



生物多样性 ›› 2016, Vol. 24 ›› Issue (10): 1189-1196. DOI: 10.17520/biods.2016265 cstr: 32101.14.biods.2016265
收稿日期:2016-09-20
接受日期:2016-10-28
出版日期:2016-10-20
发布日期:2016-11-10
通讯作者:
朱耿平
基金资助:
Gengping Zhu1,*( ), Huijie Qiao2
), Huijie Qiao2
Received:2016-09-20
Accepted:2016-10-28
Online:2016-10-20
Published:2016-11-10
Contact:
Zhu Gengping
摘要:
生态位模型在入侵生物学和保护生物学中具有广泛的应用, 其中Maxent模型最为流行, 被越来越多地应用在预测物种的现实分布和潜在分布的研究中。在Maxent模型中, 多数研究者采用默认参数来构建模型, 这些默认参数源自早期对266个物种的测试, 以预测物种的现实分布为目的。近期研究发现, Maxent模型采用复杂机械学习算法, 对采样偏差敏感, 易产生过度拟合, 模型转移能力仅在低阈值情况下较好。基于默认参数的Maxent模型不仅预测结果不可靠, 而且有时很难解释。在本研究中, 作者以入侵害虫茶翅蝽(Halyomorpha halys)为例, 采用经典模型构建方案(即构建本土模型然后将其转移至入侵地来评估), 利用ENMeval数据包来调整本土Maxent模型调控倍频和特征组合参数, 分析各种参数条件下模型的复杂度, 然后选取最低复杂度的模型参数(即为最优模型), 综合比较默认参数和调整参数后Maxent模型的响应曲线和预测结果, 探讨Maxent模型复杂度对预测结果的影响及Maxent模型构建时所需注意事项, 以期对物种潜在分布进行合理的预测, 促进Maxent模型在我国的合理运用和发展。作者认为, 环境变量的选择至关重要, 需要综合分析其对所模拟物种分布的限制作用和环境变量之间的空间相关性。构建Maxent模型前需对物种分布采样偏差及模型的构建区域进行合理地判断, 模型构建时需要比较不同参数下模型的预测结果和响应曲线, 选取复杂度较低的模型参数来最终建模。在茶翅蝽的分析中, Maxent模型的默认参数和最优模型参数不同, 与Maxent模型默认参数相比, 采用调整参数后所构建的模型预测效果较好, 响应曲线较为平滑, 模型转移能力较高, 能够较为合理反映物种对环境因子的响应和准确地模拟该物种的潜在分布。
朱耿平, 乔慧捷 (2016) Maxent模型复杂度对物种潜在分布区预测的影响. 生物多样性, 24, 1189-1196. DOI: 10.17520/biods.2016265.
Gengping Zhu, Huijie Qiao (2016) Effect of the Maxent model’s complexity on the prediction of species potential distributions. Biodiversity Science, 24, 1189-1196. DOI: 10.17520/biods.2016265.
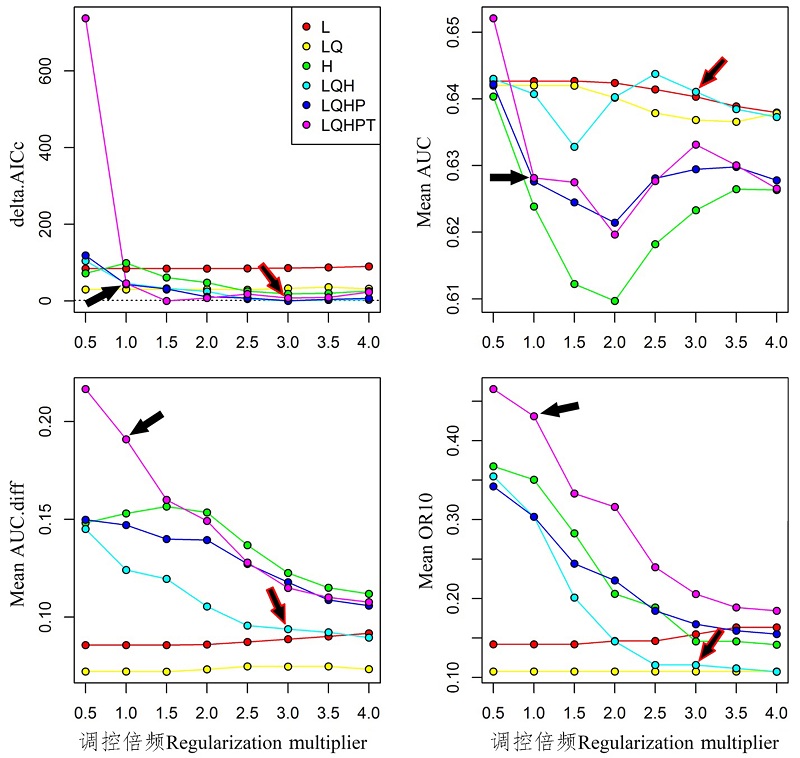
图1 不同参数下的茶翅蝽本土模型表现。黑色箭头表示Maxent默认参数, 红边箭头表示AIC值最小优化模型参数。
Fig. 1 Performances of native niche model of Halyomorpha halys under different settings. Black arrow indicates default setting, rededge arrow indicates the AICc-chosen setting.
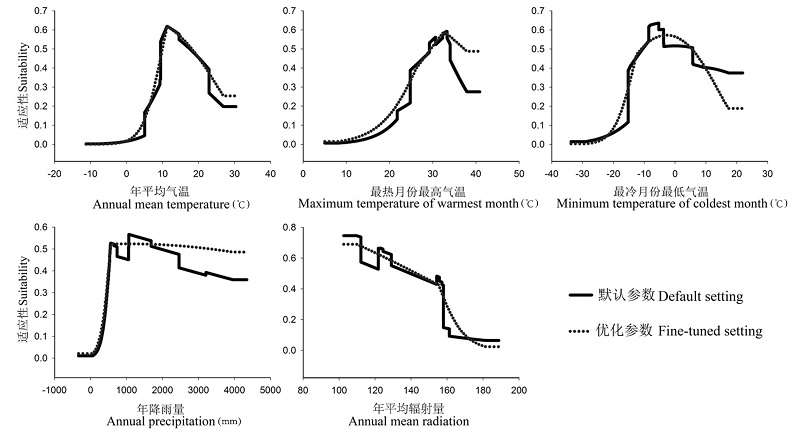
图2 基于默认参数和优化参数的本土Maxent模型中茶翅蝽对5个气候变量的响应曲线
Fig. 2 Comparison of response curves of Halyomorpha halys to five bioclimatic variables based on the default and fine-tuned Maxent settings
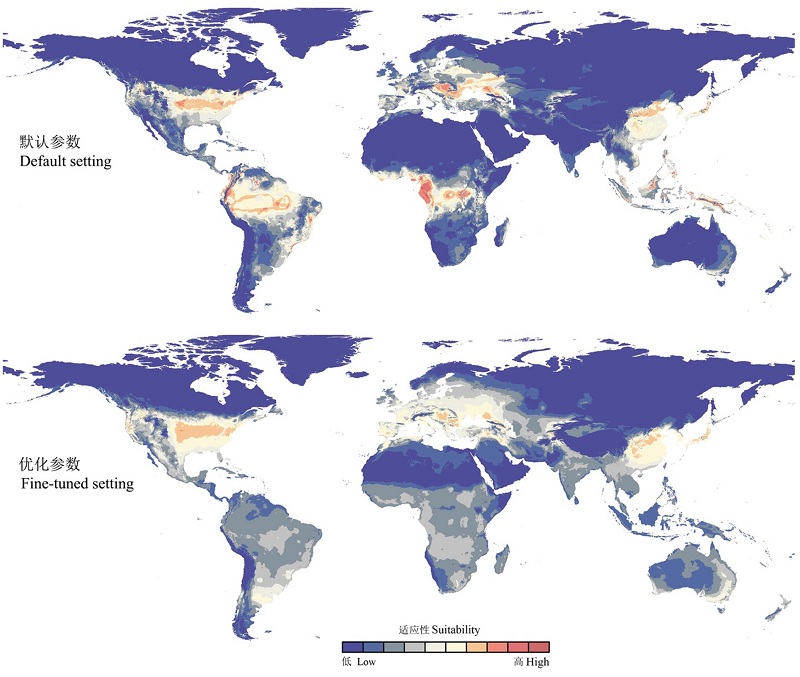
图3 基于默认参数和优化参数后的Maxent模型转移后对茶翅蝽的潜在分布预测
Fig. 3 Predictions of Halyomorpha halys based on its native Maxent models calibrated on the default and fine-tuned settings
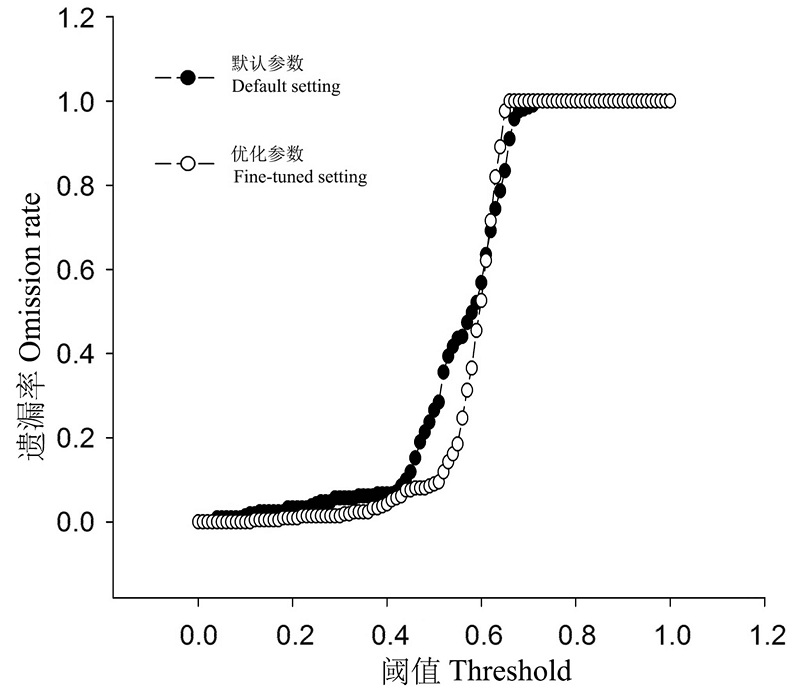
图4 基于默认参数和优化后参数的Maxent模型转移后对北美分布茶翅蝽预测的遗漏率
Fig. 4 Omission rates of North American records of Halyomorpha halys based on its native Maxent models predictions using the default and fine-tuned settings.
| 1 | Ahmed SE, McInerny G, O’Hara K, Harper R, Salido L, Emmott S, Joppa LN (2015) Scientists and software - surveying the species distribution modelling community. Diversity and Distributions, 21, 258-267. |
| 2 | Akaike H (1973) Information theory and an extension of the maximum likelihood principle. In: 2nd International Symposium on Information Theory (eds Petrov BN, Csáki F), pp. 267-281. Akadémiai Kiadó, Budapest. |
| 3 | Barbosa FG, Schneck F (2015) Characteristics of the top-cited papers in species distribution predictive models. Ecological Modelling, 313, 77-83. |
| 4 | Elith J, Phillips SJ, Hastie T, Dudík D, Chee YE, Yates CJ (2010) A statistical explanation of MaxEnt for ecologists. Diversity and Distributions, 17, 43-57. |
| 5 | Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965-1978. |
| 6 | Jiménez-Valverde A, Peterson AT, Soberón J, Overton JM, Aragón P, Lobo JM (2011) Use of niche models in invasive species risk assessments. Biological Invasions, 13, 2785-2797. |
| 7 | Kearney MR, Wintle BA, Porter WP (2010) Correlative and mechanistic models of species distribution provide congruent forecasts under climate change. Conservation Letters, 3, 203-213. |
| 8 | Kriticos DJ, Webber BL, Leriche A, Ota N, Macadam I, Bathols J, Scott JK (2011) CliMond: global high resolution historical and future scenario climate surfaces for bioclimatic modeling. Methods in Ecology and Evolution, 3, 53-64. |
| 9 | Muscarella R, Galante PJ, Soley-Guardia M, Boria RA, Kass JM, Uriarte M, Anderson RP (2014) ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for MAXENT ecological niche models. Methods in Ecology and Evolution, 5, 1198-1205. |
| 10 | Peterson AT, Papeş M, Soberón J (2008) Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecological Modelling, 213, 63-72. |
| 11 | Peterson AT, Soberón J (2012) Species distribution modeling and ecological niche modeling: getting the concepts right. Natureza & Conservacao, 10, 102-107. |
| 12 | Peterson AT, Soberón J, Pearson RG, Anderson RP, Nakamura M, Martínez-Meyer E, Araújo MB (2011) Ecological Niches and Geographical Distributions. Princeton University Press, New Jersey. |
| 13 | Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecological Modelling, 190, 231-259. |
| 14 | Phillips SJ, Dudík MM (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography, 31, 161-175. |
| 15 | Qiao HJ, Hu JH, Huang JH (2013) Theoretical basis, future directions, and challenges for ecological niche models. Scientia Sinica Vitae, 43, 915-927. (in Chinese with English abstract) |
| [乔慧捷, 胡军华, 黄继红 (2013) 生态位模型的理论基础、发展方向与挑战. 中国科学: 生命科学, 43, 915-927.] | |
| 16 | Qiao HJ, Soberón J, Peterson AT (2015) No silver bullets in correlative ecological niche modeling: insights from testing among many potential algorithms for niche estimation. Methods in Ecology and Evolution, 6, 1126-1136. |
| 17 | Soberón J, Peterson AT (2005) Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodiversity Informatics, 2, 1-10. |
| 18 | Vaz UL, Cunha HF, Nabout JC (2015) Trends and biases in global scientific literature about ecological niche models. Brazilian Journal of Biology, 75, 17-24. |
| 19 | Warren DL, Seifert SN (2011) Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecology Applications, 21, 335-342. |
| 20 | Warren DL, Wright AN, Seifert SN, Shaffer HB (2014) Incorporating model complexity and spatial sampling bias into ecological niche models of climate change risks faced by 90 California vertebrate species of concern. Diversity and Distributions, 20, 334-343. |
| 21 | Zhu GP, Bu WJ, Gao YB, Liu GQ (2012) Potential geographic distribution of brown marmorated stink bug invasion (Halyomorpha halys). PLoS ONE, 7, e31246. |
| 22 | Zhu GP, Gao YB, Zhu L (2013) Delimiting the coastal geographic background to predict potential distribution of Spartina alterniflora. Hydrobiologia, 717, 177-187. |
| 23 | Zhu GP, Redei D, Kment P, Bu WJ (2014) Effect of geographic background and equilibrium state on niche model transferability: predicting areas of invasion of Leptoglossus occidentalis. Biological Invasions, 16, 1069-1081. |
| 24 | Zhu GP, Gariepy TD, Haye T, Bu WJ (2016) Patterns of niche filling and expansion across the invaded ranges of Halyomorpha halys in North America and Europe. Journal of Pest Science, doi:10.1007/s10340-016-0786-z. |
| 25 | Zhu GP, Liu GQ, Bu WJ, Gao YB (2013) Ecological niche modeling and its applications in biodiversity conservation. Biodiversity Science, 21, 90-98. (in Chinese with English abstract) |
| [朱耿平, 刘国卿, 卜文俊, 高玉葆 (2013) 生态位模型的基本原理及其在生物多样性保护中的应用. 生物多样性, 21, 90-98.] | |
| 26 | Zhu GP, Liu Q, Gao YB (2014) Improving ecological niche model transferability to predict the potential distribution of invasive exotic species. Biodiversity Science, 22, 223-230. (in Chinese with English abstract) |
| [朱耿平, 刘强, 高玉葆 (2014) 提高生态位模型转移能力来模拟入侵物种的潜在分布. 生物多样性, 22, 223-230.] |
| [1] | 付梦娣, 朱彦鹏, 任月恒, 李爽, 秦乐, 谢正君, 王清春, 张立博. 新疆野生动物通道空间布局优化[J]. 生物多样性, 2025, 33(3): 24346-. |
| [2] | 原雪姣, 张渊媛, 张衍亮, 胡璐祎, 桑卫国, 杨峥, 陈颀. 基于飞机草历史分布数据拟合的物种分布模型及其预测能力[J]. 生物多样性, 2024, 32(11): 24288-. |
| [3] | 刘志发, 王新财, 龚粤宁, 陈道剑, 张强. 基于红外相机监测的广东南岭国家级自然保护区鸟兽多样性及其垂直分布特征[J]. 生物多样性, 2023, 31(8): 22689-. |
| [4] | 张琼悦, 邓卓迪, 胡学斌, 丁志锋, 肖荣波, 修晨, 吴政浩, 汪光, 韩东晖, 张语克, 梁健超, 胡慧建. 粤港澳大湾区城市化进程对区域内鸟类分布及栖息地连通性的影响[J]. 生物多样性, 2023, 31(3): 22161-. |
| [5] | 魏博, 刘林山, 谷昌军, 于海彬, 张镱锂, 张炳华, 崔伯豪, 宫殿清, 土艳丽. 紫茎泽兰在中国的气候生态位稳定且其分布范围仍有进一步扩展的趋势[J]. 生物多样性, 2022, 30(8): 21443-. |
| [6] | 滕继荣, 刘兴明, 何礼文, 王钧亮, 黄建, 冯杰, 王放, 翁悦. 甘肃白水江国家级自然保护区林缘社区饲养犬只对大熊猫时空节律的影响[J]. 生物多样性, 2022, 30(1): 21204-. |
| [7] | 张晨, 马伟, 陈晨, 汪沐阳, 徐文轩, 杨维康. 重大工程影响下新疆卡拉麦里山有蹄类野生动物自然保护区鹅喉羚的生境格局变化[J]. 生物多样性, 2022, 30(1): 21176-. |
| [8] | 马星, 王浩, 余蔚, 杜勇, 梁健超, 胡慧建, 邱胜荣, 刘璐. 基于MaxEnt模型分析广东省鸟类多样性热点分布及保护空缺[J]. 生物多样性, 2021, 29(8): 1097-1107. |
| [9] | 周润, 慈秀芹, 肖建华, 曹关龙, 李捷. 气候变化对亚热带常绿阔叶林优势类群樟属植物的影响及保护评估[J]. 生物多样性, 2021, 29(6): 697-711. |
| [10] | 王然, 乔慧捷. 生态位模型在流行病学中的应用[J]. 生物多样性, 2020, 28(5): 579-586. |
| [11] | 范靖宇, 李汉芃, 杨琢, 朱耿平. 基于本土最优模型模拟入侵物种水盾草在中国的潜在分布[J]. 生物多样性, 2019, 27(2): 140-148. |
| [12] | 庄鸿飞, 张殷波, 王伟, 任月恒, 刘方正, 杜金鸿, 周越. 基于最大熵模型的不同尺度物种分布概率优化热点分析: 以红色木莲为例[J]. 生物多样性, 2018, 26(9): 931-940. |
| [13] | 王波, 黄勇, 李家堂, 戴强, 王跃招, 杨道德. 西南喀斯特地貌区两栖动物丰富度分布格局与环境因子的关系[J]. 生物多样性, 2018, 26(9): 941-950. |
| [14] | 丁晨晨, 胡一鸣, 李春旺, 蒋志刚. 印度野牛在中国的分布及其栖息地适宜性分析[J]. 生物多样性, 2018, 26(9): 951-961. |
| [15] | 周中一, 刘冉, 时书纳, 苏艳军, 李文楷, 郭庆华. 基于激光雷达数据的物种分布模拟: 以美国加州内华达山脉南部区域食鱼貂分布模拟为例[J]. 生物多样性, 2018, 26(8): 878-891. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2026 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()