



Biodiv Sci ›› 2022, Vol. 30 ›› Issue (6): 22034. DOI: 10.17520/biods.2022034 cstr: 32101.14.biods.2022034
• Technology and Methodology • Previous Articles Next Articles
Xiaofeng Niu1,2, Xiaomei Wang2,3, Yan Zhang2, Zhipeng Zhao2,*( ), Enyuan Fan2,*(
), Enyuan Fan2,*( )
)
Received:2022-01-15
Accepted:2022-04-19
Online:2022-06-20
Published:2022-06-20
Contact:
Zhipeng Zhao,Enyuan Fan
Xiaofeng Niu, Xiaomei Wang, Yan Zhang, Zhipeng Zhao, Enyuan Fan. Integration and application of sturgeon identification methods[J]. Biodiv Sci, 2022, 30(6): 22034.
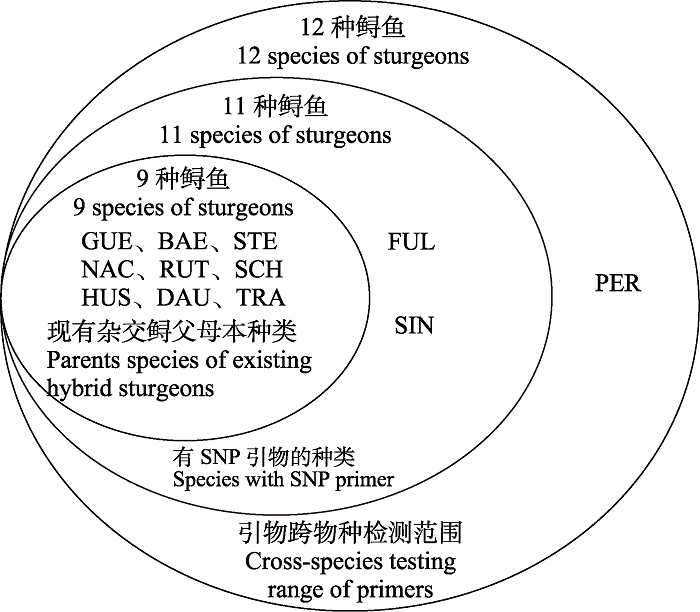
Fig. 1 Range of sturgeon species identified using known SNP primers. GUE, A. gueldenstaedtii; BAE, A. baerii; STE, A. stellatus; NAC, A. naccarii; RUT, A. ruthenus; SCH, A. schrenckii; HUS, Huso huso; DAU, H. dauricus; TRA, A. transmontanus; FUL, A. fulvescens; SIN, A. sinensis; PER, A. persicus.
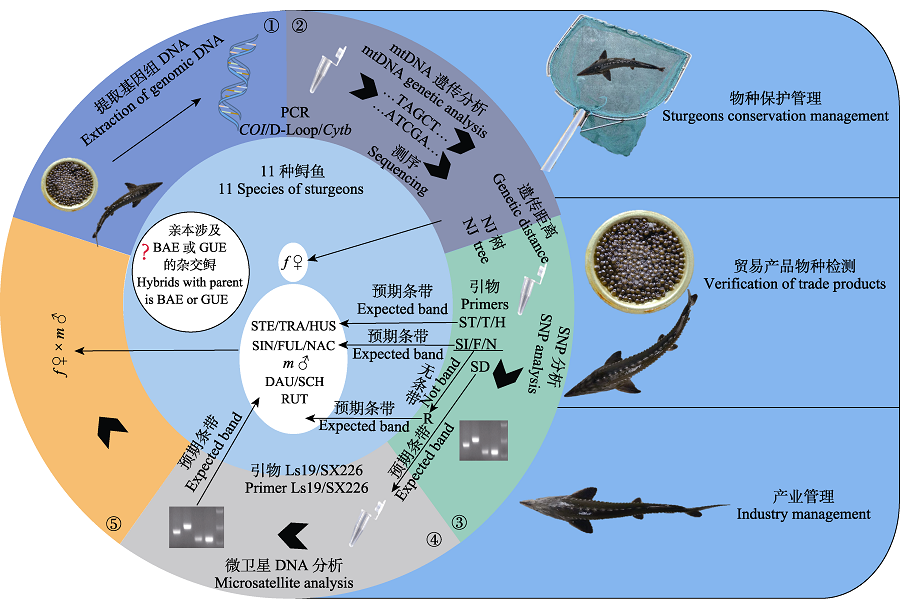
Fig. 2 Combination of methods in sturgeon identification and the application scenarios. f and m can be the same species, the primer ST, T, H, SI, F, N, and SD can be used out of order; Please refer to Fig. 1 for the full names of species abbreviation in the figure.
| 种类a Species | 拉丁名 Latin name | 鉴定可行性b Identification feasibility | 备注 Comment | |||
|---|---|---|---|---|---|---|
| 西伯利亚鲟(BAE) | Acipenser baerii | N | 缺乏可用于种类鉴定的基因标记 Lack of markers for species identification | |||
| 西伯利亚鲟 × 俄罗斯鲟 (BAE × GUE) | Acipenser baerii × Acipenser gueldenstaedtii | N | 缺乏可用于种类鉴定的基因标记 Lack of markers for species identification | |||
| 西伯利亚鲟 × 纳氏鲟 (BAE × NAC) | Acipenser baerii × Acipenser naccarii | N | 正反杂交种均不可鉴定, 但贸易中实际为NAC♀ × BAE♂ Reciprocal hybrids cannot be identified, but in trade it is NAC♀ × BAE♂ | |||
| 西伯利亚鲟 × 施氏鲟 (BAE × SCH) | Acipenser baerii × Acipenser schrenckii | N | 正反杂交种均不可鉴定 Reciprocal hybrids cannot be identified | |||
| 西伯利亚鲟 × 小体鲟 (BAE × RUT) | Acipenser baerii × Acipenser ruthenus | N | 正反杂交种均不可鉴定 Reciprocal hybrids cannot be identified | |||
| 俄罗斯鲟(GUE) | Acipenser gueldenstaedtii | N | 缺乏可用于种类鉴定的基因标记 Lack of markers for species identification | |||
| 俄罗斯鲟 × 西伯利亚鲟 (GUE × BAE) | Acipenser gueldenstaedtii × Acipenser baerii | N | 缺乏可用于种类鉴定的基因标记 Lack of markers for species identification | |||
| 俄罗斯鲟 × 闪光鲟 (GUE × STE) | Acipenser gueldenstaedtii × Acipenser stellatus | N | 正反杂交种均不可鉴定 Reciprocal hybrids cannot be identified | |||
| 纳氏鲟(NAC) | Acipenser naccarii | N | 无法区分NAC和NAC × BAE、NAC × GUE NAC, NAC × BAE and NAC × GUE could not be distinguished | |||
| 纳氏鲟 × 俄罗斯鲟 (NAC × GUE) | Acipenser naccarii × Acipenser gueldenstaedtii | N | 正反杂交种均不可鉴定 Reciprocal hybrids cannot be identified | |||
| 小体鲟(RUT) | Acipenser ruthenus | N | 无法区分RUT和RUT × BAE RUT and RUT × BAE could not be distinguished | |||
| 小体鲟 × 欧洲鳇 (RUT × HUS) | Acipenser ruthenus × Huso huso | Y | 正反杂交均可鉴定, 但贸易中实际为HUS♀ × RUT♂ Both positive and negative hybrids could be identified, but in trade it is HUS♀ × RUT♂ | |||
| 施氏鲟(SCH) | Acipenser schrenckii | N | 无法区分SCH和SCH × BAE SCH and SCH × BAE could not be distinguished | |||
| 闪光鲟(STE) | Acipenser stellatus | N | 无法区分STE和STE × GUE STE and STE × GUE could not be distinguished | |||
| 高首鲟(TRA) | Acipenser transmontanus | Y | 尽管贸易中没有TRA × BAE、TRA × GUE, 但无法区分TRA与两种杂交鲟, 仅可在现有贸易情况下判定其为纯种TRA TRA, TRA × BAE and TRA × GUE could not be distinguished although the two hybrids did not exist in trade, so TRA only be considered pure under the current trade conditions | |||
| 高首鲟 × 欧洲鳇 (TRA × HUS) | Acipenser transmontanus × Huso huso | Y | 正反杂交均可鉴定 Reciprocal hybrids could be identified | |||
| 达氏鳇(DAU) | Huso dauricus | Y | 尽管贸易中没有DAU × BAE、DAU × GUE, 但无法区分DAU与两种杂交鲟, 仅可在现有贸易情况下判定其为纯种DAU DAU, DAU × BAE and DAU × GUE could not be distinguished although the two hybrids did not exist in trade, so DAU only be considered pure under the current trade conditions | |||
| 达氏鳇 × 施氏鲟 (DAU × SCH) | Huso dauricus × Acipenser schrenckii | Y | 正反杂交均可鉴定, 但贸易中实际为DAU♀ × SCH♂ Reciprocal hybrids could be identified, but in trade it is DAU♀ × SCH♂ | |||
| 欧洲鳇(HUS) | Huso huso | N | 无法区分HUS和HUS × BAE HUS and HUS × BAE could not be distinguished | |||
| 欧洲鳇 × 纳氏鲟 (HUS × NAC) | Huso huso × Acipenser naccarii | Y | 正反杂交种均可鉴定 Reciprocal hybrids could be identified | |||
| 欧洲鳇 × 西伯利亚鲟 (HUS × BAE) | Huso huso × Acipenser baerii | N | 正反杂交种均不可鉴定 Reciprocal hybrids can not be identified | |||
| 欧洲鳇 × 施氏鲟 (HUS × SCH) | Huso huso × Acioenser schrenckii | Y | 正反杂交种均可鉴定 Reciprocal hybrids could be identified | |||
| 欧洲鳇 × 闪光鲟 (HUS × STE) | Huso huso × Acipenser stellatus | Y | 正反杂交种均可鉴定 Reciprocal hybrids could be identified | |||
| 富氏鲟(FUL) | Acipenser fulvescens | Y | 尽管贸易中没有FUL × BAE、FUL × GUE、FUL × RUT, 但无法区分FUL与三种杂交鲟, 仅可在现有贸易情况下判定其为纯种FUL FUL, FUL × BAE, FUL × GUE and FUL × RUT could not be distinguished although the three hybrids did not exist in trade, so FUL only be considered pure under the current trade conditions | |||
| 中华鲟(SIN) | Acipenser sinensis | Y | 尽管贸易中没有SIN × BAE、SIN × GUE、SIN × RUT, 但无法区分SIN与三种杂交鲟, 仅可在现有贸易情况下判定其为纯种SIN SIN, SIN × BAE, SIN × GUE and SIN × RUT could not be distinguished although the three hybrids did not exist in trade, so SIN only be considered pure under the current trade conditions | |||
| 尖吻鲟(OXY) | Acipenser oxyrhynchus | Y | 贸易中暂无其杂交鲟, 仅可通过现有引物扩增判断其是否与11种鲟鱼中除BAE和GUE之外的种类杂交产生杂交鲟; 仅可在现有贸易情况下判定其为纯种 These species have not produced hybrid sturgeon at present, and the existing primers can only judge whether these species produce hybrid sturgeon with other species except BAE and GUE among the 11 sturgeon species. So these species can only be considered pure under current trade conditions | |||
| 铲鲟(PLA) | Scaphirhynchus platorynchus | Y | ||||
| 匙吻鲟(SPA) | Polyodon spathula | Y | ||||
| 短吻鲟(BRE) | Acipenser brevirostrum | Y | ||||
| 波斯鲟(PER) | Acipenser persicus | Y | ||||
| 中吻鲟(MED) | Acipenser medirostris | Y | ||||
| 大西洋鲟(STU) | Acipenser sturio | Y | ||||
| 裸腹鲟(NUD) | Acipenser nudiventris | Y | ||||
| 白铲鲟(ALB) | Scaphirhynchus albus | Y | ||||
| 太平洋鲟(MIK) | Acipenser mikadoi | Y | ||||
| 阿姆河大拟铲鲟(KAU) | Pseudoscaphirhynchus kaufmanni | Y | ||||
Table 1 Species and identification feasibility of sturgeons in trade
| 种类a Species | 拉丁名 Latin name | 鉴定可行性b Identification feasibility | 备注 Comment | |||
|---|---|---|---|---|---|---|
| 西伯利亚鲟(BAE) | Acipenser baerii | N | 缺乏可用于种类鉴定的基因标记 Lack of markers for species identification | |||
| 西伯利亚鲟 × 俄罗斯鲟 (BAE × GUE) | Acipenser baerii × Acipenser gueldenstaedtii | N | 缺乏可用于种类鉴定的基因标记 Lack of markers for species identification | |||
| 西伯利亚鲟 × 纳氏鲟 (BAE × NAC) | Acipenser baerii × Acipenser naccarii | N | 正反杂交种均不可鉴定, 但贸易中实际为NAC♀ × BAE♂ Reciprocal hybrids cannot be identified, but in trade it is NAC♀ × BAE♂ | |||
| 西伯利亚鲟 × 施氏鲟 (BAE × SCH) | Acipenser baerii × Acipenser schrenckii | N | 正反杂交种均不可鉴定 Reciprocal hybrids cannot be identified | |||
| 西伯利亚鲟 × 小体鲟 (BAE × RUT) | Acipenser baerii × Acipenser ruthenus | N | 正反杂交种均不可鉴定 Reciprocal hybrids cannot be identified | |||
| 俄罗斯鲟(GUE) | Acipenser gueldenstaedtii | N | 缺乏可用于种类鉴定的基因标记 Lack of markers for species identification | |||
| 俄罗斯鲟 × 西伯利亚鲟 (GUE × BAE) | Acipenser gueldenstaedtii × Acipenser baerii | N | 缺乏可用于种类鉴定的基因标记 Lack of markers for species identification | |||
| 俄罗斯鲟 × 闪光鲟 (GUE × STE) | Acipenser gueldenstaedtii × Acipenser stellatus | N | 正反杂交种均不可鉴定 Reciprocal hybrids cannot be identified | |||
| 纳氏鲟(NAC) | Acipenser naccarii | N | 无法区分NAC和NAC × BAE、NAC × GUE NAC, NAC × BAE and NAC × GUE could not be distinguished | |||
| 纳氏鲟 × 俄罗斯鲟 (NAC × GUE) | Acipenser naccarii × Acipenser gueldenstaedtii | N | 正反杂交种均不可鉴定 Reciprocal hybrids cannot be identified | |||
| 小体鲟(RUT) | Acipenser ruthenus | N | 无法区分RUT和RUT × BAE RUT and RUT × BAE could not be distinguished | |||
| 小体鲟 × 欧洲鳇 (RUT × HUS) | Acipenser ruthenus × Huso huso | Y | 正反杂交均可鉴定, 但贸易中实际为HUS♀ × RUT♂ Both positive and negative hybrids could be identified, but in trade it is HUS♀ × RUT♂ | |||
| 施氏鲟(SCH) | Acipenser schrenckii | N | 无法区分SCH和SCH × BAE SCH and SCH × BAE could not be distinguished | |||
| 闪光鲟(STE) | Acipenser stellatus | N | 无法区分STE和STE × GUE STE and STE × GUE could not be distinguished | |||
| 高首鲟(TRA) | Acipenser transmontanus | Y | 尽管贸易中没有TRA × BAE、TRA × GUE, 但无法区分TRA与两种杂交鲟, 仅可在现有贸易情况下判定其为纯种TRA TRA, TRA × BAE and TRA × GUE could not be distinguished although the two hybrids did not exist in trade, so TRA only be considered pure under the current trade conditions | |||
| 高首鲟 × 欧洲鳇 (TRA × HUS) | Acipenser transmontanus × Huso huso | Y | 正反杂交均可鉴定 Reciprocal hybrids could be identified | |||
| 达氏鳇(DAU) | Huso dauricus | Y | 尽管贸易中没有DAU × BAE、DAU × GUE, 但无法区分DAU与两种杂交鲟, 仅可在现有贸易情况下判定其为纯种DAU DAU, DAU × BAE and DAU × GUE could not be distinguished although the two hybrids did not exist in trade, so DAU only be considered pure under the current trade conditions | |||
| 达氏鳇 × 施氏鲟 (DAU × SCH) | Huso dauricus × Acipenser schrenckii | Y | 正反杂交均可鉴定, 但贸易中实际为DAU♀ × SCH♂ Reciprocal hybrids could be identified, but in trade it is DAU♀ × SCH♂ | |||
| 欧洲鳇(HUS) | Huso huso | N | 无法区分HUS和HUS × BAE HUS and HUS × BAE could not be distinguished | |||
| 欧洲鳇 × 纳氏鲟 (HUS × NAC) | Huso huso × Acipenser naccarii | Y | 正反杂交种均可鉴定 Reciprocal hybrids could be identified | |||
| 欧洲鳇 × 西伯利亚鲟 (HUS × BAE) | Huso huso × Acipenser baerii | N | 正反杂交种均不可鉴定 Reciprocal hybrids can not be identified | |||
| 欧洲鳇 × 施氏鲟 (HUS × SCH) | Huso huso × Acioenser schrenckii | Y | 正反杂交种均可鉴定 Reciprocal hybrids could be identified | |||
| 欧洲鳇 × 闪光鲟 (HUS × STE) | Huso huso × Acipenser stellatus | Y | 正反杂交种均可鉴定 Reciprocal hybrids could be identified | |||
| 富氏鲟(FUL) | Acipenser fulvescens | Y | 尽管贸易中没有FUL × BAE、FUL × GUE、FUL × RUT, 但无法区分FUL与三种杂交鲟, 仅可在现有贸易情况下判定其为纯种FUL FUL, FUL × BAE, FUL × GUE and FUL × RUT could not be distinguished although the three hybrids did not exist in trade, so FUL only be considered pure under the current trade conditions | |||
| 中华鲟(SIN) | Acipenser sinensis | Y | 尽管贸易中没有SIN × BAE、SIN × GUE、SIN × RUT, 但无法区分SIN与三种杂交鲟, 仅可在现有贸易情况下判定其为纯种SIN SIN, SIN × BAE, SIN × GUE and SIN × RUT could not be distinguished although the three hybrids did not exist in trade, so SIN only be considered pure under the current trade conditions | |||
| 尖吻鲟(OXY) | Acipenser oxyrhynchus | Y | 贸易中暂无其杂交鲟, 仅可通过现有引物扩增判断其是否与11种鲟鱼中除BAE和GUE之外的种类杂交产生杂交鲟; 仅可在现有贸易情况下判定其为纯种 These species have not produced hybrid sturgeon at present, and the existing primers can only judge whether these species produce hybrid sturgeon with other species except BAE and GUE among the 11 sturgeon species. So these species can only be considered pure under current trade conditions | |||
| 铲鲟(PLA) | Scaphirhynchus platorynchus | Y | ||||
| 匙吻鲟(SPA) | Polyodon spathula | Y | ||||
| 短吻鲟(BRE) | Acipenser brevirostrum | Y | ||||
| 波斯鲟(PER) | Acipenser persicus | Y | ||||
| 中吻鲟(MED) | Acipenser medirostris | Y | ||||
| 大西洋鲟(STU) | Acipenser sturio | Y | ||||
| 裸腹鲟(NUD) | Acipenser nudiventris | Y | ||||
| 白铲鲟(ALB) | Scaphirhynchus albus | Y | ||||
| 太平洋鲟(MIK) | Acipenser mikadoi | Y | ||||
| 阿姆河大拟铲鲟(KAU) | Pseudoscaphirhynchus kaufmanni | Y | ||||
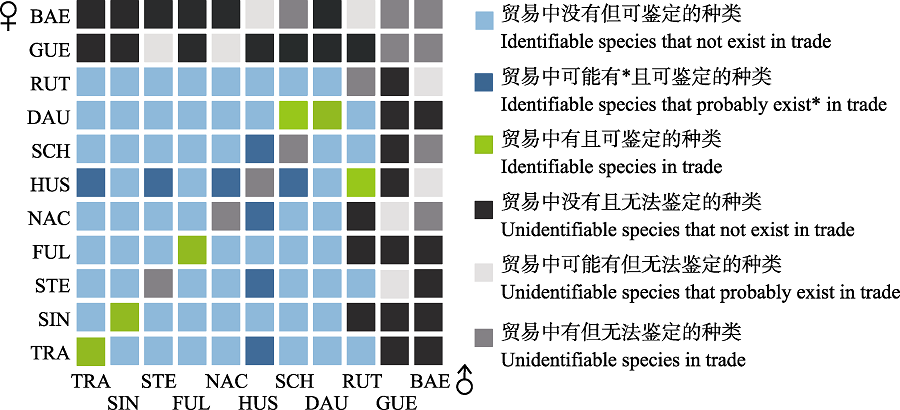
Fig. 3 Identifiability of sturgeons with parents from the 11 species. Probably exist* means that only the parent species of this hybrid is recorded in the CITES trade database, but the male parent and female parent are not marked, so it is uncertain if this hybrid is existing in actual trade. Please refer to Fig. 1 for the full names of species abbreviation in the figure.
| [1] |
Artyukhin EN, Vecsei P, Peterson DL (2007) Morphology and ecology of Pacific sturgeons. Environmental Biology of Fishes, 79, 369-381.
DOI URL |
| [2] | Barmintseva AE, Mugue NS (2013) The use of microsatellite loci for identification of sturgeon species (Acipenseridae) and hybrid forms. Russian Journal of Genetics, 49, 1093- 1105. |
| [3] |
Birstein VJ, Desalle R, Doukakis P, Hanner R, Ruban GI, Wong E (2009) Testing taxonomic boundaries and the limit of DNA barcoding in the Siberian sturgeon, Acipenser baerii. Mitochondrial DNA, 20, 110-118.
DOI PMID |
| [4] |
Birstein VJ, Doukakis P, DeSalle R (2000) Polyphyly of mtDNA lineages in the Russian sturgeon, Acipenser gueldenstaedtii: Forensic and evolutionary implications. Conservation Genetics, 1, 81-88.
DOI URL |
| [5] |
Birstein VJ, Doukakis P, Sorkin B, Desalle R (2010) Population aggregation analysis of three caviar‐producing species of sturgeons and implications for the species identification of Black caviar. Conservation Biology, 12, 766-775.
DOI URL |
| [6] |
Boscari E, Barmintseva A, Pujolar JM, Doukakis P, Mugue N, Congiu L (2014) Species and hybrid identification of sturgeon caviar: A new molecular approach to detect illegal trade. Molecular Ecology Resources, 14, 489-498.
DOI PMID |
| [7] |
Boscari E, Barmintseva A, Zhang SY, Yue HM, Li CJ, Shedko SV, Lieckfeldt D, Ludwig A, Wei QW, Mugue NS, Congiu L (2017) Genetic identification of the caviar-producing Amur and Kaluga sturgeons revealed a high level of concealed hybridization. Food Control, 82, 243-250.
DOI URL |
| [8] | Chen GZ, Zheng HX, Zhu CQ (2018) Establishment of the detection method of TaqMan real-time PCR for Acipenser sinensis. Jiangsu Journal of Agricultural Sciences, 34, 399- 403. (in Chinese with English abstract) |
| [陈光哲, 郑红霞, 祝长青 (2018) 中华鲟TaqMan实时荧光PCR检测方法的建立. 江苏农业学报, 34, 399-403.] | |
| [9] | Chen XH (2007) Biology and Resources Status of Acipenseriformes Species, China Ocean Press, Beijing. (in Chinese) |
| [陈细华 (2007) 鲟形目鱼类生物学与资源现状. 海洋出版社, 北京.] | |
| [10] | Cheng PL, Yu D, Liu HZ, Du H, Wei QW (2021) Molecular phylogeny of Acipenserformes based on complete mitochondrial genome sequence. Acta Hydrobiologica Sinica, 45, 487-495. (in Chinese with English abstract) |
| [程佩琳, 俞丹, 刘焕章, 杜浩, 危起伟 (2021) 基于线粒体基因组全序列的鲟形目鱼类(Pisces: Acipenseriformes)的分子系统发育重建. 水生生物学报, 45, 487-495.] | |
| [11] | CITES (Convention on International Trade in Endangered Species of Wild Fauna and Flora)(2001) The CITES identification guide-sturgeons and paddlefish. https://cites.org/sites/default/files/eng/notif/2001/094.shtml. (accessed on 2021-07-13) |
| [12] | CITES (Convention on International Trade in Endangered Species of Wild Fauna and Flora)(2020) AC31 Doc.16: Identification and Traceability of Sturgeons and Paddlefish (Acipenseriformes spp.). https://cites.org/sites/default/files/eng/com/ac/31/Docs/E-AC31-16.pdf. (accessed on 2021-07- 13) |
| [13] | CITES (Convention on International Trade in Endangered Species of Wild Fauna and Flora)(2021) International sturgeon Trade Data 2010-2020. https://trade.cites.org/en/cites_trade/download?filters%5Btime_range_start%5D=2010&filters%5Btime_range_end%5D=2020&filters%5Bexporters_ids%5D%5B%5D=all_exp&filters%5Bimporters_ids%5D%5B%5D=all_imp&filters%5Bsources_ids%5D%5B%5D=all_sou&filters%5Bpurposes_ids%5D%5B%5D=all_pur&filters%5Bterms_ids%5D%5B%5D=all_ter&filters%5Btaxon_concepts_ids%5D%5B%5D=102&filters%5Breset%5D=&filters%5Bselection_taxon%5D=taxonomic_cascade&web_disabled=true. (accessed on 2021-06-10) |
| [14] |
Comincini S, Lanfredi M, Rossi R, Fontana F (1998) Use of RAPD markers to determine the genetic relationships among sturgeons (Acipenseridae, Pisces). Fisheries Science, 64, 35-38.
DOI URL |
| [15] |
Congiu L, Dupanloup I, Patarnello T, Fontana F, Rossi R, Arlati G, Zane L (2010) Identification of interspecific hybrids by amplified fragment length polymorphism: The case of sturgeon. Molecular Ecology, 10, 2355-2359.
DOI URL |
| [16] |
Debus L, Winkler M, Billard R (2002) Structure of micropyle surface on oocytes and caviar grains in sturgeons. International Review of Hydrobiology, 87, 585-603.
DOI URL |
| [17] |
Desalle R, Birstein VJ (1996) PCR identification of black caviar. Nature, 381, 197-198.
DOI URL |
| [18] | Ding J, Liu SY, Su W, Sun XF, Lin CJ, Wu B, Jiang D (2018) A rapid quantitative real-time PCR method for identification of Acipenser ruthenus. Journal of Molecular Science, 34, 144-148. (in Chinese with English abstract) |
| [丁健, 刘淑艳, 苏旺, 孙晓飞, 林长军, 吴斌, 蒋丹 (2018) 利用实时荧光 PCR鉴定小体鲟物种的快速方法. 分子科学学报, 34, 144-148.] | |
| [19] | Dong CJ, Liu YY, Liu XY, Song YN, Xu P, Sun XW (2014) PCR-RFLP analysis of mtDNA to identify four kinds of sturgeons. Biotechnology Bulletin, (12), 78-85. (in Chinese with English abstract) |
|
[董传举, 刘园园, 刘晓勇, 宋迎楠, 徐鹏, 孙效文 (2014) 四种鲟鱼线粒体PCR-RFLP鉴定方法的研究. 生物技术通报, (12), 78-85.]
DOI |
|
| [20] |
Doukakis P, Pikitch EK, Rothschild A, DeSalle R, Amato G, Kolokotronis SO (2012) Testing the effectiveness of an international conservation agreement: Marketplace forensics and CITES caviar trade regulation. PLoS ONE, 7, e40907.
DOI URL |
| [21] | Gilkolaei R (1997) Molecular Population Genetic Studies of Sturgeon Species in the South Caspian Sea. PhD dissertation, Swansea University, Swansea, England. |
| [22] | Guo XH, Dong Y, Hu HX, Zhao YZ (2014) Research progresses of germplasm identification methods in sturgeons. Biotechnology Bulletin, (2), 56-63. (in Chinese with English abstract) |
| [郭向贺, 董颖, 胡红霞, 赵艳珍 (2014) 鲟鱼种质鉴定方法研究进展. 生物技术通报, (2), 56-63.] | |
| [23] |
Havelka M, Boscari E, Sergeev A, Mugue N, Congiu L, Arai K (2019) A new marker, isolated by ddRAD sequencing, detects Siberian and Russian sturgeon in hybrids. Animal Genetics, 50, 115-116.
DOI PMID |
| [24] |
Havelka M, Fujimoto T, Hagihara S, Adachi S, Arai K (2017) Nuclear DNA markers for identification of Beluga and Sterlet sturgeons and their interspecific Bester hybrid. Scientific Reports, 7, 1694.
DOI PMID |
| [25] | Hu J, Wang DQ, Wei QW, Shen L (2010) Molecular identification of Amur sturgeon (Acipenser schrenckii), Kaluga (Huso dauricus) and their reciprocal hybrids. Journal of Fishery Sciences of China, 17(1), 21-30. (in Chinese with English abstract) |
| [胡佳, 汪登强, 危起伟, 沈丽 (2010) 施氏鲟、达氏鳇及其杂交子代的分子鉴定. 中国水产科学, 17(1), 21-30.] | |
| [26] | Hu Y, Liu X, Yang J, Xiao K, Du H (2019) Development of twenty-two novel cross-species microsatellite markers for Amur sturgeon (Acipenser schrenckii) from Chinese sturgeon (Acipenser sinensis) via next-generation sequencing. Turkish Journal of Fisheries and Aquatic Sciences, 19, 85- 88. |
| [27] | Hubert N, Hanner R, Holm E, Mandrak NE, Taylor E, Burridge M, Watkinson D, Dumont P, Curry A, Bentzen P (2008) Identifying Canadian freshwater fishes through DNA barcodes. PLoS Biology, 3, e2490. |
| [28] |
Jenneckens I, Meyer JN, Horstgen-Schwarkk G, May B, Debus L, Wedekind H, Ludwig A (2001) A fixed allele at microsatellite locus LS-39 exhibiting species‐specificity for the black caviar producer Acipenser stellatus. Journal of Applied Ichthyology, 17, 39-42.
DOI URL |
| [29] |
Johnson TA, Iyengar A (2015) Phylogenetic evidence for a case of misleading rather than mislabeling in caviar in the United Kingdom. Journal of Forensic Sciences, 60, S248-S253.
DOI URL |
| [30] | Kong J, Li SK, Wang JL, Zeng SY, Liu T (2020) Application of germplasm identification in Amur sturgeon (Acipenser schrenckii), Siberian sturgeon (Acipenser baerii), and Kaluga (Huso dauricus), and their hybrids. Journal of Fishery Sciences of China, 27, 771-778. (in Chinese with English abstract) |
| [孔杰, 李世凯, 王金乐, 曾诗雨, 刘霆 (2020) 施氏鲟、西伯利亚鲟、达氏鳇及其杂交种种质鉴定方法的建立及其应用. 中国水产科学, 27, 771-778.] | |
| [31] | Liu Y, Lu CY, Li C, Cheng L, Wu WH, Zhao YF, Sun XW (2015) Genetic identification of Amur sturgeon, Kaluga and their hybrids based on microsatellite markers. Chinese Journal of Fisheries, 28(1), 18-23. (in Chinese with English abstract) |
| [刘洋, 鲁翠云, 李超, 程磊, 吴文化, 赵元凤, 孙效文 (2015) 应用微卫星标记鉴别施氏鲟、达氏鳇及其杂交子代. 水产学杂志, 28(1), 18-23.] | |
| [32] |
Ludwig A (2010) Identification of Acipenseriformes species in trade. Journal of Applied Ichthyology, 24, 2-19.
DOI URL |
| [33] | Ludwig A, Boner M, Bonscari E, Congiu L, Gessner J, Jahri J, Striebel B (2021) Identification of species and hybrids, source and geographical origin of sturgeon and paddlefish (Acipenseriformes spp.) specimens and products in trade. https://cites.org/sites/default/files/eng/com/ac/31/Docs/E-AC31-16-Add.pdf. (accessed on 2021-07-13) |
| [34] |
Ludwig A, Lieckfeldt D, Jahrl J (2015) Mislabeled and counterfeit sturgeon caviar from Bulgaria and Romania. Journal of Applied Ichthyology, 31, 587-591.
DOI URL |
| [35] |
Ludwig A, Lutz D, Jenneckens I (2002) A molecular approach to control the international trade in black caviar. International Review of Hydrobiology, 87, 661-674.
DOI URL |
| [36] |
Ludwig A, May B (2000) Heteroplasmy in the mtDNA control region of sturgeon (Acipenser, Huso and Scaphirhynchus). Genetics, 156, 1933-1947.
DOI PMID |
| [37] | May B, Krueger CC, Kincaid HL (1997) Genetic variation at microsatellite loci in sturgeon: Primer sequence homology in Acipenser and Scaphirhynchus. Canadian Journal of Fisheries & Aquatic Sciences, 54, 1542-1547. |
| [38] |
Mugue NS, Barmintseva AE, Rastorguev SM, Mugue VN, Barmintsev VA (2008) Polymorphism of the mitochondrial DNA control region in eight sturgeon species and development of a system for DNA-based species identification. Russian Journal of Genetics, 44, 793-798.
DOI URL |
| [39] | Peng T, Wang NM, Tong GX, Qu QZ, Qi Q, Suo L, Sun DJ (2009) Cross-amplifications of microsatellite markers of lake sturgeon in three kinds of sturgeons and their hybrids. Chinese Journal of Fisheries, 22(2), 12-16. (in Chinese with English abstract) |
| [彭涛, 王念民, 佟广香, 曲秋芝, 齐茜, 索力, 孙大江 (2009) 湖鲟微卫星引物在三种鲟鱼及杂交子代的通用性研究. 水产学杂志, 22(2), 12-16.] | |
| [40] | Rehbein H, Gonzales-Sotelo C, Perez-Martin RI, Quinteiro J, Santos AT (1999) Differentiation of sturgeon caviar by single strand conformation polymorphism (PCR-SSCP) analysis. Archiv Fur Lebensmittelhygiene, 50, 13-17. |
| [41] | Shao ZJ, Zhao N, Zhu B, Zhou FL, Chang JB (2002) Applicability of microsatellite primers developed from shovelnose sturgeon in Chinese sturgeon. Acta Hydrobiologica Sinica, 26, 577-584. (in Chinese with English abstract) |
| [邵昭君, 赵娜, 朱滨, 周发林, 常剑波 (2002) 铲鲟微卫星引物对中华鲟的适用性研究. 水生生物学报, 26, 577-584.] | |
| [42] | Wan C, Yang AF, Dai D, Qu F, Xu JY, Jia Y, Wang L, Yang Y(2021) A double fluorescence PCR system and its application for simultaneous detection of Acipenser baerii and Acipenser stellatus: China, CN112831571A. (in Chinese) |
| [万超, 杨爱馥, 代弟, 屈菲, 徐君怡, 贾赟, 王雷, 杨宇 (2021) 一种同时检测闪光鲟、西伯利亚鲟物种双染料荧光PCR特异性检测体系及应用: 中国, CN1128 31571A.] | |
| [43] |
Wang BY, Wang B, Chen W, Li BS (2020) Current situation and developmental trend of sturgeon culture industry in China. Open Journal of Fisheries Research, 7, 107-114. (in Chinese with English abstract)
DOI URL |
| [王保友, 王斌, 陈玮, 李宝山 (2020) 我国鲟鱼养殖产业现状及发展趋势. 水产研究, 7, 107-114.] | |
| [44] |
Wolf C, Hübner P, Lüthy J (1999) Differentiation of sturgeon species by PCR-RFLP. Food Research International, 32, 699-705.
DOI URL |
| [45] | Yang SY, Zhao ZM, Liu Z, Huang XL, Miao Y, Du ZJ, Luo W, Chen H, Chen DF, Feng Y(2018) Identification of Acipenser baerii and hybrid: Acipenser baerii ♀ × Acipenser schrenckii ♂: China, CN108660223A. (in Chinese) |
| [杨世勇, 赵仲孟, 刘钊, 黄小丽, 苗懿, 杜宗君, 罗伟, 陈虎, 陈德芳, 冯杨 西伯利亚鲟及以西杂的鉴定方法: 中国, CN108660223A.] | |
| [46] |
Zhang X, Tinacci L, Xie SZ, Wang JH, Ying XG, Wen J, Armani A (2021) Caviar products sold on Chinese business to customer (B2C) online platforms: Labelling assessment supported by molecular identification. Food Control, 131, 108370.
DOI URL |
| [47] | Zhang Y, Liu XY, Qu QZ, Sun DJ (2012) Comparative discriminate analysis of morphological traits among Acipenser schrenckii, Huso dauricus and their hybrid (A. schrenckii (♂) × H. dauricus (♀)). Freshwater Fisheries, 42, 27-32. (in Chinese with English abstract) |
| [张颖, 刘晓勇, 曲秋芝, 孙大江 (2012) 达氏鳇, 施氏鲟及其杂交种(施氏鲟♂ × 达氏鳇♀)形态差异与判别分析. 淡水渔业, 42, 27-32.] |
| [1] | Keyi Wu, Wenda Ruan, Difeng Zhou, Qingchen Chen, Chengyun Zhang, Xinyuan Pan, Shang Yu, Yang Liu, Rongbo Xiao. Syllable clustering analysis-based passive acoustic monitoring technology and its application in bird monitoring [J]. Biodiv Sci, 2023, 31(1): 22370-. |
| [2] | Xiangxiang Chen, Zhongshuai Gai, Juntuan Zhai, Jindong Xu, Peipei Jiao, Zhihua Wu, Zhijun Li. Genetic diversity and construction of core conservation units of the natural populations of Populus euphratica in Northwest China [J]. Biodiv Sci, 2021, 29(12): 1638-1649. |
| [3] | Jiaxin Kong, Zhaochen Zhang, Jian Zhang. Classification and identification of plant species based on multi-source remote sensing data: Research progress and prospect [J]. Biodiv Sci, 2019, 27(7): 796-812. |
| [4] | Jian Zhang, Lu Dong, Yanyun Zhang. Population genetic structure of Adélie penguins (Pygoscelis adeliae) from Inexpressible Island, Antarctica, using SNP markers [J]. Biodiv Sci, 2019, 27(12): 1291-1297. |
| [5] | Morigengaowa , Hui Shang, Baodong Liu, Ming Kang, Yuehong Yan. One or more species? GBS sequencing and morphological traits evidence reveal species diversification of Sphaeropteris brunoniana in China [J]. Biodiv Sci, 2019, 27(11): 1196-1204. |
| [6] | Qingqing Liu, Zhijun Dong. Population genetic structure of Gonionemus vertens based on the mitochondrial COI sequence [J]. Biodiv Sci, 2018, 26(11): 1204-1211. |
| [7] | Yun Cao, Wenjing Shen, Lian Chen, Feilong Hu, Lei Zhou, Haigen Xu. Application of metabarcoding technology in studies of fungal diversity [J]. Biodiv Sci, 2016, 24(8): 932-939. |
| [8] | Jie Yang, Jia He, Danbi Wang, En Shi, Wenyu Yang, Qifang Geng, Zhongsheng Wang*. Progress in research and application of InDel markers [J]. Biodiv Sci, 2016, 24(2): 237-243. |
| [9] | Tai Wang, Yanping Zhang, Lihong Guan, Yanyan Du, Zhongyu Lou, Wenlong Jiao. Current freshwater fish resources and the application of DNA barcoding in species identification in Gansu Province [J]. Biodiv Sci, 2015, 23(3): 306-313. |
| [10] | Dangni Zhang, Lianming Zheng, Jinru He, Wenjing Zhang, Yuanshao Lin, Yang Li. DNA barcoding of hydromedusae in northern Beibu Gulf for species identification [J]. Biodiv Sci, 2015, 23(1): 50-60. |
| [11] | Hui Wang, Baowei Zhang, Wenbo Shi, Xia Luo, Lizhi Zhou, Demin Han, Qing Chang. Structural characteristics of di-nucleotide/tetra-nucleotide repeat microsatellite DNA in Pachyhynobius shangchengensisgenomes and its effect on isolation [J]. Biodiv Sci, 2012, 20(1): 51-58. |
| [12] | Chaolun Li, Minxiao Wang, Fangping Cheng, Song Sun. DNA barcoding and its application to marine zooplankton ecology [J]. Biodiv Sci, 2011, 19(6): 805-814. |
| [13] | Haifeng Gu, Tingting Liu, Dongzhao Lan. Progress of dinoflagellate cyst research in the China seas [J]. Biodiv Sci, 2011, 19(6): 779-786. |
| [14] | Yan Liu, Jianxiu Wang, Xuejun Ge, Tong Cao. The rps4 locus as an alternative marker for barcoding bryophytes: eva- luation based on data mining from GenBank [J]. Biodiv Sci, 2011, 19(3): 311-318. |
| [15] | Lingliang Cao, Lizhi Zhou, Baowei Zhang. Genetic patterns of an invasive Procambarus clarkii population in the three river basins of Anhui Province [J]. Biodiv Sci, 2010, 18(4): 398-407. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()