



生物多样性 ›› 2018, Vol. 26 ›› Issue (7): 738-748. DOI: 10.17520/biods.2018017 cstr: 32101.14.biods.2018017
收稿日期:2018-01-16
接受日期:2018-03-26
出版日期:2018-07-20
发布日期:2018-09-11
通讯作者:
田国忠
作者简介:# 共同第一作者
基金资助:
Shaoshuai Yu, Caili Lin, Shengjie Wang, Wenxin Zhang, Guozhong Tian*( )
)
Received:2018-01-16
Accepted:2018-03-26
Online:2018-07-20
Published:2018-09-11
Contact:
Tian Guozhong
About author:# Co-first authors
摘要:
植原体寄主种类多, 危害范围广, 开展其遗传多样性、关键基因调控等方面研究有助于提高该病害综合防治水平。通过长片段PCR引物扩增我国PaWB-sdyz、PaWB-fjfz和LY-fjya1植原体株系tuf基因及其上游6个基因的片段, 进行植原体基因启动子保守区域序列特征和多位点序列分析。利用启动子探针载体pSUPV4检测植原体tuf基因上游序列的启动子活性。扩增获得PaWB-sdyz、PaWB-fjfz、LY-fjya1株系tuf基因上游12,745-12,748 bp序列, 比较分析发现PaWB-sdyz、PaWB-fjfz、LY-fjya1、OY-M、AYWB、PAa、SLY、AT植原体株系tuf与其上游6个基因的结构顺序皆为5’-rplL-rpoB-rpoC-rps12-rps7-fusA-tuf-3’。推测出可能的植原体启动子保守区域模式序列: T90T100G92T75G67A85 (-35区); T90A96T92A98T73T90 (-10区)。基于8个植原体株系的rplL-tuf核苷酸序列编码基因、非编码序列、氨基酸序列的多位点序列分析可将不同植原体株系以较高的支持率清晰地区分, 不同植原体株系rplL-tuf核苷酸非编码区变异水平更高。16SrI组植原体tuf基因上游序列存在3种变异类型, 其代表株系PaWB-fjfz、LY-fjya1 tuf基因上游130 bp片段和CWB-hnsy1 tuf基因上游129 bp片段皆具有启动子活性。
于少帅, 林彩丽, 王圣洁, 张文鑫, 田国忠 (2018) 植原体tuf基因与其上游部分基因结构和相关基因启动子保守区域特征及活性分析. 生物多样性, 26, 738-748. DOI: 10.17520/biods.2018017.
Shaoshuai Yu, Caili Lin, Shengjie Wang, Wenxin Zhang, Guozhong Tian (2018) Structures of the tuf gene and its upstream part genes and characteristic analysis of conserved regions and activity from related gene promoters of a phytoplasma. Biodiversity Science, 26, 738-748. DOI: 10.17520/biods.2018017.
| 引物 Primer | 序列 Sequence (5’-3’) | 长度 Length (nt) | Tm (℃) | G+C (%) | 扩增长度 Length amplified (bp) |
|---|---|---|---|---|---|
| op1 | 5'-GATTGACATGGCTAAGTTAACG-3' | 22 | 54.9 | 40.9 | 3,676 |
| op2 | 5'-TACACCTTTGTTTCCGTGGC-3' | 20 | 58.4 | 50.0 | |
| op3 | 5'-AGGTAGCGGTCAAGAAGAAAT-3' | 21 | 55.4 | 42.9 | 6,965 |
| op4 | 5'-GAACCGCAAAGAACTGGG-3' | 18 | 56.0 | 55.6 | |
| op5 | 5'-AATATTATTGACACTCCCGGAC-3' | 22 | 55.5 | 40.9 | 2,661 |
| op6 | 5'-ACTCTACCAGTAACAACAGTTCCTC-3' | 25 | 56.1 | 44.0 | |
| op7 | 5'-CACATTTTATTAGCGCGCC-3' | 19 | 57.2 | 47.4 | 1,519 |
| op8 | 5'-AAAACCTAACGCAATCATGG-3' | 20 | 55.5 | 40.0 |
表1 长片段DNA扩增引物信息
Table 1 Information of the primers used to amplify the large DNA fragments
| 引物 Primer | 序列 Sequence (5’-3’) | 长度 Length (nt) | Tm (℃) | G+C (%) | 扩增长度 Length amplified (bp) |
|---|---|---|---|---|---|
| op1 | 5'-GATTGACATGGCTAAGTTAACG-3' | 22 | 54.9 | 40.9 | 3,676 |
| op2 | 5'-TACACCTTTGTTTCCGTGGC-3' | 20 | 58.4 | 50.0 | |
| op3 | 5'-AGGTAGCGGTCAAGAAGAAAT-3' | 21 | 55.4 | 42.9 | 6,965 |
| op4 | 5'-GAACCGCAAAGAACTGGG-3' | 18 | 56.0 | 55.6 | |
| op5 | 5'-AATATTATTGACACTCCCGGAC-3' | 22 | 55.5 | 40.9 | 2,661 |
| op6 | 5'-ACTCTACCAGTAACAACAGTTCCTC-3' | 25 | 56.1 | 44.0 | |
| op7 | 5'-CACATTTTATTAGCGCGCC-3' | 19 | 57.2 | 47.4 | 1,519 |
| op8 | 5'-AAAACCTAACGCAATCATGG-3' | 20 | 55.5 | 40.0 |
| 引物 Primer | 引物序列 Primer sequences (5’-3’) | 引物大小 Primer size (bp) | Tm (℃) | 引入酶切位点 Restriction site added |
|---|---|---|---|---|
| TPf | cccaagcttACAACCTTACACTAAAAAAC | 29 | 45 | Hind III |
| TP4f | cccaagcttACAACCTTAAACTAAAAAAC | 29 | 45 | Hind III |
| TPr | cgcggatcCATTTTTCAAAGGCCTC | 25 | 45 | BamH I |
表2 tuf基因启动子扩增引物
Table 2 Primers used to amplify tuf gene promoters
| 引物 Primer | 引物序列 Primer sequences (5’-3’) | 引物大小 Primer size (bp) | Tm (℃) | 引入酶切位点 Restriction site added |
|---|---|---|---|---|
| TPf | cccaagcttACAACCTTACACTAAAAAAC | 29 | 45 | Hind III |
| TP4f | cccaagcttACAACCTTAAACTAAAAAAC | 29 | 45 | Hind III |
| TPr | cgcggatcCATTTTTCAAAGGCCTC | 25 | 45 | BamH I |
| 株系 Strain | 级别 Group | 扩增序列长度 Sequence amplified length (bp) | 编码区长度 Coding region length (bp) | 非编码区长度 Non-coding region length (bp) | 氨基酸长度/个 Amino acid sequence length |
|---|---|---|---|---|---|
| PaWB-sdyz | 16SrI-D | 12,746 | 12,307 | 439 | 4,096 |
| PaWB-fjfz | 16SrI-D | 12,745 | 12,307 | 438 | 4,096 |
| LY-fjya1 | 16SrI-B | 12,748 | 12,307 | 441 | 4,096 |
| OY-M | 16SrI-B | 12,745 | 12,307 | 438 | 4,096 |
| AYWB | 16SrI-A | 12,735 | 12,343 | 392 | 4,107 |
| PAa | 16SrXII | 12,611 | 12,211 | 400 | 4,063 |
| SLY | 16SrXII | 12,611 | 12,211 | 400 | 4,063 |
| AT | 16SrX | 12,835 | 12,301 | 534 | 4,093 |
| PG-8A | 4,587 | 4,158 | 429 | 1,382 |
表3 植原体扩增、编码区、非编码区序列和编码氨基酸序列及参照株系对应片段长度
Table 3 Length of sequence amplified, coding region, non-coding region and amino acid sequence of phytoplasmas as well as reference strains
| 株系 Strain | 级别 Group | 扩增序列长度 Sequence amplified length (bp) | 编码区长度 Coding region length (bp) | 非编码区长度 Non-coding region length (bp) | 氨基酸长度/个 Amino acid sequence length |
|---|---|---|---|---|---|
| PaWB-sdyz | 16SrI-D | 12,746 | 12,307 | 439 | 4,096 |
| PaWB-fjfz | 16SrI-D | 12,745 | 12,307 | 438 | 4,096 |
| LY-fjya1 | 16SrI-B | 12,748 | 12,307 | 441 | 4,096 |
| OY-M | 16SrI-B | 12,745 | 12,307 | 438 | 4,096 |
| AYWB | 16SrI-A | 12,735 | 12,343 | 392 | 4,107 |
| PAa | 16SrXII | 12,611 | 12,211 | 400 | 4,063 |
| SLY | 16SrXII | 12,611 | 12,211 | 400 | 4,063 |
| AT | 16SrX | 12,835 | 12,301 | 534 | 4,093 |
| PG-8A | 4,587 | 4,158 | 429 | 1,382 |
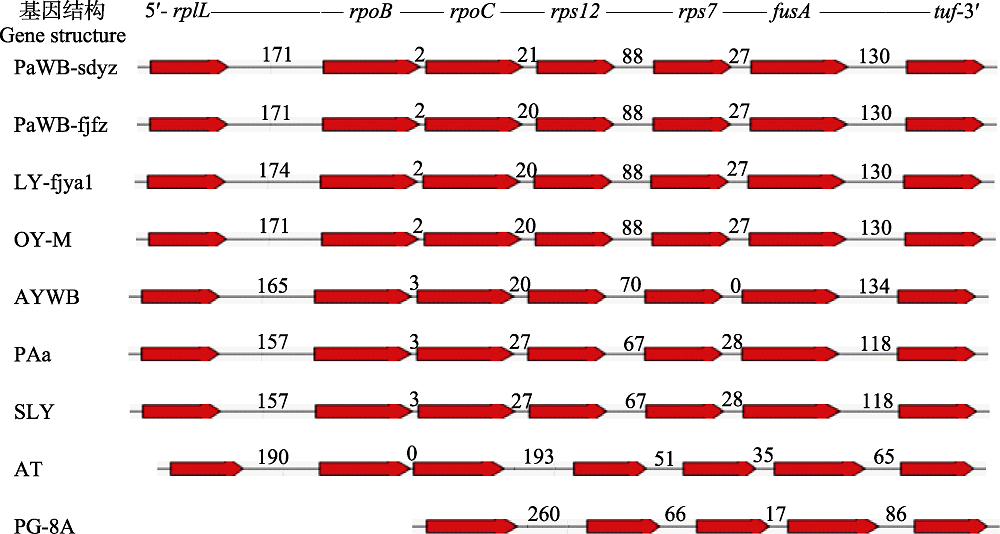
图1 不同植原体tuf基因及其上游基因结构示意图。图中样品代号见正文“1.1”和“1.2.3”; 数字代表基因间区序列长度(bp)。
Fig. 1 Gene structure diagram of tuf gene and its upstream genes from different phytoplasmas. The codes of samples in the figure were shown in “1.1” and “1.2.3”. Numbers represented the length of intergenic sequences (bp).
| 株系 Strains | rplL-rpoB | rpoC-rps12 | rps12-rps7 | fusA-tuf | ||||
|---|---|---|---|---|---|---|---|---|
| -35 | -10 | -35 | -10 | -35 | -10 | -35 | -10 | |
| PaWB-sdyz | TTGCAT | TATACC | - | - | ATAAAA | AAAAAT | TTGTGA | TATATT |
| PaWB-fjfz | TTGCAT | TATACC | - | - | ATAAAA | AAAAAT | TTGTGA | TATATT |
| LY-fjya1 | TTGCAT | TATACC | - | - | ATAAAA | AAAAAT | TTGTAA | TATATT |
| OY-M | TTGAAT | TATAAC | - | - | ATAAAA | AAAAAT | TTGTGA | TATATT |
| AYWB | TTGCAT | TATACC | - | - | - | - | TTGTGA | TATTAT |
| PAa | TTGTAT | TTTAAT | - | - | - | - | TTGATA | TATATT |
| SLY | TTGTAT | TTTAAT | - | - | - | - | TTGATA | TATATT |
| AT | ATGATA | AATAAT | TTGACT | TATAAT | - | - | - | - |
| PG-8A | - | - | TTGACA | TATAAT | - | - | ATGATA | TATTGT |
表4 植原体rplL-tuf基因间区序列相关基因启动子保守区域特征
Table 4 Characteristics of conserved region of relevant gene promoter of phytoplasma rplL-tuf intergenic sequences
| 株系 Strains | rplL-rpoB | rpoC-rps12 | rps12-rps7 | fusA-tuf | ||||
|---|---|---|---|---|---|---|---|---|
| -35 | -10 | -35 | -10 | -35 | -10 | -35 | -10 | |
| PaWB-sdyz | TTGCAT | TATACC | - | - | ATAAAA | AAAAAT | TTGTGA | TATATT |
| PaWB-fjfz | TTGCAT | TATACC | - | - | ATAAAA | AAAAAT | TTGTGA | TATATT |
| LY-fjya1 | TTGCAT | TATACC | - | - | ATAAAA | AAAAAT | TTGTAA | TATATT |
| OY-M | TTGAAT | TATAAC | - | - | ATAAAA | AAAAAT | TTGTGA | TATATT |
| AYWB | TTGCAT | TATACC | - | - | - | - | TTGTGA | TATTAT |
| PAa | TTGTAT | TTTAAT | - | - | - | - | TTGATA | TATATT |
| SLY | TTGTAT | TTTAAT | - | - | - | - | TTGATA | TATATT |
| AT | ATGATA | AATAAT | TTGACT | TATAAT | - | - | - | - |
| PG-8A | - | - | TTGACA | TATAAT | - | - | ATGATA | TATTGT |
| 保守区域 Conserved region | -35区 -35 region | -10区 -10 region | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 序列特征 Sequence characteristic | A(10) | A(0) | A(8) | A(17) | A(25) | A(85) | A(10) | A(96) | A(8) | A(98) | A(19) | A(0) |
| T(90) | T(100) | T(0) | T(75) | T(6) | T(15) | T(90) | T(4) | T(92) | T(2) | T(73) | T(90) | |
| G(0) | G(0) | G(92) | G(0) | G(67) | G(0) | G(0) | G(0) | G(0) | G(0) | G(0) | G(0) | |
| C(0) | C(0) | C(0) | C(8) | C(2) | C(0) | C(0) | C(0) | C(0) | C(0) | C(8) | C(10) | |
表5 部分植原体基因启动子保守区域不同位置核苷酸种类及出现频率
Table 5 Nucleotide and its frequency in different sites of conserved regions of some phytoplasma gene promoters
| 保守区域 Conserved region | -35区 -35 region | -10区 -10 region | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 序列特征 Sequence characteristic | A(10) | A(0) | A(8) | A(17) | A(25) | A(85) | A(10) | A(96) | A(8) | A(98) | A(19) | A(0) |
| T(90) | T(100) | T(0) | T(75) | T(6) | T(15) | T(90) | T(4) | T(92) | T(2) | T(73) | T(90) | |
| G(0) | G(0) | G(92) | G(0) | G(67) | G(0) | G(0) | G(0) | G(0) | G(0) | G(0) | G(0) | |
| C(0) | C(0) | C(0) | C(8) | C(2) | C(0) | C(0) | C(0) | C(0) | C(0) | C(8) | C(10) | |
| 株系 Strain | PaWB-sdyz | PaWB-fjfz | LY-fjya1 | OY-M | AYWB | PAa | SLY | AT | PG-8A |
|---|---|---|---|---|---|---|---|---|---|
| PaWB-sdyz | 100 | ||||||||
| PaWB-fjfz | 99.7 | 100 | |||||||
| LY-fjya1 | 99.3 | 99.3 | 100 | ||||||
| OY-M | 99.6 | 99.6 | 99.4 | 100 | |||||
| AYWB | 95.9 | 95.9 | 95.9 | 96.0 | 100 | ||||
| PAa | 81.1 | 81.1 | 81.2 | 81.1 | 81.1 | 100 | |||
| SLY | 81.1 | 81.1 | 81.2 | 81.1 | 81.1 | 99.9 | 100 | ||
| AT | 72.7 | 72.8 | 72.9 | 72.8 | 73.3 | 73.6 | 73.6 | 100 | |
| PG-8A | 71.2 | 71.3 | 71.4 | 71.3 | 71.4 | 71.3 | 71.3 | 70.9 | 100 |
表6 MLSA分析rplL-tuf核苷酸序列编码区同源比对
Table 6 Homology matrix of rplL-tuf nucleotide sequence coding region in MLSA analysis
| 株系 Strain | PaWB-sdyz | PaWB-fjfz | LY-fjya1 | OY-M | AYWB | PAa | SLY | AT | PG-8A |
|---|---|---|---|---|---|---|---|---|---|
| PaWB-sdyz | 100 | ||||||||
| PaWB-fjfz | 99.7 | 100 | |||||||
| LY-fjya1 | 99.3 | 99.3 | 100 | ||||||
| OY-M | 99.6 | 99.6 | 99.4 | 100 | |||||
| AYWB | 95.9 | 95.9 | 95.9 | 96.0 | 100 | ||||
| PAa | 81.1 | 81.1 | 81.2 | 81.1 | 81.1 | 100 | |||
| SLY | 81.1 | 81.1 | 81.2 | 81.1 | 81.1 | 99.9 | 100 | ||
| AT | 72.7 | 72.8 | 72.9 | 72.8 | 73.3 | 73.6 | 73.6 | 100 | |
| PG-8A | 71.2 | 71.3 | 71.4 | 71.3 | 71.4 | 71.3 | 71.3 | 70.9 | 100 |
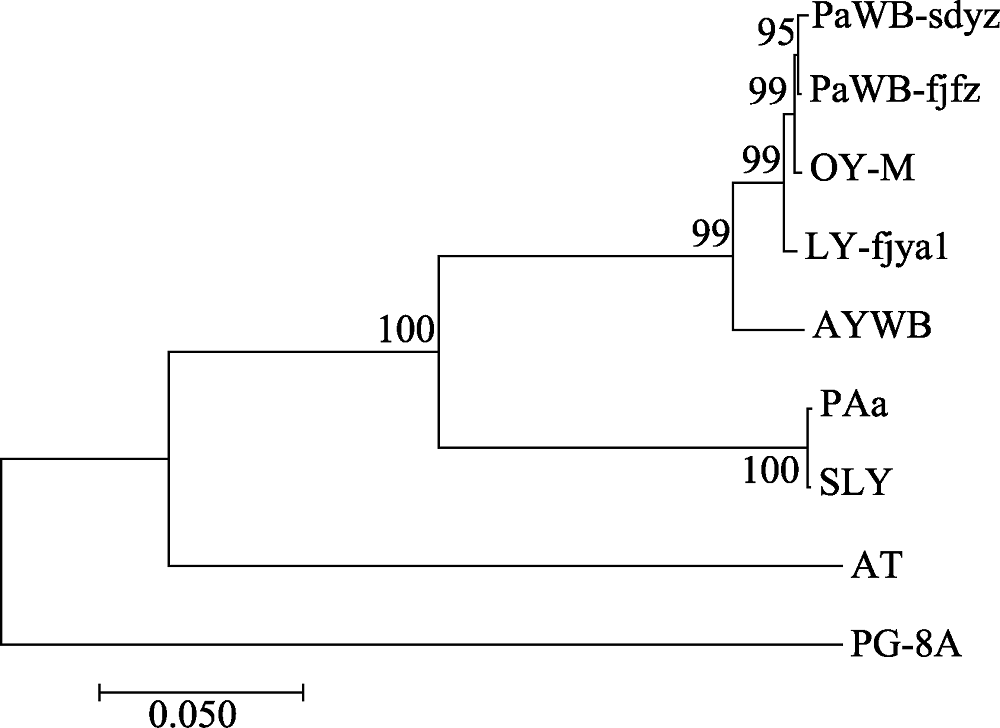
图2 基于rplL-tuf核苷酸序列编码区整合序列构建的植原体系统发育树。图中代号见“1.1”和“1.2.3”。
Fig. 2 Phylogenetic tree of the phytoplasma strains reconstructed based on the concatenated gene sequences data set of rplL-tuf nucleotide sequence coding region. The codes in the figure were shown in “1.1” and “1.2.3”.
| 株系 Strain | PaWB-sdyz | PaWB-fjfz | LY-fjya1 | OY-M | AYWB | PAa | SLY | AT | PG-8A |
|---|---|---|---|---|---|---|---|---|---|
| PaWB-sdyz | 100 | ||||||||
| PaWB-fjfz | 99.3 | 100 | |||||||
| LY-fjya1 | 99.3 | 98.6 | 100 | ||||||
| OY-M | 99.3 | 98.6 | 98.6 | 100 | |||||
| AYWB | 94.6 | 94.3 | 94.1 | 94.1 | 100 | ||||
| PAa | 71.9 | 71.8 | 71.3 | 72.3 | 73.2 | 100 | |||
| SLY | 71.9 | 71.8 | 71.3 | 72.3 | 73.2 | 100 | 100 | ||
| AT | 50.7 | 51.3 | 51.4 | 51.3 | 51.1 | 51.4 | 51.4 | 100 | |
| PG-8A | 41.8 | 41.3 | 41.5 | 41.8 | 41.3 | 40.2 | 40.2 | 44.8 | 100 |
表7 MLSA分析rplL-tuf核苷酸序列非编码区同源比对
Table 7 Homology matrix of rplL-tuf nucleotide sequence non-coding region in MLSA analysis
| 株系 Strain | PaWB-sdyz | PaWB-fjfz | LY-fjya1 | OY-M | AYWB | PAa | SLY | AT | PG-8A |
|---|---|---|---|---|---|---|---|---|---|
| PaWB-sdyz | 100 | ||||||||
| PaWB-fjfz | 99.3 | 100 | |||||||
| LY-fjya1 | 99.3 | 98.6 | 100 | ||||||
| OY-M | 99.3 | 98.6 | 98.6 | 100 | |||||
| AYWB | 94.6 | 94.3 | 94.1 | 94.1 | 100 | ||||
| PAa | 71.9 | 71.8 | 71.3 | 72.3 | 73.2 | 100 | |||
| SLY | 71.9 | 71.8 | 71.3 | 72.3 | 73.2 | 100 | 100 | ||
| AT | 50.7 | 51.3 | 51.4 | 51.3 | 51.1 | 51.4 | 51.4 | 100 | |
| PG-8A | 41.8 | 41.3 | 41.5 | 41.8 | 41.3 | 40.2 | 40.2 | 44.8 | 100 |
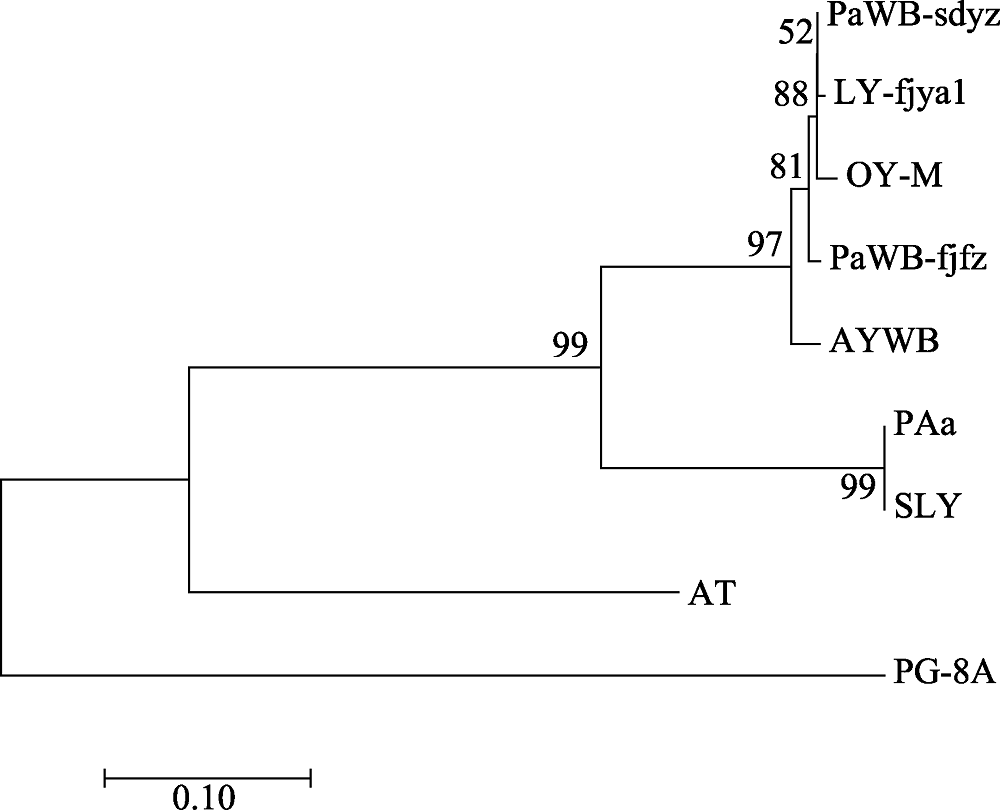
图3 基于rplL-tuf核苷酸序列非编码区整合序列构建的植原体系统发育树。图中株系代号见正文“1.1”和“1.2.3”。
Fig. 3 Phylogenetic tree of the phytoplasma strains reconstructed based on the concatenated gene sequences data set of rplL-tuf nucleotide sequence non-coding region. The codes in the figure are shown in “1.1” and “1.2.3”.
| 株系 Strain | PaWB-sdyz | PaWB-fjfz | LY-fjya1 | OY-M | AYWB | PAa | SLY | AT | PG-8A |
|---|---|---|---|---|---|---|---|---|---|
| PaWB-sdyz | 100 | ||||||||
| PaWB-fjfz | 99.6 | 100 | |||||||
| LY-fjya1 | 98.9 | 99.0 | 100 | ||||||
| OY-M | 99.4 | 99.5 | 99.2 | 100 | |||||
| AYWB | 97.8 | 97.9 | 98.0 | 98.0 | 100 | ||||
| PAa | 83.8 | 83.9 | 84.1 | 83.9 | 83.7 | 100 | |||
| SLY | 83.8 | 83.9 | 84.1 | 83.9 | 83.7 | 99.8 | 100 | ||
| AT | 69.8 | 69.9 | 70.0 | 69.9 | 69.8 | 69.0 | 69.0 | 100 | |
| PG-8A | 74.9 | 75.1 | 74.9 | 75.0 | 74.8 | 74.1 | 73.9 | 70.4 | 100 |
表8 MLSA分析RplL-TUF蛋白氨基酸序列同源比对
Table 8 Homology matrix of RplL-TUF amino acid sequence in MLSA analysis
| 株系 Strain | PaWB-sdyz | PaWB-fjfz | LY-fjya1 | OY-M | AYWB | PAa | SLY | AT | PG-8A |
|---|---|---|---|---|---|---|---|---|---|
| PaWB-sdyz | 100 | ||||||||
| PaWB-fjfz | 99.6 | 100 | |||||||
| LY-fjya1 | 98.9 | 99.0 | 100 | ||||||
| OY-M | 99.4 | 99.5 | 99.2 | 100 | |||||
| AYWB | 97.8 | 97.9 | 98.0 | 98.0 | 100 | ||||
| PAa | 83.8 | 83.9 | 84.1 | 83.9 | 83.7 | 100 | |||
| SLY | 83.8 | 83.9 | 84.1 | 83.9 | 83.7 | 99.8 | 100 | ||
| AT | 69.8 | 69.9 | 70.0 | 69.9 | 69.8 | 69.0 | 69.0 | 100 | |
| PG-8A | 74.9 | 75.1 | 74.9 | 75.0 | 74.8 | 74.1 | 73.9 | 70.4 | 100 |
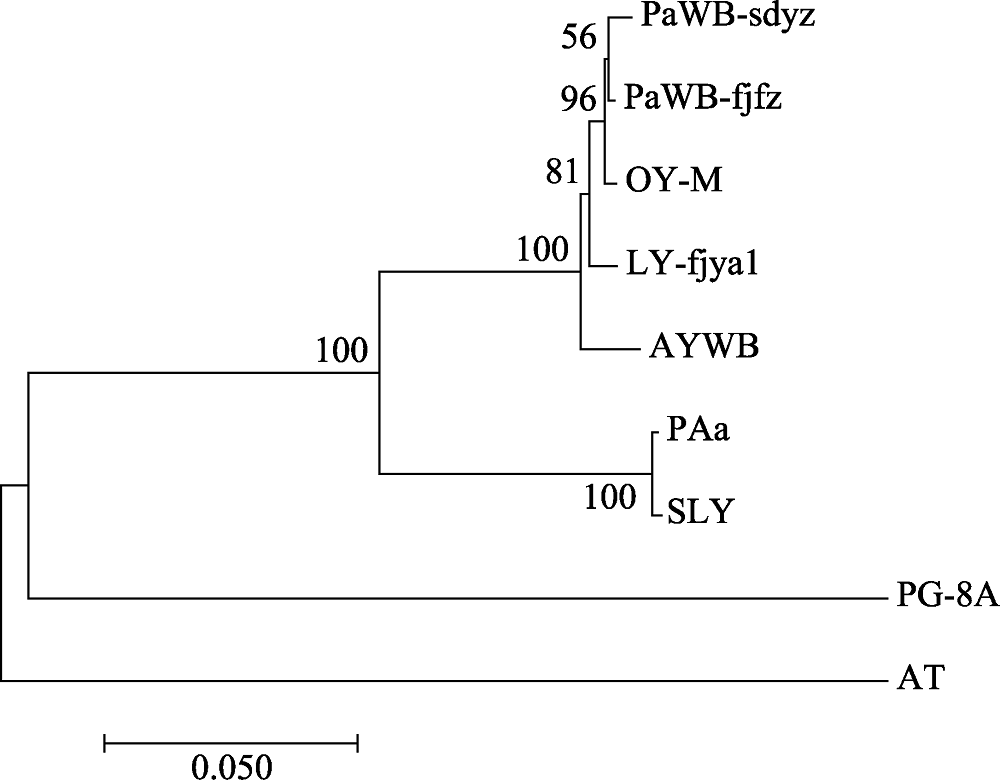
图4 基于RplL-TUF氨基酸整合序列构建的植原体系统发育树。图中代号见正文“1.1”和“1.2.3”。
Fig. 4 Phylogenetic tree of the phytoplasma strains reconstructed based on the concatenated sequences data set of RplL-TUF amino acid sequence. The codes in the figure are shown in “1.1” and “1.2.3”.
| [1] | Andersen MT, Liefting LW, Havukkala I, Beever RE (2013) Comparison of the complete genome sequence of two closely related isolates of ‘Candidatus phytoplasma australiense’ reveals genome plasticity. BMC Genomics, 14, 529. |
| [2] | Bai XD, Zhang JH, Ewing A, Miller SA, Radek AJ, Shevchenko DV, Tsukerman K, Walunas T, Lapidus A, Campbell JW, Hogenhout SA (2006) Living with genome instability: The adaptation of phytoplasma to diverse environments of their insect and plant hosts. Journal of Bacteriology, 188, 3682-3696. |
| [3] | Du HT, Zhu HY, Wang JM, Zhao W, Tao XL, Ba CF, Tian YM, Su YH (2014) Single-nucleotide polymorphisms and activity analysis of the promoter and enhancer of the pig lactase gene. Gene, 545, 56-60. |
| [4] | Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39, 783-791. |
| [5] | Ishii Y, Kakizawa S, Hoshi A, Maejima K, Kagiwada S, Yamaji Y, Oshima K, Namba S (2009) In the non-insect- transmissible line of onion yellows phytoplasma (OY-NIM), the plasmid-encoded transmembrane protein ORF3 lacks the major promoter region. Microbiology, 155, 2058-2067. |
| [6] | Kube M, Schneider B, Kuhl H, Dandekar T, Heitmann K, Migdoll AM, Reinhardt R, Seemüller E (2008) The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus phytoplasma mali’. BMC Genomics, 9, 306. |
| [7] | Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870-1874. |
| [8] | Lee IM, Hammond RW, Davis RE, Gundersen DE (1993) Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasma like organisms. Phytopathology, 83, 834-842. |
| [9] | Li MG (2004) Advanced Molecular Genetics. Science Press, Beijing. (in Chinese) |
| [李明刚 (2004) 高级分子遗传学. 科学出版社, 北京.] | |
| [10] | Liu Z, Zhang L, Xue C, Fang H, Zhao J, Liu M (2017) Genome-wide identification and analysis of MAPK and MAPKK gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genomics, 18, 855. |
| [11] | Miyata S, Furuki K, Oshima K, Sawayanagi T, Nishigawa H, Jung HY, Ugaki M, Namba S (2002a) Complete nucleotide sequence of the S10-spc operon of phytoplasma: Gene organization and genetic code resemble those of Bacillus subtilis. DNA and Cell Biology, 21, 527-534. |
| [12] | Miyata S, Furuki K, Sawayanagi T, Oshima K, Kuboyama T, Tsuchizaki T, Ugaki M, Namba S (2002b) Gene arrangement and sequence of str operon of phytoplasma resemble those of Bacillus more than those of Mycoplasma. Journal of General Plant Pathology, 68, 62-67. |
| [13] | Oshima K, Kakizawa S, Nishigawa H, Jung HY, Wei W, Suzuki S, Arashida R, Nakata D, Miyata S, Ugaki M, Namba S (2004) Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nature Genetics, 36, 27-29. |
| [14] | Post LE, Nomura M (1980) DNA sequences from the str operon of Escherichia coli. The Journal of Biological Chemistry, 255, 4660-4666. |
| [15] | Ren ZG (2014) The Qualitative and Quantitative Detection and Identification of Several Important Phytoplasmas in China. Postdoctoral Research Report, Chinese Academy of Forestry, Beijing. (in Chinese with English abstract) |
| [任争光 (2014) 我国几种重要植原体定性和定量检测鉴定研究.博士后研究报告, 中国林业科学研究院, 北京.] | |
| [16] | Salehi M, Hosseini SAE, Salehi E, Bertaccini A (2017) Genetic diversity and vector transmission of phytoplasmas associated with sesame phyllody in Iran. Folia Microbiologica, 62, 99-109. |
| [17] | Sanangelantoni AM, Tiboni O (1993) The chromosomal location of genes for elongation factor Tu and ribosomal protein S10 in the cyanobacterium Spirulina platensis provides clues to the ancestral organization of the str and S10 operons in prokaryotes. Journal of General Microbiology, 139, 2579-2584. |
| [18] | Tran-Nguyen LTT, Kube M, Schneider B, Reinhardt R, Gibb KS (2008) Comparative genome analysis of ‘Candidatus phytoplasma australiense’ (subgroup tuf-Australia I; rp-A) and ‘Ca. phytoplasma asteris’ strains OY-M and AY-WB. Journal of Bacteriology, 190, 3979-3991. |
| [19] | Turner PC (translated by Liu JY, Liu WY) (2010) Molecular Biology, 3rd edn. Science Press, Beijing. (in Chinese) |
| [特纳(著), 刘进元, 刘文颖 (译) (2010) 分子生物学, 第3版. 科学出版社, 北京.] | |
| [20] | Wang SJ (2017) Establishment of Molecular Detection Technology and Genetic Diversity Analysis of Plant Phytoplasma in China. PhD dissertation, Chinese Academy of Forestry, Beijing. (in Chinese with English abstract) |
| [王圣洁 (2017) 重要林木植原体分子检测技术的研发和遗传多样性研究. 博士学位论文, 中国林业科学研究院, 北京.] | |
| [21] | Yang YH (2008) Molecular Biology of the Gene. Higher Education Press, Beijing. (in Chinese) |
| [杨业华 (2008) 基因的分子生物学. 高等教育出版社, 北京.] | |
| [22] | Yu SS, Xu QC, Lin CL, Wang SJ, Tian GZ (2016a) Genetic diversity of phytoplasmas: Research status and prospects. Biodiversity Science, 24, 205-215. (in Chinese with English abstract) |
| [于少帅, 徐启聪, 林彩丽, 王圣洁, 田国忠 (2016a) 植原体遗传多样性研究现状与展望. 生物多样性, 24, 205-215.] | |
| [23] | Yu SS, Lin CL, Pan J, Ren ZG, Piao CG, Wang LF, Guo MW, Tian GZ (2016b) Comparative analysis of structure, function and genetic variation of upstream sequences adjoining tuf gene in paulownia and jujube witches’-broom phytoplasmas. Microbiology China, 43, 1060-1069. (in Chinese with English abstract) |
| [于少帅, 林彩丽, 潘皎, 任争光, 朴春根, 汪来发, 郭民伟, 田国忠 (2016b) 泡桐丛枝和枣疯病植原体tuf基因上游序列结构、功能和遗传变异比较分析. 微生物学通报, 43, 1060-1069.] | |
| [24] | Zhang Y, Han Q, Li C, Li W, Fan H, Xing Q, Yan B (2014) Genetic analysis of the TBX1 gene promoter in indirect inguinal hernia. Gene, 535, 290-293. |
| [25] | Zhu YX, Li Y (2002)Modern Molecular Biology, 2nd edn. Higher Education Press, Beijing. (in Chinese) |
| [朱玉贤, 李毅 (2002)现代分子生物学, 第2版. 高等教育出版社, 北京.] | |
| [26] | Zurawski G, Zurawski SM (1985) Structure of the Escherichia coli S10 ribosomal protein operon. Nucleic Acids Research, 13, 4521-4526. |
| [1] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [2] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [3] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [4] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [5] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [6] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [7] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [8] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [9] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [10] | 崔静, 徐明芳, 章群, 李瑶, 曾晓舒, 李莎. 基于3种线粒体标记探讨中日沿海角木叶鲽遗传多样性差异[J]. 生物多样性, 2022, 30(5): 21485-. |
| [11] | 孙军, 宋煜尧, 施义锋, 翟键, 燕文卓. 近十年中国海洋生物多样性研究进展[J]. 生物多样性, 2022, 30(10): 22526-. |
| [12] | 栗冬梅, 杨卫红, 李庆多, 韩茜, 宋秀平, 潘虹, 冯云. 巴尔通体在滇西南蝙蝠中高度流行并具有丰富的遗传变异特征[J]. 生物多样性, 2021, 29(9): 1245-1255. |
| [13] | 姚志, 郭军, 金晨钟, 刘勇波. 中国纳入一级保护的极小种群野生植物濒危机制[J]. 生物多样性, 2021, 29(3): 394-408. |
| [14] | 叶俊伟, 田斌. 中国西南地区重要木本油料植物扁核木的遗传结构及成因[J]. 生物多样性, 2021, 29(12): 1629-1637. |
| [15] | 向登高, 李跃飞, 李新辉, 陈蔚涛, 马秀慧. 多基因联合揭示海南鲌的遗传结构与遗传多样性[J]. 生物多样性, 2021, 29(11): 1505-1512. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn