



生物多样性 ›› 2021, Vol. 29 ›› Issue (5): 586-595. DOI: 10.17520/biods.2020263 cstr: 32101.14.biods.2020263
收稿日期:2020-06-30
接受日期:2020-09-10
出版日期:2021-05-20
发布日期:2021-02-01
通讯作者:
骆文华
作者简介:* E-mail: luowenhua2004@163.com基金资助:
Qifeng Lu1, Zhihuan Huang2, Wenhua Luo1,*( )
)
Received:2020-06-30
Accepted:2020-09-10
Online:2021-05-20
Published:2021-02-01
Contact:
Wenhua Luo
摘要:
广西火桐(Firmiana kwangsiensis)和丹霞梧桐(F. danxiaensis)是我国南方特有物种, 其分布范围狭窄, 种群数量少。为了解其叶绿体基因组结构及系统发生关系, 本文通过高通量测序方法获得广西火桐和丹霞梧桐的浅层基因组数据, 通过生物信息学方法对叶绿体全基因组进行组装, 并对其结构特征进行分析。结果表明: 广西火桐和丹霞梧桐的叶绿体基因组大小分别为160,836 bp和161,253 bp, 具有典型被子植物叶绿体基因组环状四分体结构, 包含长度分别为89,700 bp、90,142 bp的大单拷贝区(large single copy, LSC), 长度分别为19,970 bp、20,067 bp的小单拷贝区(small single copy, SSC)及长度分别为25,583 bp、25,522 bp的2个反向重复序列区(inverted repeat sequence, IR)。两个物种的叶绿体基因组共注释得到131个基因, 包括86个蛋白编码基因、37个tRNA基因和8个rRNA基因。广西火桐的叶绿体基因组中共检测出26个正向重复序列、2个反向重复序列、21个回文重复序列、21个串联重复序列和98个简单重复序列; 丹霞梧桐叶绿体基因组中共检测出23个正向重复序列、5个反向重复序列、21个回文重复序列、30个串联重复序列和107个简单重复序列。系统发生分析结果表明5种梧桐属(Firmiana)植物构成两个强烈支持的分支(支持率100%), 一个分支为广西火桐、美丽火桐(F. pulcherrima)和火桐(F. colorata), 其中广西火桐与美丽火桐构成姐妹群; 另一分支是互为姐妹群的丹霞梧桐和云南梧桐(F. major)。综上所述, 广西火桐和丹霞梧桐的叶绿体基因组结构、基因排列及重复序列具有较高的相似性, 系统进化树将5种梧桐属物种分为两个分支, 其中广西火桐和美丽火桐最近; 而丹霞梧桐与云南梧桐关系最近。本研究鉴定的SSR位点可为梧桐属物种系统发生、进化关系的研究提供遗传信息。
陆奇丰, 黄至欢, 骆文华 (2021) 极小种群濒危植物广西火桐、丹霞梧桐的叶绿体基因组特征. 生物多样性, 29, 586-595. DOI: 10.17520/biods.2020263.
Qifeng Lu, Zhihuan Huang, Wenhua Luo (2021) Characterization of complete chloroplast genome in Firmiana kwangsiensis and F. danxiaensis with extremely small populations. Biodiversity Science, 29, 586-595. DOI: 10.17520/biods.2020263.
| 基因组特征 Genome characteristics | 广西火桐 F. kwangsiensis | 丹霞梧桐 F. danxiaensis |
|---|---|---|
| 序列全长 Genome size (bp) | 160,836 | 161,253 |
| LSC区长度 LSC length (bp) | 89,700 | 90,142 |
| SSC区长度 SSC length (bp) | 19,970 | 20,067 |
| IR区长度 IR length (bp) | 25,583 | 25,522 |
| GC含量 GC content (%) | 37.04 | 36.87 |
| LSC区GC含量 GC content of LSC (%) | 34.92 | 34.68 |
| SSC区GC含量 GC content of SSC (%) | 31.46 | 31.24 |
| IR区GC含量 GC content of IR (%) | 42.93 | 42.97 |
| A碱基占比 A base content (%) | 30.98 | 31.13 |
| T碱基占比 T base content (%) | 31.98 | 31.99 |
| C碱基占比 C base content (%) | 18.87 | 18.61 |
| G碱基占比 G base content (%) | 18.17 | 18.26 |
| 基因总数 Total genes | 131 | 131 |
| 编码蛋白基因数 No. of protein coding sequences | 86 | 86 |
| tRNA | 37 | 37 |
| rRNA | 8 | 8 |
表1 广西火桐和丹霞梧桐的叶绿体基因组特征比较
Table 1 Comparison of chloroplast genome characteristics of Firmiana kwangsiensis and F. danxiaensis
| 基因组特征 Genome characteristics | 广西火桐 F. kwangsiensis | 丹霞梧桐 F. danxiaensis |
|---|---|---|
| 序列全长 Genome size (bp) | 160,836 | 161,253 |
| LSC区长度 LSC length (bp) | 89,700 | 90,142 |
| SSC区长度 SSC length (bp) | 19,970 | 20,067 |
| IR区长度 IR length (bp) | 25,583 | 25,522 |
| GC含量 GC content (%) | 37.04 | 36.87 |
| LSC区GC含量 GC content of LSC (%) | 34.92 | 34.68 |
| SSC区GC含量 GC content of SSC (%) | 31.46 | 31.24 |
| IR区GC含量 GC content of IR (%) | 42.93 | 42.97 |
| A碱基占比 A base content (%) | 30.98 | 31.13 |
| T碱基占比 T base content (%) | 31.98 | 31.99 |
| C碱基占比 C base content (%) | 18.87 | 18.61 |
| G碱基占比 G base content (%) | 18.17 | 18.26 |
| 基因总数 Total genes | 131 | 131 |
| 编码蛋白基因数 No. of protein coding sequences | 86 | 86 |
| tRNA | 37 | 37 |
| rRNA | 8 | 8 |
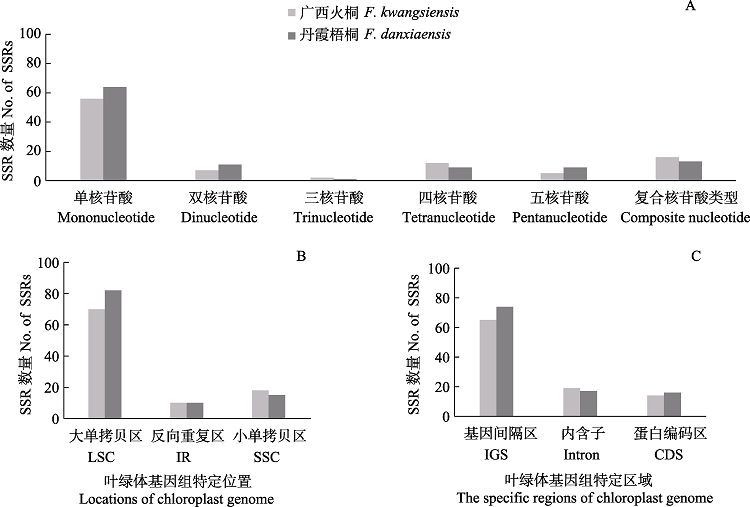
图2 广西火桐和丹霞梧桐的叶绿体基因组SSR分析。(A)不同SSR类型的数量; (B)大单拷贝区(LSC); 反向重复区(IR)和小单拷贝区(SSC)的SSR数量; (C)基因间隔区(IGS)、基因编码区(CDS)及内含子(intron)的SSR数量。
Fig. 2 SSR analysis of the chloroplast genomes of Firmiana kwangsiensis and F. danxiaensis. (A) Number of SSRs in different types; (B) Number of SSRs in the LSC (large single copy), SSC (small single copy), and IR (inverted repeat region); (C) Number of SSRs in the intergenic regions (IGS), protein-coding gene (CDS) and introns.
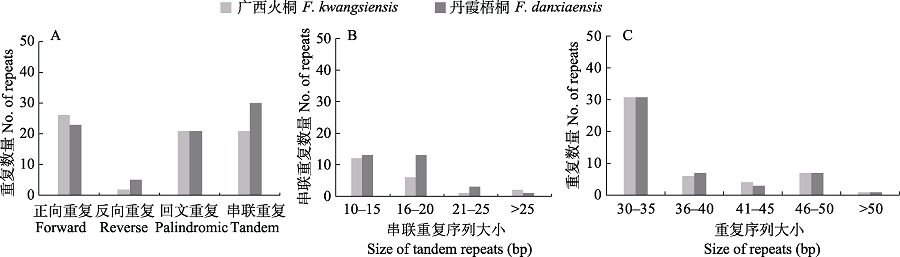
图3 广西火桐和丹霞梧桐的叶绿体基因组重复序列分析。(A)不同重复序列类型的分布数量; (B)串联重复序列长度的分布范围; (C)正向、反向和回文重复序列长度的分布范围。
Fig. 3 Repeat sequences analysis of Firmiana kwangsiensis and F. danxiaensis chloroplast genome. (A) Number of different types of repeats; (B) Size of tandem repeats in specific range; (C) Size of forward, reverse and palindromic repeats in specific range.
| [1] |
Abdullah , Shahzadi I, Mehmood F, Ali Z, Malik MS, Waseem S, Mirza B, Ahmed I, Waheed MT (2019) Comparative analyses of chloroplast genomes among three Firmiana species: Identification of mutational hotspots and phylogenetic relationship with other species of Malvaceae. Plant Gene, 19, 100199.
DOI URL |
| [2] | Ajaib M, Wahla SQ, Khan KM (2014) Firmiana simplex: A potential source of antimicrobials. Journal of the Chemical Society of Pakistan, 36, 744-753. |
| [3] |
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology, 19, 455-477.
DOI URL |
| [4] |
Beier S, Thiel T, Münch T, Scholz U, Mascher M (2017) MISA-web: A web server for microsatellite prediction. Bioinformatics, 33, 2583-2585.
DOI URL |
| [5] |
Benson G (1999) Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Research, 27, 573-580.
DOI URL |
| [6] | Birky CW (1995) Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proceedings of the National Academy of Sciences, USA, 92, 11331-11338. |
| [7] | Chen L, Zhou H, Wang MQ, Wu XT, Lin XY, Zhang Y, Wen YF (2018) Community characteristics comparison of Firmiana danxiaensis species. Chinese Wild Plant Resources, 37(2), 46-49. (in Chinese with English abstract) |
| [ 陈璐, 周宏, 王敏求, 武星彤, 林雪莹, 张原, 文亚峰 (2018) 丹霞梧桐群落特征比较研究. 中国野生植物资源, 37(2), 46-49.] | |
| [8] |
Chen Q, Wu XB, Zhang DQ (2019) Phylogenetic analysis of Fritillaria cirrhosa D. Don and its closely related species based on complete chloroplast genomes. PeerJ, 7, e7480.
DOI URL |
| [9] |
Chen SF, Li MW, Hou RF, Liao WB, Zhou RC, Fan Q (2014) Low genetic diversity and weak population differentiation in Firmiana danxiaensis, a tree species endemic to Danxia landform in northern Guangdong, China. Biochemical Systematics and Ecology, 55, 66-72.
DOI URL |
| [10] |
Chen SF, Zhou YQ, Chen YR, Gu J (2018) Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34, i884-i890.
DOI URL |
| [11] |
Chen SF, Li MW, Jing HJ, Zhou RC, Yang GL, Wu W, Fan Q, Liao WB (2015) De novo transcriptome assembly in Firmiana danxiaensis, a tree species endemic to the Danxia landform. PLoS ONE, 10, e0139373.
DOI URL |
| [12] |
Cheng Y, Zhang LM, Qi JM, Zhang LW (2020) Complete chloroplast genome sequence of Hibiscus cannabinus and comparative analysis of the Malvaceae family. Frontiers in Genetics, 11, 227.
DOI URL |
| [13] |
Chmielewski M, Meyza K, Chybicki IJ, Dzialuk A, Litkowiec M, Burczyk J (2015) Chloroplast microsatellites as a tool for phylogeographic studies: The case of white oaks in Poland. iForest, 8, 765-771.
DOI URL |
| [14] |
Daniell H, Lin CS, Yu M, Chang WJ (2016) Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biology, 17, 134.
DOI PMID |
| [15] | Doyle JJ (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19, 11-15. |
| [16] |
Du QZ, Wang BW, Wei ZZ, Zhang DQ, Li BL (2012) Genetic diversity and population structure of Chinese white poplar (Populus tomentosa) revealed by SSR markers. Journal of Heredity, 103, 853-862.
DOI URL |
| [17] | Fan Q, Guo W, Chen SF, Liao WB (2011) Phylogeny of Firmiana (Sterculiaceae) based on nrDNA ITS analysis. In: The 10th Youth Academic Seminar of National System and Evolutionary Botany (ed. Botanic Society of Yunnan), p. 68. Kunming. (in Chinese with English abstract) |
| [ 凡强, 郭微, 陈素芳, 廖文波 (2011) ITS序列探讨梧桐属的系统发育. 见: 2011年全国系统与进化植物学暨第十届青年学术研讨会论文集(云南植物学会编), 68页. 昆明.] | |
| [18] |
Gao L, Yi X, Yang YX, Su YJ, Wang T (2009) Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: Insights into evolutionary changes in fern chloroplast genomes. BMC Evolutionary Biology, 9, 130.
DOI URL |
| [19] |
Haberle RC, Fourcade HM, Boore JL, Jansen RK (2008) Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. Journal of Molecular Evolution, 66, 350-361.
DOI URL |
| [20] |
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11, 119.
DOI PMID |
| [21] | Ivanova Z, Sablok G, Daskalova E, Zahmanova G, Apostolova E, Yahubyan G, Baev V (2017) Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Frontiers in Plant Science, 8, 204. |
| [22] | Jiang M, Wang JF, Ying MH, Yang RM, Ma JY (2020) Assembly and sequence analysis of Tetrastigma hemsleyanum chloroplast genome. Chinese Traditional and Herbal Drugs, 51, 461-468. (in Chinese with English abstract) |
| [ 蒋明, 王军峰, 应梦豪, 杨如棉, 马佳莹 (2020) 三叶崖爬藤叶绿体基因组的组装与序列分析. 中草药, 51, 461-468.] | |
| [23] |
Kim JW, Yang H, Cho N, Kim B, Kim YC, Sung SH (2015) Hepatoprotective constituents of Firmiana simplex stem bark against ethanol insult to primary rat hepatocytes. Pharmacognosy Magazine, 11(41), 55-60.
DOI URL |
| [24] |
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001) REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research, 29, 4633-4642.
PMID |
| [25] |
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nature Methods, 9, 357-359.
DOI PMID |
| [26] |
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Research, 32, 11-16.
PMID |
| [27] | Li Q, Guo QQ, Gao C, Li HE (2020) Characterization of complete chloroplast genome of Camellia weiningensis in Weining, Guizhou Province. Acta Horticulturae Sinica, 47, 779-787. (in Chinese with English abstract) |
| [ 李倩, 郭其强, 高超, 李慧娥 (2020) 贵州威宁红花油茶的叶绿体基因组特征分析. 园艺学报, 47, 779-787.] | |
| [28] |
Liang CL, Wang L, Lei J, Duan BZ, Ma WS, Xiao SM, Qi HJ, Wang Z, Liu YQ, Shen XF, Guo S, Hu HY, Xu J, Chen SL (2019) A comparative analysis of the chloroplast genomes of four Salvia medicinal plants. Engineering, 5, 907-915.
DOI URL |
| [29] | Lim SY, Subedi L, Shin D, Kim CS, Lee KR, Kim SY (2017) A new neolignan derivative, balanophonin isolated from Firmiana simplex delays the progress of neuronal cell death by inhibiting microglial activation. Biomolecules & Therapeutics, 25, 519-527. |
| [30] | Luo WH, Mao SZ, Ding L, Huang YS, Deng T (2010) Study on community characteristics of endangered species Erythropsis kwangsiensis. Journal of Fujian Forestry Science and Technology, 37(4), 6-10. (in Chinese with English abstract) |
| [ 骆文华, 毛世忠, 丁莉, 黄俞淞, 邓涛 (2010) 濒危植物广西火桐群落特征研究. 福建林业科技, 37(4), 6-10.] | |
| [31] | Luo WH, Dai WJ, Liu J, Hu XH, Li XJ, Huang SX (2015) Comparison of genetic diversity of natural and ex-situ conservation populations of Erythropsis kwangsiensis. Journal of Central South University of Forestry & Technology, 35(2), 66-71. (in Chinese with English abstract) |
| [ 骆文华, 代文娟, 刘建, 胡兴华, 李祥军, 黄仕训 (2015) 广西火桐自然种群和迁地保护种群的遗传多样性比较. 中南林业科技大学学报, 35(2), 66-71.] | |
| [32] |
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research, 41, W575-W581.
DOI URL |
| [33] | National Forestry Administration, Ministry of Agriculture (1999) List of Key State Protected Wild Plants (The first bacth). Plant Journal, (5), 4-11. |
| [ 国家林业局, 农业部 (1999) 国家重点保护野生植物名录(第一批). 植物杂志, (5), 4-11.] | |
| [34] |
Nie XJ, Lü SZ, Zhang YX, Du XH, Wang L, Biradar SS, Tan XF, Wan FH, Song WN (2012) Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS ONE, 7, e36869.
DOI URL |
| [35] | Qiao YG, He JX, Wang YF, Cao YP, Jia MJ, Zhang XR, Liang JP, Song Y (2019) Analysis of chloroplast genome and its characteristics of medicinal plant Sophora flavescens. Acta Pharmaceutica Sinica, 54, 2106-2112. (in Chinese with English abstract) |
| [ 乔永刚, 贺嘉欣, 王勇飞, 曹亚萍, 贾孟君, 张鑫瑞, 梁建萍, 宋芸 (2019) 药用植物苦参的叶绿体基因组及其特征分析. 药学学报, 54, 2106-2112.] | |
| [36] |
Somaratne Y, Guan DL, Wang WQ, Zhao L, Xu SQ (2019) Complete chloroplast genome sequence of Xanthium sibiricum provides useful DNA barcodes for future species identification and phylogeny. Plant Systematics and Evolution, 305, 949-960.
DOI |
| [37] |
Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312-1313.
DOI URL |
| [38] |
Tan W, Gao H, Zhang HY, Yu XL, Tian XX, Jiang WL, Zhou K (2020) The complete chloroplast genome of Chinese medicine (Psoralea corylifolia): Molecular structures, barcoding and phylogenetic analysis. Plant Gene, 21, 100216.
DOI URL |
| [39] |
Wang JH, Cai YC, Zhao KK, Zhu ZX, Zhou RC, Wang HF (2018) Characterization of the complete chloroplast genome sequence of Firmiana pulcherrima (Malvaceae). Conservation Genetics Resources, 10, 445-448.
DOI URL |
| [40] | Wang S, Xie Y (2004) China Species Red List (Vol. 1). Higher Education Press, Beijing. (in Chinese) |
| [ 汪松, 解焱 (2004) 中国物种红色名录(第一卷). 高等教育出版社, 北京.] | |
| [41] |
Wilkie P, Clark A, Pennington RT, Cheek M, Bayer C, Wilcock CC (2006) Phylogenetic relationships within the subfamily Sterculioideae (Malvaceae/Sterculiaceae-Sterculieae) using the chloroplast gene ndhF. Systematic Botany, 31, 160-170.
DOI URL |
| [42] | Woo KW, Choi SU, Kim KH, Lee KR (2015) Ursane saponins from the stems of Firmiana simplex and their cytotoxic activity. Journal of the Brazilian Chemical Society, 26, 1450-1456. |
| [43] |
Wu ST, Zhu ZW, Fu LM, Niu BF, Li WZ (2011) WebMGA: A customizable web server for fast metagenomic sequence analysis. BMC Genomics, 12, 444.
DOI URL |
| [44] |
Wu XT, Chen L, Wang MQ, Zhang Y, Lin XY, Li XY, Zhou H, Wen YF (2018) Population structure and genetic divergence in Firmiana danxiaensis. Biodiversity Science, 26, 1168-1179. (in Chinese with English abstract)
DOI URL |
| [ 武星彤, 陈璐, 王敏求, 张原, 林雪莹, 李鑫玉, 周宏, 文亚峰 (2018) 丹霞梧桐群体遗传结构及其遗传分化. 生物多样性, 26, 1168-1179.] | |
| [45] |
Ya JD, Yu ZX, Yang YQ, Zhang SD, Zhang ZR, Cai J, Yang JB, Yu WB (2018) Complete chloroplast genome of Firmiana major (Malvaceae), a critically endangered species endemic to southwest China. Conservation Genetics Resources, 10, 713-715.
DOI URL |
| [46] | Yu T, Zhang YY, Gao J, Ke L, Ma WB, Li JQ (2019) Complete chloroplast genome sequence of Betula halophila, a plant species with extremely small populations. Scientia Silvae Sinicae, 55(2), 41-49. (in Chinese with English abstract) |
| [ 于涛, 张宇阳, 高健, 柯蕾, 马文宝, 李俊清 (2019) 极小种群濒危植物盐桦叶绿体基因组特征分析. 林业科学, 55(2), 41-49.] | |
| [47] | Zhang MY, Wang XF, Gao J, Liu AP, Yan YG, Yang XJ, Zhang G (2020) Complete chloroplast genome of Paeonia mairei H. Lév.: Characterization and phylogeny. Acta Pharmaceutica Sinica, 55, 168-176. (in Chinese with English abstract) |
| [ 张明英, 王西芳, 高静, 刘阿萍, 颜永刚, 杨新杰, 张岗 (2020) 美丽芍药叶绿体全基因组解析及系统发育分析. 药学学报, 55, 168-176.] | |
| [48] |
Zhao XQ, Yan M, Ding Y, Huo Y, Yuan ZH (2019) Characterization and comparative analysis of the complete chloroplast genome sequence from Prunus avium ‘Summit’. PeerJ, 7, e8210.
DOI URL |
| [1] | 罗正明, 刘晋仙, 张变华, 周妍英, 郝爱华, 杨凯, 柴宝峰. 不同退化阶段亚高山草甸土壤原生生物群落多样性特征及驱动因素[J]. 生物多样性, 2023, 31(8): 23136-. |
| [2] | 毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌. 毛竹扩张对杉木林土壤细菌群落的影响[J]. 生物多样性, 2023, 31(6): 22659-. |
| [3] | 赵雯, 王丹丹, 热依拉·木民, 黄开钏, 刘顺, 崔宝凯. 阿尔山地区兴安落叶松林土壤微生物群落结构[J]. 生物多样性, 2023, 31(2): 22258-. |
| [4] | 夏凡, 杨婧, 李建, 史洋, 盖立新, 黄文华, 张经纬, 杨南, 高福利, 韩莹莹, 鲍伟东. 北京地区四个豹猫亚种群肠道菌群的组成[J]. 生物多样性, 2022, 30(9): 22103-. |
| [5] | 孙翌昕, 李英滨, 李玉辉, 李冰, 杜晓芳, 李琪. 高通量测序技术在线虫多样性研究中的应用[J]. 生物多样性, 2022, 30(12): 22266-. |
| [6] | 高程, 郭良栋. 微生物物种多样性、群落构建与功能性状研究进展[J]. 生物多样性, 2022, 30(10): 22429-. |
| [7] | 夏呈强, 李毅, 党延茹, 察倩倩, 贺晓艳, 秦启龙. 中印度洋与南海西部表层海水细菌多样性[J]. 生物多样性, 2022, 30(1): 21407-. |
| [8] | 王楠, 黄菁华, 霍娜, 杨盼盼, 张欣玥, 赵世伟. 宁南山区不同植被恢复方式下土壤线虫群落特征:形态学鉴定与高通量测序法比较[J]. 生物多样性, 2021, 29(11): 1513-1529. |
| [9] | 靳新影, 张肖冲, 金多, 陈韵, 李靖宇. 腾格里沙漠东南缘不同生物土壤结皮细菌多样性及其季节动态特征[J]. 生物多样性, 2020, 28(6): 718-726. |
| [10] | 韩本凤, 周欣, 张雪. 基因组学技术在病毒鉴定与宿主溯源中的应用[J]. 生物多样性, 2020, 28(5): 587-595. |
| [11] | 张全建, 杨彪, 付强, 王磊, 龚旭, 张远彬. 邛崃山系水鹿的冬季食性[J]. 生物多样性, 2020, 28(10): 1192-1201. |
| [12] | 陆琪,胡强,施小刚,金森龙,李晟,姚蒙. 基于分子宏条形码分析四川卧龙国家级自然保护区雪豹的食性[J]. 生物多样性, 2019, 27(9): 960-969. |
| [13] | 刘君, 王宁, 崔岱宗, 卢磊, 赵敏. 小兴安岭大亮子河国家森林公园不同生境下土壤细菌多样性和群落结构[J]. 生物多样性, 2019, 27(8): 911-918. |
| [14] | 刘山林. DNA条形码参考数据集构建和序列分析相关的新兴技术[J]. 生物多样性, 2019, 27(5): 526-533. |
| [15] | 张雪, 李兴安, 苏秦之, 曹棋钠, 李晨伊, 牛庆生, 郑浩. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn