lee MY, Eo SH (2018) Microsatellite DNA analysis reveals lower than expected genetic diversity in the threatened leopard cat (Prionailurus bengalensis) in South Korea. Genes & Genomics, 40, 521-530. |
| [19] | Kong FL, Zhao JC, Han SS, Zeng B, Yang JD, Si XH, Yang BQ, Yang MY, Xu HL, Li Y (2014) Characterization of the gut microbiota in the red panda (Ailurus fulgens). PLoS ONE, 9, e87885. |
| [20] |
Konopka A (2009) What is microbial community ecology? The ISME Journal, 3, 1223-1230.
DOI URL |
| [21] |
Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U (2011) Genotype is a stronger determinant than sex of the mouse gut microbiota. Microbial Ecology, 61, 423-428.
DOI PMID |
| [22] |
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI (2008) Worlds within worlds: Evolution of the vertebrate gut microbiota. Nature Reviews Microbiology, 6, 776-788.
DOI PMID |
| [23] | Luan XF, Li DQ, Li GL (2011) Study on Biodiversity and Protection of Beijing Yunfengshan Nature Reserve. China Land Press, Beijing. (in Chinese) |
| [栾晓峰, 李迪强, 李广良 (2011) 北京云峰山自然保护区生物多样性及保护研究. 中国大地出版社, 北京.] | |
| [24] | Masuoka H, Shimada K, Kiyosue-Yasuda T, Kiyosue M, Oishi Y, Kimura S, Ohashi Y, Fujisawa T, Hotta K, Yamada A, Hirayama K (2017) Transition of the intestinal microbiota of cats with age. PLoS ONE, 12, e0181739. |
| [25] |
Mittal P, Saxena R, Gupta A, Mahajan S, Sharma VK (2020) The gene catalog and comparative analysis of gut microbiome of big cats provide new insights on Panthera species. Frontiers in Microbiology, 11, 1012.
DOI PMID |
| [26] | Mozejko-Ciesielska J, Pokoj T, Ciesielski S (2018) Transcriptome remodeling of Pseudomonas putida KT 2440 during mcl-PHAs synthesis: Effect of different carbon sources and response to nitrogen stress. Journal of Industrial Microbiology & Biotechnology, 45, 433-446. |
| [27] |
Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI (2011) Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science, 332, 970-974.
DOI PMID |
| [28] | Ning Y (2020) Study on Amur Tiger Dispersal, Inbreeding and Gut Microbiota Based on DNA Analysis. PhD dissertation, Northeast Forestry University, Harbin. (in Chinese with English abstract) |
| [宁瑶 (2020) 基于DNA分析的东北虎扩散状况、近交以及肠道菌群的研究. 博士学位论文, 东北林业大学, 哈尔滨.] | |
| [29] |
Phillips CD, Phelan G, Dowd SE, McDonough MM, Ferguson AW, Hanson JD, Siles L, Ordonez-Garza N, San Francisco M, Baker RJ (2012) Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Molecular Ecology, 21, 2617-2627.
DOI PMID |
| [30] |
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: A versatile open source tool for metagenomics. PeerJ, 4, e2584.
DOI URL |
| [31] |
Saka T, Nishita Y, Masuda R (2018) Low genetic variation in the MHC class II DRB gene and MHC-linked microsatellites in endangered island populations of the leopard cat (Prionailurus bengalensis) in Japan. Immunogenetics, 70, 115-124.
DOI URL |
| [32] |
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60.
DOI URL |
| [33] |
Sheppard SK, Harwood JD (2005) Advances in molecular ecology: Tracking trophic links through predator prey food-webs. Functional Ecology, 19, 751-762.
DOI URL |
| [34] |
Sicuro FL, Oliveira LFB (2015) Variations in leopard cat (Prionailurus bengalensis) skull morphology and body size: Sexual and geographic influences. PeerJ, 3, e1309.
DOI URL |
| [35] |
Sinha T, Vich VA, Garmaeva S, Jankipersadsing SA, Imhann F, Collij V, Bonder MJ, Jiang XF, Gurry T, Alm EJ, D’Amato M, Weersma RK, Scherjon S, Wijmenga C, Fu JY, Kurilshikov A, Zhernakova A (2019) Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles. Gut Microbes, 10, 358-366.
DOI PMID |
| [36] |
Sommer F, Bäckhed F (2013) The gut microbiota—Masters of host development and physiology. Nature Reviews Microbiology, 11, 227-238.
DOI PMID |
| [37] |
Sun BH, Wang X, Bernstein S, Huffman MA, Xia DP, Gu ZY, Chen R, Sheeran LK, Wagner RS, Li JH (2016) Marked variation between winter and spring gut microbiota in free-ranging Tibetan macaques (Macaca thibetana). Scientific Reports, 6, 26035.
DOI URL |
| [38] |
Tamada T, Siriaroonrat B, Subramaniam V, Hamachi M, Lin LK, Oshida T, Rerkamnuaychoke W, Masuda R (2008) Molecular diversity and phylogeography of the Asian leopard cat, Felis bengalensis, inferred from mitochondrial and Y-Chromosomal DNA sequences. Zoological Science, 25, 154-163.
DOI URL |
| [39] |
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI (2009) A core gut microbiome in obese and lean twins. Nature, 457, 480-484.
DOI URL |
| [40] |
Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M (2018) Glycan utilization and cross-feeding activities by bifidobacteria. Trends in Microbiology, 26, 339-350.
DOI PMID |
| [41] | Wang Y, Sheng HF, He Y, Wu JY, Jiang YX, Tam NFY, Zhou HW (2012) Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of Illumina tags. Applied & Environmental Microbiology, 78, 8264-8271. |
| [42] |
Wasimuddin, Menke S, Melzheimer J, Thalwitzer S, Heinrich S, Wachter B, Sommer S (2017) Gut microbiomes of free-ranging and captive Namibian cheetahs: Diversity, putative functions and occurrence of potential pathogens. Molecular Ecology, 26, 5515-5527.
DOI PMID |
| [43] |
Wei FW, Wang X, Wu Q (2015) The giant panda gut microbiome. Trends in Microbiology, 23, 450-452.
DOI PMID |
| [44] | Xu XM, Zhang ZB (2021) Sex- and age-specific variation of gut microbiota in Brandt’s voles. PeerJ, 9, e11434. |
| [45] |
Zhang HH, Liu GS, Chen L, Sha WL (2015) Composition and diversity of the bacterial community in snow leopard (Uncia uncia) distal gut. Annals of Microbiology, 65, 703-711.
DOI URL |
| [46] |
Zhang TH, Yang Y, Liang Y, Jiao X, Zhao CH (2018) Beneficial effect of intestinal fermentation of natural polysaccharides. Nutrients, 10, 1055.
DOI URL |
| [47] | Zhao B (2005) Studies on Plant Diversity in Mountain Areas in Beijing. PhD dissertation, Beijing Forestry University, Beijing. (in Chinese with English abstract) |
| [赵勃 (2005) 北京山区植物多样性研究. 博士学位论文, 北京林业大学, 北京.] |
| [1] | Zhiyu Liu, Xin Ji, Guohui Sui, Ding Yang, Xuankun Li. Invertebrate diversity in buffalo grass and weedy lawns at Beijing Capital International Airport [J]. Biodiv Sci, 2025, 33(4): 24456-. |
| [2] | Sicheng Han, Daowei Lu, Yuchen Han, Ruohan Li, Jing Yang, Ge Sun, Lu Yang, Junwei Qian, Xiang Fang, Shu-Jin Luo. Distribution of leopard cats in the nearest mountains to urban Beijing and its affecting environmental factors [J]. Biodiv Sci, 2024, 32(8): 24138-. |
| [3] | Fei Duan, Mingzhang Liu, Hongliang Bu, Le Yu, Sheng Li. Effects of urbanization on bird community composition and functional traits: A case study of the Beijing-Tianjin-Hebei region [J]. Biodiv Sci, 2024, 32(8): 23473-. |
| [4] | Zhengming Luo, Jinxian Liu, Bianhua Zhang, Yanying Zhou, Aihua Hao, Kai Yang, Baofeng Chai. Diversity characteristics and driving factors of soil protist communities in subalpine meadow at different degradation stages [J]. Biodiv Sci, 2023, 31(8): 23136-. |
| [5] | Yinger Mao, Xiumei Zhou, Nan Wang, Xiuxiu Li, Yuke You, Shangbin Bai. Impact of Phyllostachys edulis expansion to Chinese fir forest on the soil bacterial community [J]. Biodiv Sci, 2023, 31(6): 22659-. |
| [6] | Luqin Yin, Cheng Wang, Wenjing Han. Food source characteristics and diversity of birds based on feeding behavior in residential areas of Beijing [J]. Biodiv Sci, 2023, 31(5): 22473-. |
| [7] | Yexi Zhao, Jiayu Zhang, Zihan Li, Qinmijia Xie, Xin Deng, Nan Wang. Use of native and alien plants during night roosting by urban birds in Beijing [J]. Biodiv Sci, 2023, 31(3): 22399-. |
| [8] | Wen Zhao, Dandan Wang, Mumin Reyila, Kaichuan Huang, Shun Liu, Baokai Cui. Soil microbial community structure of Larix gmelinii forest in the Aershan area [J]. Biodiv Sci, 2023, 31(2): 22258-. |
| [9] | Xueqin Deng, Tong Liu, Tianshi Liu, Kai Xu, Song Yao, Xiaoqun Huang, Zhishu Xiao. Seasonal variation of daily activity rhythm of leopard cats (Prionailurus bengalensis) and their potential prey in Neixiang Baotianman National Nature Reserve of Henan Province, China [J]. Biodiv Sci, 2022, 30(9): 22263-. |
| [10] | Xia Cui, Quanru Liu, Chaoran Wu, Yufei He, Jinshuang Ma. The alien invasive plants in Beijing-Tianjin-Hebei [J]. Biodiv Sci, 2022, 30(8): 21497-. |
| [11] | Qinwen Lin, Chaoran Wu, Xia Cui, Jinshuang Ma. Thirty-year changes of vascular plants in Beijing [J]. Biodiv Sci, 2022, 30(6): 22107-. |
| [12] | Cui Xiao, Bing Liu, Chaoran Wu, Jinshuang Ma, Jianfei Ye, Xiaofei Xia, Qinwen Lin. A dataset on inventory and geographical distributions of vascular plants in Beijing, China [J]. Biodiv Sci, 2022, 30(6): 22064-. |
| [13] | Yixin Sun, Yingbin Li, Yuhui Li, Bing Li, Xiaofang Du, Qi Li. Application of high-throughput sequencing technique in the study of nematode diversity [J]. Biodiv Sci, 2022, 30(12): 22266-. |
| [14] | Cheng Gao, Liang-Dong Guo. Progress on microbial species diversity, community assembly and functional traits [J]. Biodiv Sci, 2022, 30(10): 22429-. |
| [15] | Qifeng Lu, Zhihuan Huang, Wenhua Luo. Characterization of complete chloroplast genome in Firmiana kwangsiensis and F. danxiaensis with extremely small populations [J]. Biodiv Sci, 2021, 29(5): 586-595. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn