



生物多样性 ›› 2018, Vol. 26 ›› Issue (5): 445-456. DOI: 10.17520/biods.2018058 cstr: 32101.14.biods.2018058
所属专题: 传粉生物学; 物种形成与系统进化; 昆虫多样性与生态功能
收稿日期:2018-02-15
接受日期:2018-05-11
出版日期:2018-05-20
发布日期:2018-09-11
通讯作者:
周欣
作者简介:# 共同第一作者
基金资助:
Dandan Lang1,2, Min Tang1,3, Xin Zhou1,3,*( )
)
Received:2018-02-15
Accepted:2018-05-11
Online:2018-05-20
Published:2018-09-11
Contact:
Zhou Xin
About author:# Co-first authors
摘要:
传粉者是重要的生态功能提供者, 在维持稳定的生态系统和高效的农业生产力中发挥着重要作用。因此, 传粉网络的构建和监测工作对评价生态系统平衡和调控农业生产至关重要。该工作的基础就是通过对传粉者及植物的物种鉴定构建其相关性。传统的形态学物种鉴定对分类学专家的专业知识、时间和经验都提出较高的要求。DNA条形码和高通量测序技术(high-throughput sequencing, HTS)的发展及其在传粉网络研究中的应用, 提供了高效、准确鉴定传粉者与植物的方法, 大大提高了传粉网络构建的效率。本文阐述了传粉网络研究相关的研究方法和技术进展, 并提出利用高通量测序技术结合无PCR扩增(PCR-free)的“超级条形码”技术, 有望实现以更高的灵敏度和分辨率对混合物种样品进行定性及相对定量的监测。该方法的有效性已在其他生物多样性研究中得以验证, 在传粉网络研究中虽处于初始阶段, 但应用前景广阔。
郎丹丹, 唐敏, 周欣 (2018) 传粉网络构建的定性定量分子研究: 应用与展望. 生物多样性, 26, 445-456. DOI: 10.17520/biods.2018058.
Dandan Lang,Min Tang,Xin Zhou (2018) Qualitative and quantitative molecular construction of plant-pollinator network: Application and prospective. Biodiversity Science, 26, 445-456. DOI: 10.17520/biods.2018058.
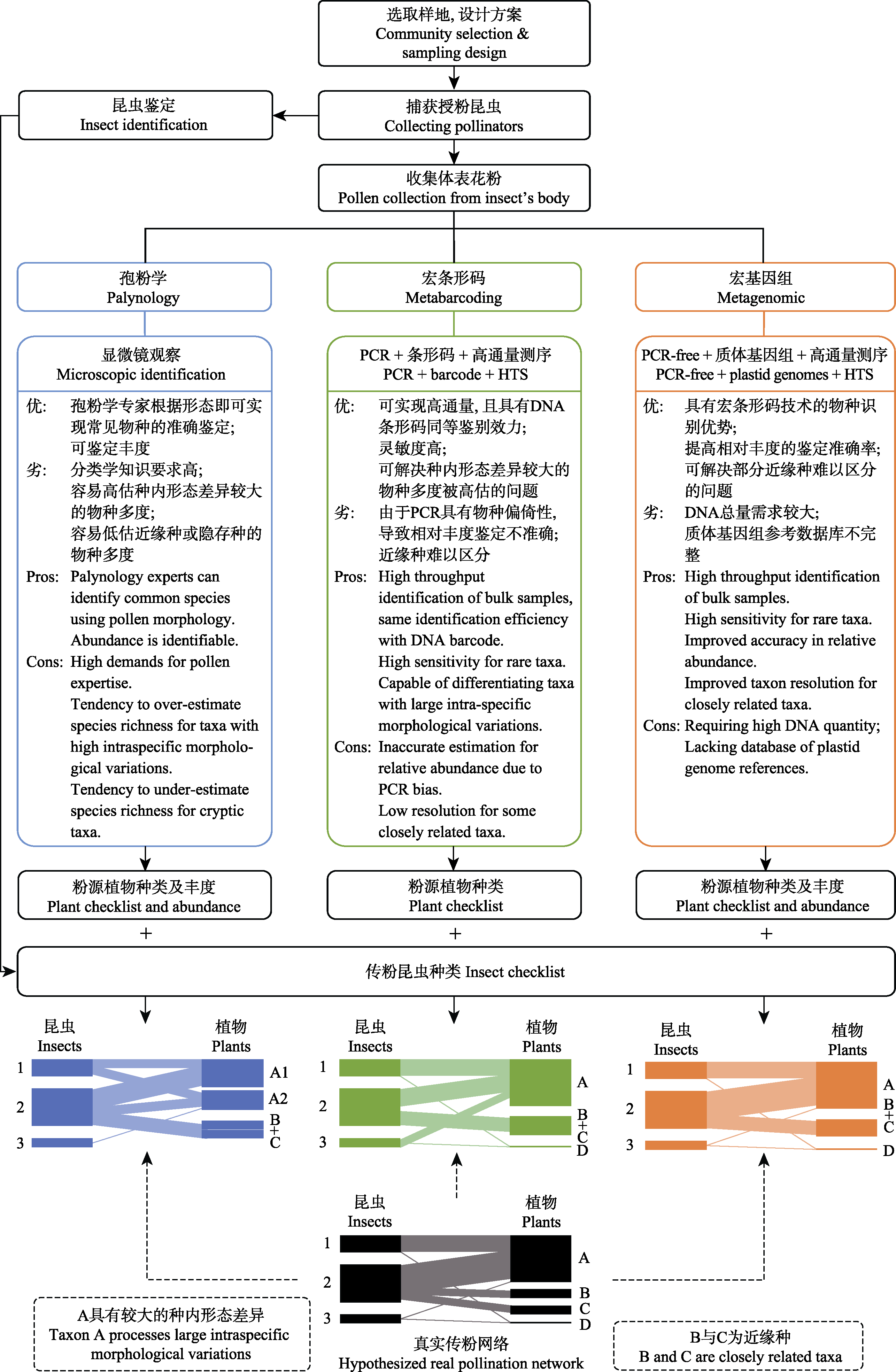
图1 传粉网络构建流程及混合花粉组成分析方法比较。图中蓝色框和绿色框中内容分别为形态学与宏条形码方法, 橙色框中内容为宏基因组方法。底部黑色网络为真实传粉网络模型, 蓝、绿和橙色网络模型分别代表经对应方法分析可能得到的网络结构。由于各种方法都有其局限性, 所得网络均与真实网络存在一定差异。宏条形码和宏基因组技术可解决孢粉学种内形态差异的问题, 但仍存在部分近缘种难以区分的情况。相对于宏条形码技术, 宏基因组技术可避免PCR带来的物种偏倚性, 提高相对丰度的准确率。
Fig. 1 Construction of pollination network and comparison of analysis methods of mixed pollen composition. The contents of the blue and the green boxes refer to morphological and molecular methods, respectively. The content in the orange box is the PCR-free genome-skimming (metagenomics) approach proposed in this paper. The black network model at the bottom represents the real pollination network, and the blue, green and orange network models represent network structures constructed by the corresponding methods. Due to the limitations of various methods, the constructed networks are potentially deviated from the real network. Metabarcoding and metagenomic techniques can alleviate issues caused by intra-specific morphological variations; but some closely related species remain difficult to differentiate. Compared to metabarcoding, the metagenomic technology can reduce species bias caused by PCR and improve the accuracy in relative abundance.
| 物种 Species | NCBI序列号 NCBI accession number | 物种 Species | NCBI序列号 NCBI accession number |
|---|---|---|---|
| Apis andreniformis | KF736157.1 | Bombus lapidarius | KT164641.1 |
| Apis cerana | NC_014295.1 | Bombus lucorum | KT164681.1 |
| Apis dorsata | KC294229.1 | Bombus pascuorum | KT164630.1 |
| Apis florea | NC_021401.1 | Bombus terrestris | KT368150.1 |
| Apis mellifera sahariensis | NC_035883.1 | Melipona bicolor | NC_004529.1 |
| Apis nigrocincta | KY799147.1 | Melipona scutellaris | NC_026198.1 |
| Bombus breviceps | MF478986.1 | Nomada fabriciana | KT164663.1 |
| Bombus consobrinus | MF995069.1 | Nomada flava | KT164670.1 |
| Bombus hypocrita sapporensis | NC_011923.1 | Nomada flavoguttata | KT164617.1 |
| Bombus ignitus | NC_010967.1 | Nomada goodeniana | KT164660.1 |
表1 蜜蜂科20个物种(包括蜜蜂属6个物种)列表及其线粒体基因组NCBI序列号
Table 1 The list of 20 species of Apidae (including six species of Apis) and their mitochondrial genomes’ accession numbers in NCBI
| 物种 Species | NCBI序列号 NCBI accession number | 物种 Species | NCBI序列号 NCBI accession number |
|---|---|---|---|
| Apis andreniformis | KF736157.1 | Bombus lapidarius | KT164641.1 |
| Apis cerana | NC_014295.1 | Bombus lucorum | KT164681.1 |
| Apis dorsata | KC294229.1 | Bombus pascuorum | KT164630.1 |
| Apis florea | NC_021401.1 | Bombus terrestris | KT368150.1 |
| Apis mellifera sahariensis | NC_035883.1 | Melipona bicolor | NC_004529.1 |
| Apis nigrocincta | KY799147.1 | Melipona scutellaris | NC_026198.1 |
| Bombus breviceps | MF478986.1 | Nomada fabriciana | KT164663.1 |
| Bombus consobrinus | MF995069.1 | Nomada flava | KT164670.1 |
| Bombus hypocrita sapporensis | NC_011923.1 | Nomada flavoguttata | KT164617.1 |
| Bombus ignitus | NC_010967.1 | Nomada goodeniana | KT164660.1 |
| 物种 Species | NCBI序列号 NCBI accession number | 物种 Species | NCBI序列号 NCBI accession number |
|---|---|---|---|
| Anoectochilus emeiensis | NC_033895.1 | Dendrobium parciflorum | NC_035334.1 |
| Apostasia odorata | NC_030722.1 | Dendrobium parishii | NC_035339.1 |
| Bletilla ochracea | NC_029483.1 | Dendrobium pendulum | NC_029705.1 |
| Bletilla striata | NC_028422.1 | Dendrobium primulinum | NC_035321.1 |
| Calanthe triplicata | NC_024544.1 | Dendrobium salaccense | NC_035332.1 |
| Cattleya crispata | NC_026568.1 | Dendrobium spatella | NC_035333.1 |
| Cattleya liliputana | NC_032083.1 | Dendrobium strongylanthum | NC_027691.1 |
| Cephalanthera longifolia | NC_030704.1 | Dendrobium wardianum | NC_035329.1 |
| Cymbidium aloifolium | NC_021429.1 | Dendrobium wilsonii | NC_035330.1 |
| Cymbidium ensifolium | NC_028525.1 | Dendrobium xichouense | NC_035341.1 |
| Cymbidium faberi | NC_027743.1 | Elleanthus sodiroi | NC_027266.1 |
| Cymbidium goeringii | NC_028524.1 | Epipactis mairei | NC_030705.1 |
| Cymbidium kanran | NC_029711.1 | Epipactis veratrifolia | NC_030708.1 |
| Cymbidium lancifolium | NC_029712.1 | Erycina pusilla | NC_018114.1 |
| Cymbidium macrorhizon | NC_029713.1 | Gastrochilus fuscopunctatus | NC_035830.1 |
| Cymbidium mannii | NC_021433.1 | Gastrochilus japonicus | NC_035833.1 |
| Cymbidium sinense | NC_021430.1 | Goodyera fumata | NC_026773.1 |
| Cymbidium tortisepalum | NC_021431.1 | Goodyera procera | NC_029363.1 |
| Cymbidium tracyanum | NC_021432.1 | Goodyera schlechtendaliana | NC_029364.1 |
| Cypripedium formosanum | NC_026772.1 | Goodyera velutina | NC_029365.1 |
| Cypripedium macranthos | NC_024421.1 | Habenaria pantlingiana | NC_026775.1 |
| Dendrobium aphyllum | NC_035322.1 | Habenaria radiata | NC_035834.1 |
| Dendrobium brymerianum | NC_035323.1 | Listera fugongensis | NC_030711.1 |
| Dendrobium catenatum | NC_024019.1 | Ludisia discolor | NC_030540.1 |
| Dendrobium chrysanthum | NC_035336.1 | Masdevallia coccinea | NC_026541.1 |
| Dendrobium chrysotoxum | NC_028549.1 | Masdevallia picturata | NC_026777.1 |
| Dendrobium crepidatum | NC_035331.1 | Neottia ovate | NC_030712.1 |
| Dendrobium denneanum | NC_035324.1 | Neottia pinetorum | NC_030710.1 |
| Dendrobium devonianum | NC_035325.1 | Oberonia japonica | NC_035832.1 |
| Dendrobium ellipsophyllum | NC_035340.1 | Paphiopedilum armeniacum | NC_026779.1 |
| Dendrobium exile | NC_035343.1 | Paphiopedilum niveum | NC_026776.1 |
| Dendrobium falconeri | NC_035326.1 | Pelatantheria scolopendrifolia | NC_035829.1 |
| Dendrobium fanjingshanense | NC_035344.1 | Phalaenopsis equestris | NC_017609.1 |
| Dendrobium fimbriatum | NC_035342.1 | Phragmipedium longifolium | NC_028149.1 |
| Dendrobium gratiosissimum | NC_035327.1 | Sobralia callosa | NC_028147.1 |
| Dendrobium henryi | NC_035335.1 | Thrixspermum japonicum | NC_035831.1 |
| Dendrobium hercoglossum | NC_035328.1 | Vanilla aphylla | NC_035320.1 |
| Dendrobium huoshanense | NC_028430.1 | Vanilla planifolia | NC_026778.1 |
| Dendrobium jenkinsii | NC_035337.1 | Sobralia aff. bouchei | NC_028209.1 |
| Dendrobium lohohense | NC_035338.1 | Phalaenopsis hybrid | NC_025593.1 |
| Dendrobium moniliforme | NC_035154.1 | Phalaenopsis aphrodite formosana | NC_007499.1 |
| Dendrobium nobile | NC_029456.1 | Oncidium hybrid | NC_014056.1 |
表2 兰科84个物种(包括石斛兰属31个物种)列表及其叶绿体基因组NCBI序列号
Table 2 The list of 84 species of Orchidaceae (including 31 species of Dendrobium) and their chloroplast genomes’ accession numbers in NCBI
| 物种 Species | NCBI序列号 NCBI accession number | 物种 Species | NCBI序列号 NCBI accession number |
|---|---|---|---|
| Anoectochilus emeiensis | NC_033895.1 | Dendrobium parciflorum | NC_035334.1 |
| Apostasia odorata | NC_030722.1 | Dendrobium parishii | NC_035339.1 |
| Bletilla ochracea | NC_029483.1 | Dendrobium pendulum | NC_029705.1 |
| Bletilla striata | NC_028422.1 | Dendrobium primulinum | NC_035321.1 |
| Calanthe triplicata | NC_024544.1 | Dendrobium salaccense | NC_035332.1 |
| Cattleya crispata | NC_026568.1 | Dendrobium spatella | NC_035333.1 |
| Cattleya liliputana | NC_032083.1 | Dendrobium strongylanthum | NC_027691.1 |
| Cephalanthera longifolia | NC_030704.1 | Dendrobium wardianum | NC_035329.1 |
| Cymbidium aloifolium | NC_021429.1 | Dendrobium wilsonii | NC_035330.1 |
| Cymbidium ensifolium | NC_028525.1 | Dendrobium xichouense | NC_035341.1 |
| Cymbidium faberi | NC_027743.1 | Elleanthus sodiroi | NC_027266.1 |
| Cymbidium goeringii | NC_028524.1 | Epipactis mairei | NC_030705.1 |
| Cymbidium kanran | NC_029711.1 | Epipactis veratrifolia | NC_030708.1 |
| Cymbidium lancifolium | NC_029712.1 | Erycina pusilla | NC_018114.1 |
| Cymbidium macrorhizon | NC_029713.1 | Gastrochilus fuscopunctatus | NC_035830.1 |
| Cymbidium mannii | NC_021433.1 | Gastrochilus japonicus | NC_035833.1 |
| Cymbidium sinense | NC_021430.1 | Goodyera fumata | NC_026773.1 |
| Cymbidium tortisepalum | NC_021431.1 | Goodyera procera | NC_029363.1 |
| Cymbidium tracyanum | NC_021432.1 | Goodyera schlechtendaliana | NC_029364.1 |
| Cypripedium formosanum | NC_026772.1 | Goodyera velutina | NC_029365.1 |
| Cypripedium macranthos | NC_024421.1 | Habenaria pantlingiana | NC_026775.1 |
| Dendrobium aphyllum | NC_035322.1 | Habenaria radiata | NC_035834.1 |
| Dendrobium brymerianum | NC_035323.1 | Listera fugongensis | NC_030711.1 |
| Dendrobium catenatum | NC_024019.1 | Ludisia discolor | NC_030540.1 |
| Dendrobium chrysanthum | NC_035336.1 | Masdevallia coccinea | NC_026541.1 |
| Dendrobium chrysotoxum | NC_028549.1 | Masdevallia picturata | NC_026777.1 |
| Dendrobium crepidatum | NC_035331.1 | Neottia ovate | NC_030712.1 |
| Dendrobium denneanum | NC_035324.1 | Neottia pinetorum | NC_030710.1 |
| Dendrobium devonianum | NC_035325.1 | Oberonia japonica | NC_035832.1 |
| Dendrobium ellipsophyllum | NC_035340.1 | Paphiopedilum armeniacum | NC_026779.1 |
| Dendrobium exile | NC_035343.1 | Paphiopedilum niveum | NC_026776.1 |
| Dendrobium falconeri | NC_035326.1 | Pelatantheria scolopendrifolia | NC_035829.1 |
| Dendrobium fanjingshanense | NC_035344.1 | Phalaenopsis equestris | NC_017609.1 |
| Dendrobium fimbriatum | NC_035342.1 | Phragmipedium longifolium | NC_028149.1 |
| Dendrobium gratiosissimum | NC_035327.1 | Sobralia callosa | NC_028147.1 |
| Dendrobium henryi | NC_035335.1 | Thrixspermum japonicum | NC_035831.1 |
| Dendrobium hercoglossum | NC_035328.1 | Vanilla aphylla | NC_035320.1 |
| Dendrobium huoshanense | NC_028430.1 | Vanilla planifolia | NC_026778.1 |
| Dendrobium jenkinsii | NC_035337.1 | Sobralia aff. bouchei | NC_028209.1 |
| Dendrobium lohohense | NC_035338.1 | Phalaenopsis hybrid | NC_025593.1 |
| Dendrobium moniliforme | NC_035154.1 | Phalaenopsis aphrodite formosana | NC_007499.1 |
| Dendrobium nobile | NC_029456.1 | Oncidium hybrid | NC_014056.1 |
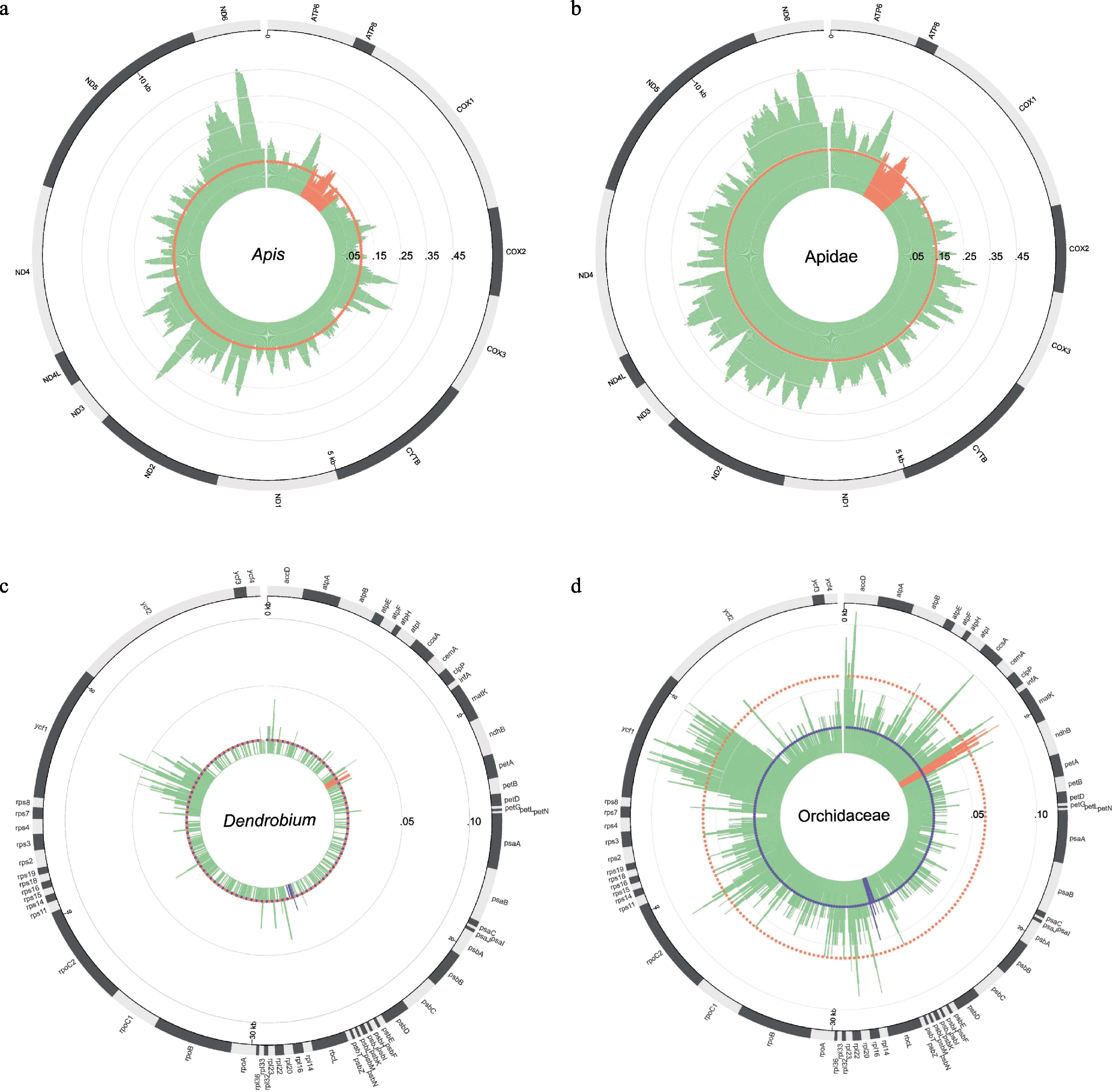
图3 线粒体基因组和叶绿体基因组种间及属间遗传距离图。(a) Apis属内6个物种线粒体基因组上13个蛋白编码基因序列P-distance图; (b) Apidae科内20个物种线粒体基因组上13个蛋白编码基因序列P-diatance图; (c) Dendrobium属内31个物种叶绿体基因组上67个蛋白编码基因序列P-distance图; (d) Orchidaceae科内84个物种叶绿体基因组上67个蛋白编码基因序列P-distance图。图a和b中, 橙色区域为COI条形码序列所在位置, 橙色虚线圆圈表示COI条形码序列P-distance中值; 图c和d中, 橙色区域为matK条形码序列所在位置, 紫色区域为rbcL条形码序列所在位置, 橙色虚线圆圈表示matK条形码序列P-distance中值, 紫色虚线圆圈表示rbcL条形码序列P-distance中值。
Fig. 3 The P-distance map among species and genera of the mitochondrial genomes and the chloroplast genomes. (a) The P-distance map of 13 protein-coding genes in the mitochondrial genomes among six species in Apis; (b) The P-distance map of 13 protein-coding genes in the mitochondrial genomes among 20 species in Apidae; (c) The P-distance map of 67 protein-coding genes in the chloroplast genomes among 31 species in Dendrobium; (d) The P-distance map of 67 protein-coding genes in the chloroplast genomes among 84 species in Orchidaceae. In panels a & b, the orange area is the location of the COI barcode, and the orange dotted circle represents the median value of the COI barcode’s P-distance. In panels c & d, the orange area is the location of the matK barcode, the purple area is the location of the rbcL barcode, and the orange dotted circle represents the median value of the matK barcode’s P-distance, the purple dotted circle represents the median value of the rbcL barcode’s P-distance.
| 1 | Aguilar R, Ashworth L, Galetto L, Aizen MA (2006) Plant reproductive susceptibility to habitat fragmentation: Review and synthesis through a meta-analysis. Ecology Letters, 9, 968-980. |
| 2 | Arif IA, Khan HA, Al Sadoon M, Shobrak M (2011) Limited efficiency of universal mini-barcode primers for DNA amplification from desert reptiles, birds and mammals. Genetics and Molecular Research, 10, 3559-3564. |
| 3 | Ashman TL, Knight TM, Steets JA, Amarasekare P, Burd M, Campbell DR, Dudash MR, Johnston MO, Mazer SJ, Mitchell RJ, Morgan MT, Wilson WG (2004) Pollen limitation of plant reproduction: Ecological and evolutionary causes and consequences. Ecology, 85, 2408-2421. |
| 4 | Aziz AN, Sauve RJ (2008) Genetic mapping of Echinacea purpurea via individual pollen DNA fingerprinting. Molecular Breeding, 21, 227-232. |
| 5 | Bagella S, Satta A, Floris I, Caria MC, Rossetti I, Podani J (2013) Effects of plant community composition and flowering phenology on honeybee foraging in Mediterranean sylvo-pastoral systems. Applied Vegetation Science, 16, 689-697. |
| 6 | Bambara SB (1991) Using pollen to identify honey. American Bee Journal, 131, 242-243. |
| 7 | Bascompte J, Jordano P (2007) Plant-animal mutualistic networks: The architecture of biodiversity. Annual Review of Ecology, Evolution and Systematics, 38, 567-593. |
| 8 | Bell KL, Julie F, Kevin SB, Emily KD, David G, Brice L, Connor M, Berry JB (2017) Applying pollen DNA metabarcoding to the study of plant-pollinator interactions. Applications in Plant Sciences, 5, 1600124. |
| 9 | Bell KL, Kevin SB, Kazufusa CO, Roman A, Berry JB (2016) Review and future prospects for DNA barcoding methods in forensic palynology. Forensic Science International: Genetics, 21, 110-116. |
| 10 | Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H (2010) ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiology, 10, 189. |
| 11 | Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E (2007) The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE, 2, e197. |
| 12 | Bosch J, González AM, Rodrigo A, Navarro D (2009) Plant- pollinator networks: Adding the pollinator’s perspective. Ecology Letters, 12, 409-419. |
| 13 | Bruni I, Galimberti A, Caridi L, Scaccabarozzi D, Mattia DF, Casiraghi M, Labra M (2015) A DNA barcoding approach to identify plant species in multiflower honey. Food Chemistry, 170, 308-315. |
| 14 | Cameron SA, Hines HM, Williams PH (2007) A comprehensive phylogeny of the bumble bees (Bombus). Biological Journal of the Linnean Society, 91, 161-188. |
| 15 | CBOL Plant Working Group (2009) A DNA barcode for land plants. Proceedings of the National Academy of Sciences, USA, 106, 12794-12797. |
| 16 | Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YJ, Ma XY, Gao T, Pang XH, Luo K, Li Y, Li XW, Jia XC, Lin YL, Leon C (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE, 5, 1-8. |
| 17 | China Plant BOL Group, Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, Yang JB, Fu CX, Zeng CX, Yan HF, Zhu YJ, Sun YS, Chen SY, Zhao L, Wang K, Yang T, Duan GW (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences, USA, 108, 19641-19646. |
| 18 | Coissac E, Hollingsworth PM, Lavergne S, Taberlet P (2016) From barcodes to genomes: Extending the concept of DNA barcoding. Molecular Ecology, 25, 1423-1428. |
| 19 | Danforth BN, Cardinal S, Praz C, Almeida EA, Michez D (2013) The impact of molecular data on our understanding of bee phylogeny and evolution. Annual Review of Entomology, 58, 57-78. |
| 20 | Escriche I, Kadar M, Juan-Borrás M, Domenech E (2011) Using flavonoids, phenolic compounds and headspace volatile profile for botanical authentication of lemon and orange honeys. Food Research International, 44, 1504-1513. |
| 21 | Fang Q, Huang SQ (2014) Progress in pollination ecology at the community level. Chinese Science Bulletin, 59, 449-458. (in Chinese with English abstract) |
| [方强, 黄双全 (2014) 群落水平上传粉生态学的研究进展. 科学通报, 59, 449-458.] | |
| 22 | Gómez-Rodríguez C, Crampton-Platt A, Timmermans MJTN, Baselga A, Vogler AP (2015) Validating the power of mitochondrial metagenomics for community ecology and phylogenetics of complex assemblages. Methods in Ecology and Evolution, 6, 883-894. |
| 23 | Hajibabaei M, Shokralla S, Zhou X, Singer GAC, Baird DJ (2011) Environmental Barcoding: A next-generation sequencing approach for biomonitoring Applications Using River Benthos. PLoS ONE, 6, e17497. |
| 24 | Hebert PD, Cywinska A, Ball SL (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences, 270, 313-321. |
| 25 | Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS ONE, 6, e19254. |
| 26 | Hollingsworth PM, Li DZ, van der Bank M, Twyford AD (2016) Telling plant species apart with DNA: From barcodes to genomes. Philosophical Transactions of the Royal Society B: Biological Sciences, 371, 20150338. |
| 27 | Holt KA, Bennett KD (2014) Principles and methods for automated palynology. New Phytologist, 203, 735-742. |
| 28 | Janzen DH, Hallwachs W, Burns J, Solis MA, Woodley NE (2009) Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity. Molecular Ecology Resources, 9, 1-26. |
| 29 | Kaiser-Bunbury CN, James M, Andrew EW, Terence V, Ronny G, Jens MO, Nico B (2017) Ecosystem restoration strengthens pollination network resilience and function. Nature, 542, 223-227. |
| 30 | Keller A, Danner N, Grimmer G, Ankenbrand M, von der Ohe K, von der Ohe W, Rost S, Härtel S, Steffan-Dewenter I (2015) Evaluating multiplexed next-generation sequencing as a method in palynology for mixed pollen samples. Plant Biology, 17, 558-566. |
| 31 | Khansari E, Zarre S, Alizadeh K, Attar F, Aghabeigi F, Salmaki Y (2012) Pollen morphology of Campanula (Campanulaceae) and allied genera in Iran with special focus on its systematic implication. Flora-Morphology, Distribution, Functional Ecology of Plants, 207, 203-211. |
| 32 | King C, Ballantyne G, Willmer PG (2013) Why flower visitation is a poor proxy for pollination: Measuring single-visit pollen deposition, with implications for pollination networks and conservation. Methods in Ecology and Evolution, 4, 811-818. |
| 33 | Klein AM, Vaissière BE, Cane JH, Steffandewenter I, Cunningham SA, Kremen C, Tscharntke T (2007) Importance of pollinators in changing landscapes for world crops. Proceedings of the Royal Society B: Biological Sciences, 274, 303-313. |
| 34 | Lahaye R, Savolainen V, Duthoit S, Maurin O, van der Bank M (2008) A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park as a model system (South Africa).. |
| 35 | Li XW, Yang Y, Henry RJ, Rossetto M, Wang YT, Chen SL (2015) Plant DNA barcoding: From gene to genome. Biological Reviews, 90, 157-166. |
| 36 | Li YW, Zhou X, Feng G, Hu HY, Niu LM, Hebert PD, Huang DW (2010) COI and ITS2 sequences delimit species, reveal cryptic taxa and host specificity of fig-associated Sycophila (Hymenoptera, Eurytomidae). Molecular Ecology Resources, 10, 31-40. |
| 37 | Liu SL, Li YY, Lu JL, Su X, Tang M, Zhang R, Zhou LL, Zhou CR, Yang Q, Ji YQ, Yu DW, Zhou X (2013) SOAPBarcode: Revealing arthropod biodiversity through assembly of Illumina shotgun sequences of PCR amplicons. Methods in Ecology and Evolution, 4, 1142-1150. |
| 38 | Liu SL, Wang X, Xie L, Tan MH, Li ZY, Su X, Zhang H, Misof B, Kjer KM, Tang M, Niehuis O, Jiang H, Zhou X (2016) Mitochondrial capture enriches mito-DNA 100 fold, enabling PCR-free mitogenomics biodiversity analysis. Molecular Ecology Resources, 16, 470. |
| 39 | Liu SL, Yang CT, Zhou CR, Zhou X (2017) Filling reference gaps via assembling DNA barcodes using high-throughput sequencing—moving toward barcoding the world. GigaScience, 6, 1-8. |
| 40 | Loreau M, Mazancourt CD (2013) Biodiversity and ecosystem stability: A synthesis of underlying mechanisms. Ecology Letters, 16, 106-115. |
| 41 | Macher JN, Zizka VMA, Weigand AM, Leese F (2017) A simple centrifugation protocol for metagenomic studies increases mitochondrial DNA yield by two orders of magnitude. Methods in Ecology and Evolution, 7, 1071-1075. |
| 42 | Matsuki Y, Isagi Y, Suyama Y (2007) The determination of multiple microsatellite genotypes and DNA sequences from a single pollen grain. Molecular Ecology Notes, 7, 194-198. |
| 43 | Murray TE, Úna F, Brown MJF, Paxton RJ (2008) Cryptic species diversity in a widespread bumble bee complex revealed using mitochondrial DNA RFLPs. Conservation Genetics, 9, 653-666. |
| 44 | Parks M, Cronn R, Liston A (2009) Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology, 7, 84. |
| 45 | Pecnikar ZF, Buzan EV (2014) 20 years since the introduction of DNA barcoding: From theory to application. Journal of Applied Genetics, 55, 43-52. |
| 46 | Piñol J, Mir G, Gomez-Polo P, Agustí N (2015) Universal and blocking primer mismatches limit the use of high-throughput DNA sequencing for the quantitative metabarcoding of arthropods. Molecular Ecology Resources, 15, 819-830. |
| 47 | Popic TJ, Wardle GM, Davila YC (2013) Flower-visitor networks only partially predict the function of pollen transport by bees. Austral Ecology, 38, 76-86. |
| 48 | Pornon A, Escaravage N, Burrus M, Holota H, Khimoun A, Mariette J, Pellizzari C, Iribar A, Etienne R, Taberlet P, Vidal M, Winterton P, Zinger L, Andalo C (2016) Using metabarcoding to reveal and quantify plant-pollinator interactions. Scientific Reports, 6, 27282. |
| 49 | Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, Kunin WE (2010) Global pollinator declines: Trends, impacts and drivers. Trends in Ecology and Evolution, 25, 345-353. |
| 50 | Potts SG, Imperatriz-Fonseca V, Ngo HT, Aizen MA, Biesmeijer JC, Breeze TD, Dicks LV, Garibaldi LA, Hill R, Settele J, Vanbergen AJ (2016) Safeguarding pollinators and their values to human well-being. Nature, 540, 220-229. |
| 51 | Rahl M (2008) Microscopic identification and purity determination of pollen grains. Humana Press, 138, 263-269. |
| 52 | Raphaël C, Tony D, Alice V, Nicolas R, Jean-Claude R, Aurélie B, Taberlet P, Pont D (2016) Spatial representativeness of environmental DNA metabarcoding signal for fish biodiversity assessment in a natural freshwater system. PLoS ONE, 11, e0157366. |
| 53 | Rasmont P, Coppée A, Michez D, Meulemeester TD (2008) An overview of the Bombus terrestris (L.1758) subspecies (Aymenoptera: Apidae). Annales-Societe Entomologique de France, 44, 243-250. |
| 54 | Richardson RT, Lin CH, Quijia JO, Riusech NS, Goodell K, Johnson RM (2015a) Rank-based characterization of pollen assemblages collected by honey bees using a multi-locus metabarcoding approach. Applications in Plant Sciences, 3, 1500043. |
| 55 | Richardson RT, Lin CH, Sponsler DB, Quijia JO, Goodell K, Johnson RM (2015b) Application of ITS2 metabarcoding to determine the provenance of pollen collected by honey bees in an agroecosystem. Applications in Plant Sciences, 3, 1400066. |
| 56 | Ricketts TH, Regetz J, Steffan-Dewenter I, Cunningham SA, Kremen C, Bogdanski A, Gemmill-Herren B, Greenleaf SS, Klein AM, Mayfield MM, Morandin LA, Ochieng A, Potts SG, Viana BF (2008) Landscape effects on crop pollination services: Are there general patterns? Ecology Letters, 11, 499-515. |
| 57 | Ruhsam M, Rai HS, Mathews S, Ross TG, Graham SW, Raubeson LA, Mei W, Thomas PI, Gardner MF, Ennos RA, Hollingsworth PM (2015) Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Molecular Ecology Resources, 15, 1067-1078. |
| 58 | Schmidt S, Schmid-Egger C, Moriniere J, Haszprunar G, Hebert PDN (2015) DNA barcoding largely supports 250 years of classical taxonomy: Identifications for Central European bees (Hymenoptera, Apoidea partim). Molecular Ecology Resources, 15, 985-1000. |
| 59 | Sheffield CS, Hebert PDN, Kevan P, Packer L (2009) DNA barcoding a regional bee (Hymenoptera: Apoidea) fauna and its potential for ecological studies. Molecular Ecology Resources, 9, 196-207. |
| 60 | Shi ZY, Yang CQ, Hao MD, Wang XY, Ward RD, Zhang AB (2018) FuzzyID2: A software package for large dataset species identification via barcoding and metabarcoding using Hidden Markov models and fuzzy set methods. Molecular Ecology Resources, 18, 666-675. |
| 61 | Smart MD, Cornman RS, Iwanowicz DD, McDermott- Kubeczko M, Pettis JS, Spivak MS, Otto CRV (2017) A comparison of honey bee-collected pollen from working agricultural lands using light microscopy and its metabarcoding. Environmental Entomology, 46, 38-49. |
| 62 | Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN (2006) DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proceedings of the National Academy of Sciences, USA, 103, 3657-3662. |
| 63 | Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E (2012) Towards next-generation biodiversity assessment using DNA metabarcoding. Molecular Ecology, 21, 2045-2050. |
| 64 | Tang M, Hardman CJ, Ji YQ, Meng GL, Liu SL, Tan MH, Yang SZ, Moss ED, Wang JX, Yang CX, Bruce C, Nevard T, Potts SG, Zhou X, Yu DW (2015) High-throughput monitoring of wild bee diversity and abundance via mitogenomics. Methods in Ecology and Evolution, 6, 1034-1043. |
| 65 | Tang M, Tan MH, Meng GL, Yang SZ, Su X, Liu SL, Song WH, Li YY, Wu Q, Zhang AB, Zhou X (2014) Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Research, 42, e166. |
| 66 | Tang M, Yi TS, Wang X, Tan MH, Zhou X (2013) The application of metabarcoding technology in identification of plant species diversity. Plant Diversity and Resources, 35, 769-773. (in Chinese with English abstract) |
| [唐敏, 伊廷双, 王欣, 谭美华, 周欣 (2013) Metabarcoding技术在植物鉴定和多样性研究中的应用. 植物分类与资源学报, 35, 769-773.] | |
| 67 | Weiner CN, Werner M, Linsenmair KE, Blüthgen N (2014) Land-use impacts on plant-pollinator networks: Interaction strength and specialization predict pollinator declines. Ecology, 95, 466-474. |
| 68 | Whitlock BA, Hale AM, Groff PA (2010) Intraspecific inversions pose a challenge for the trnH-psbA plant DNA barcode. PLoS ONE, 5, e11533. |
| 69 | Widmer A, Cozzolino S, Pellegrino G, Soliva M, Dafni A (2000) Molecular analysis of orchid pollinaria and pollinaria-remains found on insects. Molecular Ecology, 9, 1911-1914. |
| 70 | Wilson EE, Sidhu CS, LeVan KE, Holway DA (2010) Pollen foraging behavior of solitary Hawaiian bees revealed through molecular pollen analysis. Molecular Ecology, 19, 4823-4829. |
| 71 | Yamasaki YK, Nieman CC, Chang AN, Collier TC, Main BJ, Lee Y (2016) Improved tools for genomic DNA library construction of small insects. https://f1000research.com/posters/5-211. |
| 72 | Yu DW, Ji YQ, Emerson BC, Wang XY, Ye CX, Yang CY, Ding ZL (2012) Biodiversity soup: Metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods in Ecology and Evolution, 3, 613-623. |
| 73 | Zhang AB, Hao MD, Yang CQ, Shi ZY (2017) BarcodingR: An integrated R package for species identification using DNA barcodes. Methods in Ecology and Evolution, 8, 627-634. |
| 74 | Zhou X, Li YY, Liu SL, Yang Q, Su X, Zhou LL, Tang M, Fu RB, Li JG, Huang QF (2013) Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification. GigaScience, 2, 4. |
| [1] | 林珍, 向家宝, 蔡何佳奕, 高贝, 杨金涛, 李俊毅, 周青松, 黄晓磊, 邓鋆. 七种半翅目昆虫线粒体基因组数据[J]. 生物多样性, 2025, 33(2): 24434-. |
| [2] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [3] | 贺加贝, 柯可, 孙海明, 胡丽萍, 赵晓伟, 王文豪, 赵强. 基于DNA宏条形码技术分析香螺食性[J]. 生物多样性, 2025, 33(1): 24403-. |
| [4] | 巴苏艳, 赵春艳, 刘媛, 方强. 通过虫体花粉识别构建植物‒传粉者网络: 人工模型与AI模型高度一致[J]. 生物多样性, 2024, 32(6): 24088-. |
| [5] | 张飞飞, 杨天凤, 陈莉荣, 刘冬梅, 杨柳园, 杨杜宇, 鞠鹏, 陆露. 被子植物花粉颜色多样性及应用研究进展[J]. 生物多样性, 2024, 32(1): 23346-. |
| [6] | 吕晓琴, 李杨, 王顺雨, 姚仁秀, 王晓月. 中甸乌头总状花序不同位置花粉和花蜜的化学性状没有显著差异[J]. 生物多样性, 2024, 32(1): 23371-. |
| [7] | 董志远, 陈琳琳, 张乃鹏, 陈莉, 孙德斌, 倪艳梅, 李宝泉. 基于环境DNA宏条形码技术研究黄河三角洲典型潮沟系统鱼类多样性及其对水文连通性的响应[J]. 生物多样性, 2023, 31(7): 23073-. |
| [8] | 湛振杰, 张超, 陈敏豪, 王嘉栋, 富爱华, 范雨薇, 栾晓峰. 基于DNA宏条形码技术的大兴安岭北部欧亚水獭冬季食性分析[J]. 生物多样性, 2023, 31(6): 22586-. |
| [9] | 吴帆, 刘深云, 江虎强, 王茜, 陈开威, 李红亮. 中华蜜蜂和意大利蜜蜂秋冬期传粉植物多样性比较[J]. 生物多样性, 2023, 31(5): 22528-. |
| [10] | 彭步青, 陶玲, 李靖, 范荣辉, 陈顺德, 付长坤, 王琼, 唐刻意. 基于DNA宏条形码研究四川老君山国家级自然保护区6种同域共存小型哺乳动物的食性[J]. 生物多样性, 2023, 31(4): 22474-. |
| [11] | 卢聪聪, 刘倩, 黄晓磊. 三种蚜虫的线粒体基因组数据[J]. 生物多样性, 2022, 30(7): 22204-. |
| [12] | 沈梅, 郭宁宁, 罗遵兰, 郭晓晨, 孙光, 肖能文. 基于eDNA metabarcoding探究北京市主要河流鱼类分布及影响因素[J]. 生物多样性, 2022, 30(7): 22240-. |
| [13] | 翟俊杰, 赵慧峰, 商光申, 孙振钧, 张玉峰, 王兴. 蚯蚓基因组学的研究进展: 基于全基因组及线粒体基因组[J]. 生物多样性, 2022, 30(12): 22257-. |
| [14] | 胡德美, 姚仁秀, 陈燕, 游贤松, 王顺雨, 汤晓辛, 王晓月. 青篱柴通过促进亲和花粉生长而提高传粉精确性[J]. 生物多样性, 2021, 29(7): 887-896. |
| [15] | 陆奇丰, 黄至欢, 骆文华. 极小种群濒危植物广西火桐、丹霞梧桐的叶绿体基因组特征[J]. 生物多样性, 2021, 29(5): 586-595. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn