



生物多样性 ›› 2021, Vol. 29 ›› Issue (6): 722-734. DOI: 10.17520/biods.2020484 cstr: 32101.14.biods.2020484
所属专题: 物种形成与系统进化
王婷1,2,3, 夏增强1,2,4, 舒江平1,2,5, 张娇4, 王美娜1,2, 陈建兵1,2, 王慷林6, 向建英7,*( ), 严岳鸿1,2,*(
), 严岳鸿1,2,*( )
)
收稿日期:2020-12-29
接受日期:2021-04-14
出版日期:2021-06-20
发布日期:2021-04-22
通讯作者:
向建英,严岳鸿
作者简介:jy_xiang@hotmail.com基金资助:
Ting Wang1,2,3, Zengqiang Xia1,2,4, Jiangping Shu1,2,5, Jiao Zhang4, Meina Wang1,2, Jianbing Chen1,2, Kanglin Wang6, Jianying Xiang7,*( ), Yuehong Yan1,2,*(
), Yuehong Yan1,2,*( )
)
Received:2020-12-29
Accepted:2021-04-14
Online:2021-06-20
Published:2021-04-22
Contact:
Jianying Xiang,Yuehong Yan
摘要:
全基因组复制在维管植物的物种形成过程中普遍存在, 被认为是物种适应极端环境的重要机制之一。确定全基因组复制事件的发生时间对理解生物的适应性演化具有重要意义。然而, 在维管植物, 特别是蕨类植物中, 全基因组复制事件的发生时间及其演化意义仍知之甚少。本研究以蕨类植物重要基部类群——福建莲座蕨(Angiopteris fokiensis)为例, 基于不同采样点(广东、广西、上海)的3个转录组学数据, 利用同义替换率(Ks)和绝对定年的方法分析全基因组复制事件的发生时间和物种单位时间内的分子演化速率, 并对事件发生后保留下的基因进行基因功能注释和富集分析。结果表明, 福建莲座蕨在159‒165 Mya发生了一次全基因组复制事件, 该复制事件优先保留的基因主要与营养代谢、信号传导、适应调节和组织结构生长相关。另外, 福建莲座蕨的分子演化速率为1.66 × 10‒9 (同义替换/位点/年), 是除裸子植物外, 陆生植物中已知演化速率最缓慢的类群。综合以上研究结果, 我们推测福建莲座蕨全基因组复制的发生可能与裸子植物繁盛、核心被子植物集中兴起或托阿尔阶灭绝事件有关。而复制后显著保留基因可能促进了莲座蕨属(Angiopteris)植物的遗传和形态创新, 从而帮助其快速适应环境的剧烈变化。进一步对该类群植物演化速率缓慢的原因进行讨论, 推测莲座蕨属缓慢的演化速率可能与其本身世代周期长、基因组较大及其生长环境稳定有关。本研究通过分析福建莲座蕨的全基因组复制历史和复制基因的保留模式, 推测全基因组复制事件对促进演化速率较慢的植物适应极端环境变化具有重要意义, 可为理解其他陆生植物的适应性演化提供更多启发。
王婷, 夏增强, 舒江平, 张娇, 王美娜, 陈建兵, 王慷林, 向建英, 严岳鸿 (2021) 全基因组复制事件的绝对定年揭示莲座蕨属植物的迟滞演化. 生物多样性, 29, 722-734. DOI: 10.17520/biods.2020484.
Ting Wang, Zengqiang Xia, Jiangping Shu, Jiao Zhang, Meina Wang, Jianbing Chen, Kanglin Wang, Jianying Xiang, Yuehong Yan (2021) Dating whole-genome duplication reveals the evolutionary retardation of Angiopteris. Biodiversity Science, 29, 722-734. DOI: 10.17520/biods.2020484.
| 物种 Species | NCBI登录号 NCBI no. | 转录本Transcript | N50 (bp) | 鸟嘌呤/胞嘧啶 GC (%) | 编码序列数 No. of coding sequence | BUSCO (%) | 文献 References |
|---|---|---|---|---|---|---|---|
| 福建莲座蕨 Angiopteris fokiensis | SRR2103714 | 46,702 | 1,720 | 44.95 | 34,286 | 89.6 | Shen et al, |
| 福建莲座蕨 Angiopteris fokiensis | SRR5499396 | 62,608 | 2,021 | 44.29 | 39,010 | 95.1 | You et al, |
| 福建莲座蕨 Angiopteris fokiensis | SRR6920615 | 62,119 | 2,021 | 44.29 | 39,043 | 95.4 | Huang et al, |
| 披散木贼 Equisetum diffusum | SRR2103706 | 55,430 | 1,537 | 44.93 | 39,399 | 91.4 | Shen et al, |
| 桂皮紫萁 Osmundastrum cinnamomeum | SRR6727971 | 28,229 | 1,922 | 46.43 | 22,868 | 84.4 | Wolf et al, |
| 紫萁 Osmunda japonica | SRR2103721 | 42,649 | 2,005 | 46.22 | 31,414 | 92.8 | Huang et al, |
| 细叶双扇蕨 Dipteris lobbiana | SRR6920640 | 148,711 | 1,085 | 46.09 | 94,834 | 78.4 | Huang et al, |
| 芒萁 Dicranopteris pedata | SRR5490811 | 62,818 | 1,954 | 45.78 | 40,874 | 97.5 | You et al, |
| 假芒萁 Sticherus truncatus | SRR6920643 | 74,801 | 1,557 | 44.30 | 50,507 | 88.2 | Huang et al, |
表1 序列组装情况统计
Table 1 Summary of de novo assembly for transcriptome
| 物种 Species | NCBI登录号 NCBI no. | 转录本Transcript | N50 (bp) | 鸟嘌呤/胞嘧啶 GC (%) | 编码序列数 No. of coding sequence | BUSCO (%) | 文献 References |
|---|---|---|---|---|---|---|---|
| 福建莲座蕨 Angiopteris fokiensis | SRR2103714 | 46,702 | 1,720 | 44.95 | 34,286 | 89.6 | Shen et al, |
| 福建莲座蕨 Angiopteris fokiensis | SRR5499396 | 62,608 | 2,021 | 44.29 | 39,010 | 95.1 | You et al, |
| 福建莲座蕨 Angiopteris fokiensis | SRR6920615 | 62,119 | 2,021 | 44.29 | 39,043 | 95.4 | Huang et al, |
| 披散木贼 Equisetum diffusum | SRR2103706 | 55,430 | 1,537 | 44.93 | 39,399 | 91.4 | Shen et al, |
| 桂皮紫萁 Osmundastrum cinnamomeum | SRR6727971 | 28,229 | 1,922 | 46.43 | 22,868 | 84.4 | Wolf et al, |
| 紫萁 Osmunda japonica | SRR2103721 | 42,649 | 2,005 | 46.22 | 31,414 | 92.8 | Huang et al, |
| 细叶双扇蕨 Dipteris lobbiana | SRR6920640 | 148,711 | 1,085 | 46.09 | 94,834 | 78.4 | Huang et al, |
| 芒萁 Dicranopteris pedata | SRR5490811 | 62,818 | 1,954 | 45.78 | 40,874 | 97.5 | You et al, |
| 假芒萁 Sticherus truncatus | SRR6920643 | 74,801 | 1,557 | 44.30 | 50,507 | 88.2 | Huang et al, |
| 样本 Sample | 采集地 Collection sites | 加倍数目 No. of duplicates | 组件数目 No. of components | 贝叶斯信息标准 Bayesian information criterion | 中值 Median (Ks) | 方差 Variance | 比例 Proportion |
|---|---|---|---|---|---|---|---|
| 福建莲座蕨 Angiopteris fokiensis SRR2103714 | 上海 Shanghai | 3,667 | 6 | ?6,824.9400 | 0.1310 | 0.0005 | 0.0763 |
| 3,667 | 6 | ?6,824.9400 | 0.2147 | 0.0029 | 0.0577 | ||
| 3,667 | 6 | ?6,824.9400 | 0.5814 | 0.0200 | 0.2697 | ||
| 3,667 | 6 | ?6,824.9400 | 0.9172 | 0.0312 | 0.2529 | ||
| 3,667 | 6 | ?6,824.9400 | 1.3513 | 0.1683 | 0.2040 | ||
| 3,667 | 6 | ?6,824.9400 | 3.0112 | 0.8830 | 0.1393 | ||
| 福建莲座蕨 Angiopteris fokiensis SRR5499396 | 广东 Guangdong | 4,238 | 7 | ?7,662.1730 | 0.1241 | 0.0003 | 0.0680 |
| 4,238 | 7 | ?7,662.1730 | 0.2002 | 0.0023 | 0.0921 | ||
| 4,238 | 7 | ?7,662.1730 | 0.5856 | 0.0191 | 0.2940 | ||
| 4,238 | 7 | ?7,662.1730 | 0.9331 | 0.0198 | 0.1781 | ||
| 4,238 | 7 | ?7,662.1730 | 1.1911 | 0.1116 | 0.1935 | ||
| 4,238 | 7 | ?7,662.1730 | 2.3596 | 0.4994 | 0.1330 | ||
| 4,238 | 7 | ?7,662.1730 | 4.1443 | 0.2343 | 0.0412 | ||
| 福建莲座蕨 Angiopteris fokiensis SRR6920615 | 广西 Guangxi | 4,235 | 6 | ?7,662.1730 | 0.1259 | 0.0003 | 0.0753 |
| 4,235 | 6 | ?7,662.1730 | 0.1996 | 0.0022 | 0.0834 | ||
| 4,235 | 6 | ?7,662.1730 | 0.5489 | 0.0148 | 0.2082 | ||
| 4,235 | 6 | ?7,662.1730 | 0.8841 | 0.0452 | 0.3284 | ||
| 4,235 | 6 | ?7,662.1730 | 1.3642 | 0.1874 | 0.1606 | ||
| 4,235 | 6 | ?7,662.1730 | 2.9677 | 0.9873 | 0.1441 |
表2 基于混合模型拟合3个福建莲座蕨样本的同义替换率(Ks)结果(加粗表示峰值显著)
Table 2 Synonymous substitution rate (Ks) results of three Angiopteris fokiensisbased on mixture modeling (Bold indicates a significant peaks)
| 样本 Sample | 采集地 Collection sites | 加倍数目 No. of duplicates | 组件数目 No. of components | 贝叶斯信息标准 Bayesian information criterion | 中值 Median (Ks) | 方差 Variance | 比例 Proportion |
|---|---|---|---|---|---|---|---|
| 福建莲座蕨 Angiopteris fokiensis SRR2103714 | 上海 Shanghai | 3,667 | 6 | ?6,824.9400 | 0.1310 | 0.0005 | 0.0763 |
| 3,667 | 6 | ?6,824.9400 | 0.2147 | 0.0029 | 0.0577 | ||
| 3,667 | 6 | ?6,824.9400 | 0.5814 | 0.0200 | 0.2697 | ||
| 3,667 | 6 | ?6,824.9400 | 0.9172 | 0.0312 | 0.2529 | ||
| 3,667 | 6 | ?6,824.9400 | 1.3513 | 0.1683 | 0.2040 | ||
| 3,667 | 6 | ?6,824.9400 | 3.0112 | 0.8830 | 0.1393 | ||
| 福建莲座蕨 Angiopteris fokiensis SRR5499396 | 广东 Guangdong | 4,238 | 7 | ?7,662.1730 | 0.1241 | 0.0003 | 0.0680 |
| 4,238 | 7 | ?7,662.1730 | 0.2002 | 0.0023 | 0.0921 | ||
| 4,238 | 7 | ?7,662.1730 | 0.5856 | 0.0191 | 0.2940 | ||
| 4,238 | 7 | ?7,662.1730 | 0.9331 | 0.0198 | 0.1781 | ||
| 4,238 | 7 | ?7,662.1730 | 1.1911 | 0.1116 | 0.1935 | ||
| 4,238 | 7 | ?7,662.1730 | 2.3596 | 0.4994 | 0.1330 | ||
| 4,238 | 7 | ?7,662.1730 | 4.1443 | 0.2343 | 0.0412 | ||
| 福建莲座蕨 Angiopteris fokiensis SRR6920615 | 广西 Guangxi | 4,235 | 6 | ?7,662.1730 | 0.1259 | 0.0003 | 0.0753 |
| 4,235 | 6 | ?7,662.1730 | 0.1996 | 0.0022 | 0.0834 | ||
| 4,235 | 6 | ?7,662.1730 | 0.5489 | 0.0148 | 0.2082 | ||
| 4,235 | 6 | ?7,662.1730 | 0.8841 | 0.0452 | 0.3284 | ||
| 4,235 | 6 | ?7,662.1730 | 1.3642 | 0.1874 | 0.1606 | ||
| 4,235 | 6 | ?7,662.1730 | 2.9677 | 0.9873 | 0.1441 |
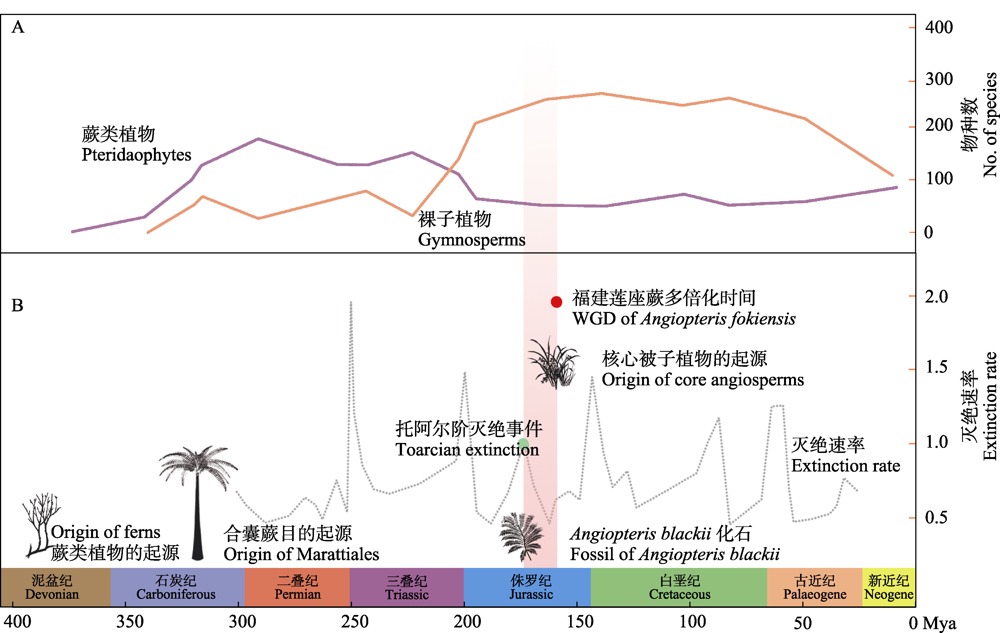
图5 蕨类植物和裸子植物不同地质年代的化石物种数(Niklas et al, 1983; Katz, 2018) (A)及蕨类植物、被子植物起源和物种灭绝速率的时间线(Lehtonen et al, 2017, 2020; Clapham & Renne, 2019; Li et al, 2019) (B)
Fig. 5 The number of fossils of species in different geological ages between pteridophytes and gymonsperms (Niklas et al, 1983; Katz, 2018) (A) and a proposed timeline for flowering plants and ferns' origin and exitinction rate of species (Lehtonen et al, 2017, 2020; Clapham & Renne, 2019; Li et al, 2019) (B)
| [1] |
Banks JA (1999) Gametophyte development in ferns. Annual Review of Plant Physiology and Plant Molecular Biology, 50,163-186.
PMID |
| [2] |
Barker MS, Vogel H, Schranz ME (2009) Paleopolyploidy in the Brassicales: Analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales . Genome Biology Evolution, 1,391-399.
DOI URL |
| [3] |
Blanc G, Wolfe KH (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. The Plant Cell, 16,1667-1678.
DOI URL |
| [4] |
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30,2114-2120.
DOI URL |
| [5] |
Bomfleur B, McLoughlin S, Vajda V (2014) Fossilized nuclei and chromosomes reveal 180 million years of genomic stasis in royal rerns. Science, 343,1376-1377.
DOI URL |
| [6] |
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology, 10,e1003537.
DOI URL |
| [7] |
Bromham L, Hua X, Lanfear R, Cowman PF (2015) Exploring the relationships between mutation rates, life history, genome size, environment, and species richness in flowering plants. The American Naturalist, 185,507-524.
DOI URL |
| [8] |
Bromham L, Penny D (2003) The modern molecular clock. Nature Reviews Genetics, 4,216-224.
DOI URL |
| [9] |
Cai LM, Xi ZX, Amorim AM, Sugumaran M, Rest JS, Liu L, Davis CC (2019) Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytologist, 221,565-576.
DOI URL |
| [10] |
Chao DY, Dilkes B, Luo HB, Douglas A, Yakubova E, Lahner B, Salt DE (2013) Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis . Science, 341,658-659.
DOI URL |
| [11] | Charlesworth B, Smith DB (1982) A computer model of speciation by founder effects . Genetical Research, 39,227-236. |
| [12] | Chen WH, Xiao WB (2008) On the development of gametophyte and sporophyte of Ceratopteris thalictroides . Journal of Hunan Agricultural University (Natural Sciences), 34,306-310, 383. (in Chinese with English abstract) |
| 陈蔚辉, 肖卫彬 (2008) 水蕨配子体和孢子体发育的研究. 湖南农业大学学报(自然科学版), 34,306-310, 383.] | |
| [13] |
Clapham ME, Renne PR (2019) Flood basalts and mass extinctions. Annual Review of Earth and Planetary Sciences, 47,275-303.
DOI URL |
| [14] |
Clark J, Hidalgo O, Pellicer J, Liu HM, Marquardt J, Robert Y, Christenhusz M, Zhang SZ, Gibby M, Leitch IJ, Schneider H (2016) Genome evolution of ferns: Evidence for relative stasis of genome size across the fern phylogeny. New Phytologist, 210,1072-1082.
DOI URL |
| [15] | Collinson ME (1996) What use are fossil ferns?-20 years on: with a review of the fossil history of extant pteridophyte families and genera. In: Pteridology in Perspective: Proceedings of the Holttum Memorial Pteridophyte Symposium, Kew, 1995 (eds Camus JM, Gibby M, Johns RJ), pp. 349-394. Whitstable Litho Ltd., Kent, |
| [16] |
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21,3674-3676.
PMID |
| [17] |
Cui LY, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, Albert VA, Ma H, dePamphilis CW (2006) Widespread genome duplications throughout the history of flowering plants. Genome Research, 16,738-749.
DOI URL |
| [18] |
De La Torre AR Li Z van de Peer Y Ingvarsson PK (2017) Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Molecular Biology and Evolution, 34,1363-1377.
DOI URL |
| [19] |
Diallo AM, Nielsen LR, Kjær ED, Petersen KK, Ræbild A (2016) Polyploidy can confer superiority to West African Acacia senegal (L.) Willd. trees . Frontiers in Plant Science, 7,821.
DOI PMID |
| [20] |
DiMichele WA, Phillips TL, Nelson WJ (2002) Place vs. time and vegetational persistence: A comparison of four tropical mires from the Illinois Basin during the height of the Pennsylvanian Ice Age. International Journal of Coal Geology, 50,43-72.
DOI URL |
| [21] |
Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32,1792-1797.
DOI URL |
| [22] |
Enright AJ, van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Research, 30,1575-1584.
PMID |
| [23] | Fawcett JA, Maere S, van de Peer Y (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proceedings of the National Academy of Sciences, USA, 106,5737-5742. |
| [24] |
Fraley C, Raftery AE (2003) Enhanced model-based clustering, density estimation, and discriminant analysis software: MCLUST. Journal of Classification, 20,263-286.
DOI URL |
| [25] | Futuyma DJ (translated by Ge S, Gu HY, Rao GY, Zhang DX, Yang J, Kong HZ, Wang YF) (2016) Evolution, Higher Education Press, Beijing. (in Chinese) |
| 葛颂, 顾红雅, 饶广远, 张德兴, 杨继, 孔宏智, 王宇飞(译) (2016) 生物进化, 高等教育出版社, 北京.] | |
| [26] | Gaut B, Yang L, Takuno S, Eguiarte LE (2011) The patterns and causes of variation in plant nucleotide substitution rates. Annual Review of Ecology, Evolution & Systematics, 42,245-266. |
| [27] | Goldman N, Yang Z (1994) A Codon-based model of nucleotide substitution for protein-coding DNA sequences . Molecular Biology and Evolution,725-736. |
| [28] |
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng QD, Chen ZH, Mauceli E, Hacohen N, Gnirke A, Rhind Ndi Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 29,644-652.
DOI PMID |
| [29] |
Hanson L, Leitch IJ (2002) DNA amounts for five pteridophyte species fill phylogenetic gaps in C-value data. Botanical Journal of the Linnean Society, 140,169-173.
DOI URL |
| [30] | Harris TM (1961) The Yorkshire Jurassic Flora. I. Thalophyta-Pteridophyta. Trustees of the British Museum (Natural History), India. |
| [31] | He ZR (2009) Systematic Study of Angiopteridaceae from China. PhD dissertation, Yunnan University, Kunming. (in Chinese with English abstract) |
| 和兆荣 (2009) 中国莲座蕨科系统学研究. 博士学位论文, 云南大学, 昆明.] | |
| [32] |
Hill CR (1987) Jurassic Angiopteris (Marattiales) from North Yorkshire . Review of Palaeobotany and Palynology, 51,65-93.
DOI URL |
| [33] |
Hu SS, Dilcher DL, Schneider H, Jarzen DM (2006) Eusporangiate ferns from the Dakota formation, Minnesota, USA. International Journal of Plant Sciences, 167,579-589.
DOI URL |
| [34] |
Huang CH, Qi X, Chen D, Qi J, Ma H (2020) Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns. Journal of Integrative Plant Biology, 62,433-455.
DOI URL |
| [35] |
Jansa SA, Forsman JF, Voss RS (2006) Different patterns of selection on the nuclear genes IRBP and DMP- 1 affect the efficiency but not the outcome of phylogeny estimation for didelphid marsupials. Molecular Phylogenetics and Evolution, 38,363-380.
DOI URL |
| [36] |
Jiao YN, Leebens-Mack J, Ayyampalayam S, Bowers JE, McKain MR, McNeal J, Rolf M, Ruzicka DR, Wafula E, Wickett NJ, Wu XL, Zhang Y, Wang J, Zhang YT, Carpenter EJ, Deyholos MK, Kutchan TM, Chanderbali AS, Soltis PS, Stevenson DW, McCombie R, Pires JC, Wong GKS, Soltis DE, Depamhilis CW (2012) A genome triplication associated with early diversification of the core eudicots. Genome Biology, 13,R3.
DOI URL |
| [37] |
Jiao YN, Li JP, Tang HB, Paterson AH (2014) Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. The Plant Cell, 26,2792-2802.
DOI URL |
| [38] |
Katoh K, Misawa K, Kuma KI, Miyata T (2002) MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30,3059-3066.
DOI URL |
| [39] |
Katz O (2018) Extending the scope of Darwin's ‘abominable mystery': Integrative approaches to understanding angiosperm origins and species richness. Annals of Botany, 121,1-8.
DOI URL |
| [40] |
Klekowski EJ, Baker HG (1966) Evolutionary significance of polyploidy in the pteridophyta. Science, 153,305-307.
DOI URL |
| [41] |
Korall P, Schuettpelz E, Pryer KM (2010) Abrupt deceleration of molecular evolution linked to the origin of arborescence in ferns. Evolution, 64,2786-2792.
DOI PMID |
| [42] |
Laird CD, McConaughy BL, McCarthy BJ (1969) Rate of fixation of nucleotide substitutions in evolution. Nature, 224,149-154.
PMID |
| [43] |
Lanfear R, Kokko H, Eyre-Walker A (2014) Population size and the rate of evolution. Trends in Ecology & Evolution, 29,33-41.
DOI URL |
| [44] |
Lehtonen S, Poczai P, Sablok G, Hyvönen J, Karger DN, Flores J (2020) Exploring the phylogeny of the marattialean ferns. Cladistics, 36,569-593.
DOI URL |
| [45] |
Lehtonen S, Silvestro D, Karger DN, Scotese C, Tuomisto H, Kessler M, Peña C, Wahlberg N, Antonelli A (2017) Environmentally driven extinction and opportunistic origination explain fern diversification patterns. Scientific Reports, 7,4831.
DOI PMID |
| [46] | Li HT, Yi TS, Gao LM, Ma PF, Zhang T, Yang JB, Gitzendanner MA, Fritsch PW, Cai J, Luo Y, Wang H, van der Bank M, Zhang SD, Wang QF, Wang J, Zhang ZR, Fu CN, Yang J, Hollingsworth PM, Chase MW, Soltis DE, Soltis PS, Li DZ, (2019) Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants, 5,461-470. |
| [47] |
Li WZ, Godzik A (2006) Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 22,1658-1659.
DOI URL |
| [48] |
Lidgard S, Crane PR (1990) Angiosperm diversification and Cretaceous floristic trends: A comparison of palynofloras and leaf macrofloras. Paleobiology, 16,77-93.
DOI URL |
| [49] | Lundblad AB (1950) Studies in the Rhaeto-Liassie Flora of Sweden, 1. Pteridophyta, Pteridospermae and Cycadophyta from the mining district of NW Scania. Kungl Svenska Vetenskap Handl, Fjärde Ser, 1,1-82. |
| [50] |
Mandel JR (2019) A Jurassic leap for flowering plants. Nature Plants, 5,455-456.
DOI URL |
| [51] | Matasci N, Hung LH, Yan ZX, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, Burleigh JG, Gitzendanner MA, Wafula E, Der JP, DePamphilis CW, Roure B, Philippe H, Ruhfel BR, Miles NW, Graham SW, Mathews S, Surek B, Melkonian M, Soltis DE, Soltis PS, Rothfels C, Pokorny L, Shaw JA, DeGironimo L, Stevenson DW, Villarreal JC, Chen T, Kutchan TM, Rolf M, Baucom RS, Deyholos MK, Samudrala R, Tian ZJ, Wu XL, Sun X, Zhang Y, Wang J, Leebens-Mack J, Wong GKS (2014) Data access for the 1,000 Plants (1KP) project. GigaScience, 3,2047-217X. |
| [52] |
Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, Otto SP (2011) Recently formed polyploid plants diversify at lower rates. Science, 333,1257.
DOI URL |
| [53] | Morgan J (1959) The morphology and anatomy of American species of the genus Psaronius . Illinois Biological Monographs, 27,1-108. |
| [54] |
Murdock AG (2008a) Phylogeny of marattioid ferns (Marattiaceae): Inferring a root in the absence of a closely related outgroup. American Journal of Botany, 95,626-641.
DOI URL |
| [55] |
Murdock AG (2008b) A taxonomic revision of the eusporangiate fern family Marattiaceae, with description of a new genus Ptisana . Taxon, 57,737-755.
DOI URL |
| [56] |
Nagalingum NS, Drinnan AN, Lupia R, McLoughlin S (2002) Fern spore diversity and abundance in Australia during the Cretaceous. Review of Palaeobotany and Palynology, 119,69-92.
DOI URL |
| [57] |
Niklas KJ, Tiffney BH, Knoll AH (1983) Patterns in vascular land plant diversification. Nature, 303,614-616.
DOI URL |
| [58] |
Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annual Review of Genetics, 34,401-437.
DOI URL |
| [59] |
PPG I (2016) A community-derived classification for extant lycophytes and ferns. Journal of Systematics and Evolution, 54,563-603.
DOI URL |
| [60] |
Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, Cranfill R (2004) Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. American Journal of Botany, 91,1582-1598.
DOI URL |
| [61] |
Qi XP, Kuo LY, Guo CC, Li H, Li ZY, Qi J, Wang LB, Hu Y, Xiang JY, Zhang CF, Guo J, Huang CH, Ma H (2018) A well-resolved fern nuclear phylogeny reveals the evolution history of numerous transcription factor families. Molecular Phylogenetics and Evolution, 127,961-977.
DOI URL |
| [62] | Rothwell GW (1996) Phylogenetic relationships of ferns: A palaeobotanical perspective. Royal Botanic Gardens, Kew,395-404. |
| [63] |
Rothwell GW, Millay MA, Stockey RA (2018) Resolving the overall pattern of marattialean fern phylogeny. American Journal of Botany, 105,1304-1314.
DOI PMID |
| [64] |
Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome, 47,868-876.
PMID |
| [65] |
Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallón S, Lupia R (2004) Ferns diversified in the shadow of angiosperms. Nature, 428,553-557.
PMID |
| [66] |
Schuettpelz E, Pryer KM (2006) Reconciling extreme branch length differences: Decoupling time and rate through the evolutionary history of filmy ferns. Systematic Biology, 55,485-502.
DOI URL |
| [67] | Schuettpelz E, Pryer KM (2009) Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proceedings of the National Academy of Sciences, USA, 106,11200-11205. |
| [68] | Shen H, Jin DM, Shu JP, Zhou XL, Lei M, Wei R, Shang H, Wei HJ, Zhang R, Liu L, Gu YF, Zhang XC, Yan YH (2018) Large-scale phylogenomic analysis resolves a backbone phylogeny in ferns. GigaScience, 7, gix116. |
| [69] |
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31,3210-3212.
DOI URL |
| [70] |
Skog JE (2001) Biogeography of Mesozoic leptosporangiate ferns related to extant ferns. Brittonia, 53,236-269.
DOI URL |
| [71] | Soltis PS, Soltis DE, Savolainen V, Crane PR, Barraclough TG (2002) Rate heterogeneity among lineages of tracheophytes: Integration of molecular and fossil data and evidence for molecular living fossils. Proceedings of the National Academy of Sciences, USA, 99,4430-4435. |
| [72] | Thomas JA, Welch JJ, Woolfit M, Bromham L (2006) There is no universal molecular clock for invertebrates, but rate variation does not scale with body size. Proceedings of the National Academy of Sciences, USA, 103,7366-7371. |
| [73] | Vandepoele K, De Vos W, Taylor JS, Meyer A, van de Peer Y (2004) Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proceedings of the National Academy of Sciences, USA, 101,1638-1643. |
| [74] |
Vanneste K, Baele G, Maere S, van de Peer Y (2014) Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Research, 24,1334-1347.
DOI PMID |
| [75] |
Vanneste K, Sterck L, Myburg AA, van de Peer Y, Mizrachi E (2015) Horsetails are ancient polyploids: Evidence from Equisetum giganteum . The Plant Cell, 27,1567-1578.
DOI PMID |
| [76] | van de Peer Y, Ashman TL, Soltis PS, Soltis DE (2021) Polyploidy: An evolutionary and ecological force in stressful times. The Plant Cell, 33,11-26. |
| [77] |
Wang H, Zhang R, Zhang J, Shen H, Dai XL, Yan YH (2019) De novo transcriptome assembly reveals the whole genome duplication events of Didymochlaena trancatula . Biodiversity Science, 27,1221-1227. (in Chinese with English abstract)
DOI |
|
汪浩, 张锐, 张娇, 沈慧, 戴锡玲, 严岳鸿 (2019) 转录组测序揭示翼盖蕨( Didymochlaena trancatula)的全基因组复制历史 . 生物多样性, 27,1221-1227.]
DOI |
|
| [78] |
Wolf PG, Robison TA, Johnson MG, Sundue MA, Testo WL, Rothfels CJ (2018) Target sequence capture of nuclear-encoded genes for phylogenetic analysis in ferns. Applications in Plant Sciences, 6,e01148.
DOI URL |
| [79] |
Wu SD, Han BC, Jiao YN (2020) Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Molecular Plant, 13,59-71.
DOI URL |
| [80] | Wu CI, Li WH (1985) Evidence for higher rates of nucleotide substitution in rodents than in man. Proceedings of the National Academy of Sciences, USA, 82,1741-1745. |
| [81] | Yan XF, Wang Y, Li YM (2007) Plant secondary metabolism and its response to environment. Acta Ecologica Sinica, 27,2554-2562. (in Chinese with English abstract) |
| 阎秀峰, 王洋, 李一蒙 (2007) 植物次生代谢及其与环境的关系. 生态学报, 27,2554-2562.] | |
| [82] |
Yang ZH (2007) PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24,1586-1591.
DOI URL |
| [83] |
You CJ, Cui J, Wang H, Qi XP, Kuo LY, Ma H, Gao L, Mo BX, Chen XM (2017) Conservation and divergence of small RNA pathways and microRNAs in land plants. Genome Biology, 18,158.
DOI URL |
| [84] |
Yu Y, Xiang QY, Manos PS, Soltis DE, Soltis PS, Song BH, Cheng SF, Liu X, Wong G (2017) Whole-genome duplication and molecular evolution in Cornus L. (Cornaceae)—Insights from transcriptome sequences . PLoS ONE, 12,e0171361.
DOI URL |
| [85] |
Zhang LS, Chen F, Zhang XT, Li Z, Zhao YY, Lohaus R, Chang XJ, Dong W, Ho SYW, Liu X, Song AX, Chen JH, Guo WL, Wang ZJ, Zhuang YY, Wang HF, Chen XQ, Hu J, Liu YH, Qin Y, Wang K, Dong SS, Liu Y, Zhang SZ, Yu XX, Wu Q, Wang LS, Yan XQ, Jiao YN, Kong HZ, Zhou XF, Yu CW, Chen YC, Li F, Wang JH, Chen W, Chen XL, Jia QD, Zhang C, Jiang YF, Zhang WB, Liu GH, Fu JY, Chen F, Ma H, van de Peer Y, Tang HB (2020a) The water lily genome and the early evolution of flowering plants. Nature, 577,79-84.
DOI URL |
| [86] | Zhang LS, Wu SD, Chang XJ, Wang XY, Zhao YP, Xia YP, Trigiano RN, Jiao YN, Chen F (2020b) The ancient wave of polyploidization events in flowering plants and their facilitated adaptation to environmental stress. Plant, Cell & Environment, 43,2847-2856. |
| [87] |
Zhang PP, Fu JM, Hu LX (2012) Effects of alkali stress on growth, free amino acids and carbohydrates metabolism in Kentucky bluegrass (Poa pratensis). Ecotoxicology, 21,1911-1918.
DOI URL |
| [88] |
Zhang R, Wang FG, Zhang J, Shang H, Liu L, Wang H, Zhao GH, Shen H, Yan YH (2019) Dating whole genome duplication in Ceratopteris thalictroides and potential adaptive values of retained gene duplicates . International Journal of Molecular Sciences, 20,1926.
DOI URL |
| [89] |
Zhong BJ, Fong R, Collins LJ, McLenachan PA, Penny D (2014) Two new fern chloroplasts and decelerated evolution linked to the long generation time in tree ferns. Genome Biology and Evolution, 6,1166-1173.
DOI URL |
| [90] |
Zhou N, Wang YD, Ya L, Porter AS, Kürschner WM, Li LQ, Lu N, McElwain JC (2020) An inter-comparison study of three stomatal-proxy methods for CO 2 reconstruction applied to Early Jurassic Ginkgoales plants . Palaeogeography, Palaeoclimatology, Palaeoecology, 542,109547.
DOI URL |
| [1] | 景昭阳, 程可光, 舒恒, 马永鹏, 刘平丽. 全基因组重测序方法在濒危植物保护中的应用[J]. 生物多样性, 2023, 31(5): 22679-. |
| [2] | 钱宏, 张健, 赵静超. 世界上已知维管植物有多少种? 基于多个全球植物数据库的整合[J]. 生物多样性, 2022, 30(7): 22254-. |
| [3] | 张淑梅, 李微, 李丁男. 辽宁省高等植物多样性编目[J]. 生物多样性, 2022, 30(6): 22038-. |
| [4] | 张军, 彭焕文, 夏富才, 王伟. 青藏高原高山区和泛北极地区种子植物多倍体比较[J]. 生物多样性, 2021, 29(11): 1470-1480. |
| [5] | 范兴科, 燕霞, 冯媛媛, 冉进华, 钱朝菊, 尹晓月, 周姗姗, 房庭舟, 马小飞. 红砂基因组大小变异及物种分化[J]. 生物多样性, 2021, 29(10): 1308-1320. |
| [6] | 孙晶琦, 陈泉, 李航宇, 常艳芬, 巩合德, 宋亮, 卢华正. 附生蕨类植物的克隆性研究进展[J]. 生物多样性, 2019, 27(11): 1184-1195. |
| [7] | 汪浩, 张锐, 张娇, 沈慧, 戴锡玲, 严岳鸿. 转录组测序揭示翼盖蕨(Didymochlaena trancatula)的全基因组复制历史[J]. 生物多样性, 2019, 27(11): 1221-1227. |
| [8] | 詹臻, 张剑锋, 曹建国, 戴锡玲. 普通针毛蕨颈卵器和卵的发育[J]. 生物多样性, 2019, 27(11): 1236-1244. |
| [9] | 张开梅, 沈羽, 周晓丽, 方炎明. 21世纪以来蕨类植物研究论文的发表情况: 基于Web of Science的数据统计[J]. 生物多样性, 2019, 27(11): 1245-1250. |
| [10] | 姬红利, 詹选怀, 张丽, 彭焱松, 周赛霞, 胡菀. 幕阜山脉石松类和蕨类植物多样性及生物地理学特征[J]. 生物多样性, 2019, 27(11): 1251-1259. |
| [11] | 杜维波, 卢元. 黄土高原石松类和蕨类植物的多样性与地理分布[J]. 生物多样性, 2019, 27(11): 1260-1267. |
| [12] | 董仕勇, 左政裕, 严岳鸿, 向建英. 中国石松类和蕨类植物的红色名录评估[J]. 生物多样性, 2017, 25(7): 765-773. |
| [13] | 常艳芬. 铁角蕨科的多倍化与物种多样性形成[J]. 生物多样性, 2017, 25(6): 621-626. |
| [14] | 王玉国. 自然杂交与物种形成[J]. 生物多样性, 2017, 25(6): 565-576. |
| [15] | 李霖锋, 刘宝. 表观遗传变异在植物杂交与多倍化过程中的作用[J]. 生物多样性, 2017, 25(6): 600-607. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()