



Biodiv Sci ›› 2019, Vol. 27 ›› Issue (4): 366-372. DOI: 10.17520/biods.2018332 cstr: 32101.14.biods.2018332
• Original Papers: Plant Diversity • Previous Articles Next Articles
Zhixiang Chen1, Xueying Yao1, R. Downie Stephen2, Qizhi Wang1,*( )
)
Received:2018-12-18
Accepted:2019-01-09
Online:2019-04-20
Published:2019-06-05
Contact:
Qizhi Wang
Zhixiang Chen, Xueying Yao, R. Downie Stephen, Qizhi Wang. Assembling and analysis of Sanicula orthacantha chloroplast genome[J]. Biodiv Sci, 2019, 27(4): 366-372.
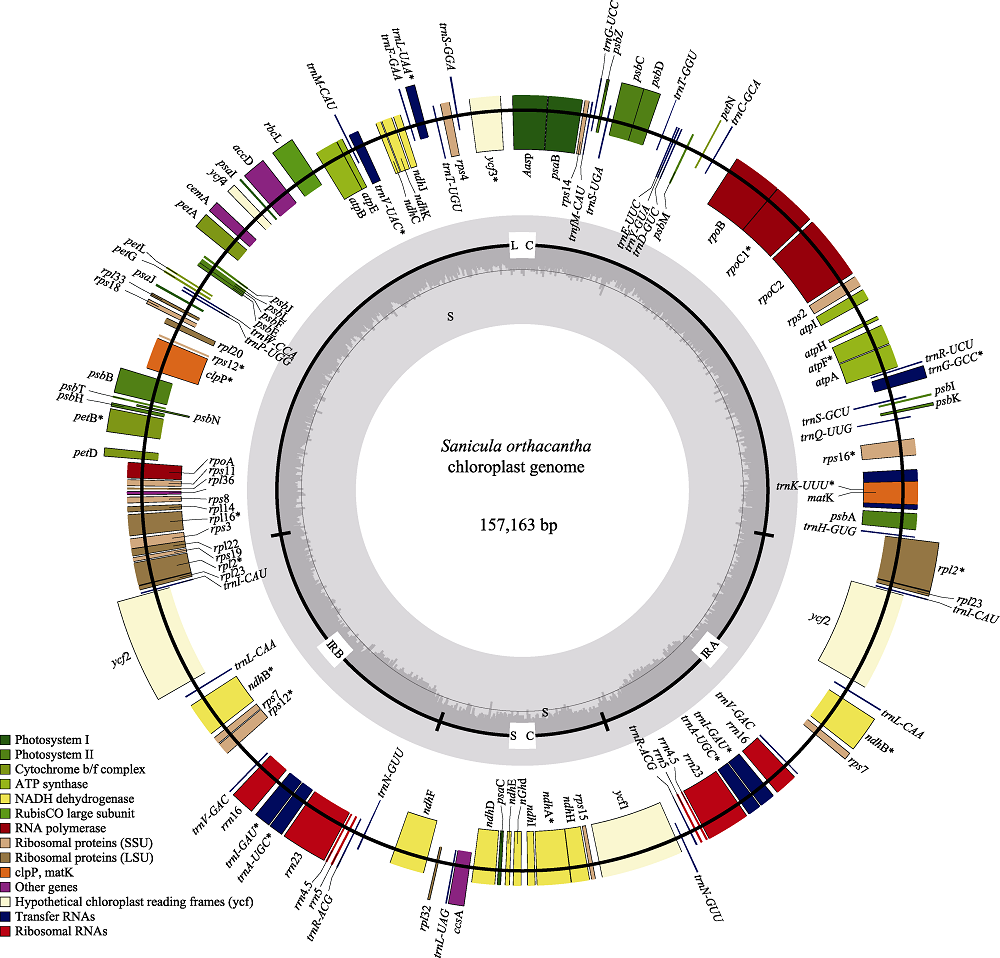
Fig. 1 Sequence map of the Sanicula orthacantha chloroplast genome. Genes drawn outside of the circle are transcribed counter-clockwise, while genes shown on the inside of the circle are transcribed clockwise. Genes belonging to different functional groups are color-coded. The dark gray in the inner circle indicates GC content, while the light gray corresponds to AT content.
| 基因分类 Category for genes | 基因分组 Group of genes | 基因名称 Name of genes |
|---|---|---|
| 表达相关基因 Self replication | 核糖体RNA基因 Ribosomal RNAs | rrn4.5(×2), rrn5(×2), rrn16(×2), rrn 23(×2) |
| 转运RNA基因 Transfer RNAs | trnA-UGC(×2), trnC-GCA, trnD-GUG, trnE-UCC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU(×2), trnI-GAU(×2), trnK- UUU, trnL-CAA(×2), trnL-UAA, trnL-UAG, trnM-CAU, trnN-GUU (×2) trnP-UGG, trnQ-UUC, trnR-ACG(×2) trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnV-UAC, trnW-CCA, trnY-GUA | |
| 核糖体小亚基基因 Ribosomal small subunit (SSU) | rps16, rps2, rps14, rps4, rps18, rps11, rps8, rps3, rps19, rps7(×2), rps12, rps15 | |
| 核糖体大亚基基因 Ribosomal large subuni (LSU) | rpl33, rpl20, rpl36, rpl14, rpl16, rpl22, rpl2(×2), rpl23(×2), rpl32 | |
| RNA聚合酶亚基基因 RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| 光合作用相关基因 Genes for photosynthesis | 光合系统I基因 Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| 光合系统II基因 Photosystem II | psbA, psbB, psbK, psbI, psbM, psbD, psbC, psbE, psbJ, psbL, psbT, psbH, psbN, psbF, psbZ, psbJ | |
| 细胞色素复合物基因 Cytochrome b/f complex | petA, petD, petG, petL, petN | |
| ATP合酶基因 ATP synthase | atpA, atpF, atpH, atpI, atpE, atpB | |
| 依赖ATP的蛋白酶单元p基因 ATP-dependent protease subunit p gene | clpP | |
| 二磷酸核酮糖羧化酶大亚基基因 RubiscoCO large subunit | rbcL | |
| NADH脱氢酶基因 NADH dehydrogenase | ndhJ, ndhK, ndhC, ndhB(×2), ndhF, ndhD, ndhE, ndhG, ndhI, ndhA, ndhH | |
| 其他基因 Other genes | 成熟酶基因 Maturase | matK |
| 包裹膜蛋白基因 Envelop membrane protein | cemA | |
| 乙酰辅酶A羧化酶亚基基因 Subunit of acetyl-CoA-carboxylase | accD | |
| c型细胞色素合成基因 c-type cytochrome synthesis ccsA gene | ccsA | |
| 转录起始因子基因 Transcription initiation factor IF-1 | InfA | |
| 未知功能基因 Genes of unknown function | 保守开放阅读框 Conserved open reading frames | ycf1, ycf2(×2), ycf3, ycf4 |
Table 1 List of genes found in Sanicula orthacantha chloroplast genome
| 基因分类 Category for genes | 基因分组 Group of genes | 基因名称 Name of genes |
|---|---|---|
| 表达相关基因 Self replication | 核糖体RNA基因 Ribosomal RNAs | rrn4.5(×2), rrn5(×2), rrn16(×2), rrn 23(×2) |
| 转运RNA基因 Transfer RNAs | trnA-UGC(×2), trnC-GCA, trnD-GUG, trnE-UCC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU(×2), trnI-GAU(×2), trnK- UUU, trnL-CAA(×2), trnL-UAA, trnL-UAG, trnM-CAU, trnN-GUU (×2) trnP-UGG, trnQ-UUC, trnR-ACG(×2) trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnV-UAC, trnW-CCA, trnY-GUA | |
| 核糖体小亚基基因 Ribosomal small subunit (SSU) | rps16, rps2, rps14, rps4, rps18, rps11, rps8, rps3, rps19, rps7(×2), rps12, rps15 | |
| 核糖体大亚基基因 Ribosomal large subuni (LSU) | rpl33, rpl20, rpl36, rpl14, rpl16, rpl22, rpl2(×2), rpl23(×2), rpl32 | |
| RNA聚合酶亚基基因 RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| 光合作用相关基因 Genes for photosynthesis | 光合系统I基因 Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| 光合系统II基因 Photosystem II | psbA, psbB, psbK, psbI, psbM, psbD, psbC, psbE, psbJ, psbL, psbT, psbH, psbN, psbF, psbZ, psbJ | |
| 细胞色素复合物基因 Cytochrome b/f complex | petA, petD, petG, petL, petN | |
| ATP合酶基因 ATP synthase | atpA, atpF, atpH, atpI, atpE, atpB | |
| 依赖ATP的蛋白酶单元p基因 ATP-dependent protease subunit p gene | clpP | |
| 二磷酸核酮糖羧化酶大亚基基因 RubiscoCO large subunit | rbcL | |
| NADH脱氢酶基因 NADH dehydrogenase | ndhJ, ndhK, ndhC, ndhB(×2), ndhF, ndhD, ndhE, ndhG, ndhI, ndhA, ndhH | |
| 其他基因 Other genes | 成熟酶基因 Maturase | matK |
| 包裹膜蛋白基因 Envelop membrane protein | cemA | |
| 乙酰辅酶A羧化酶亚基基因 Subunit of acetyl-CoA-carboxylase | accD | |
| c型细胞色素合成基因 c-type cytochrome synthesis ccsA gene | ccsA | |
| 转录起始因子基因 Transcription initiation factor IF-1 | InfA | |
| 未知功能基因 Genes of unknown function | 保守开放阅读框 Conserved open reading frames | ycf1, ycf2(×2), ycf3, ycf4 |
| [1] | Andrews S (2013) Babraham Bioinformatics FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. |
| [2] | Bayly MJ, Rigault P, Spokevicius A, Ladiges PY, Ades PK, Anderson C, Bossinger G, Merchant A, Udovicic F, Woodrow IE (2013) Chloroplast genome analysis of Australian eucalypts—Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Molecular Phylogenetics & Evolution, 69, 704-716. |
| [3] |
Clegg MT, Gaut BS, Learn G, Morton BR (1994) Rates and patterns of chloroplast DNA evolution. Proceedings of the National Academy of Sciences, USA, 91, 6795-6801.
DOI URL |
| [4] |
Daniell H, Lin CS, Ming Y, Chang WJ (2016) Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biology, 17, 134-163.
DOI URL |
| [5] |
Dierckxsens N, Mardulyn P, Smits G (2017) NOVO-Plasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Research, 45, e18.
DOI URL |
| [6] |
Downie SR, Jansen RK (2015) A comparative analysis of whole plastid genomes from the Apiales: Expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Systematic Botany, 40, 336-351.
DOI URL |
| [7] |
Ge L, Shen LQ, Chen QY, Li XM, Zhang L (2017) The complete chloroplast genome sequence of Hydrocotyle sibthorpioides (Apiales: Araliaceae). Mitochondrial DNA Part B, 2, 29-30.
DOI URL |
| [8] |
Jansen RK, Raubeson LA, Boore JL, Depamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ (2005) Methods for obtaining and analyzing whole chloroplast genome sequences. Methods in Enzymology, 395, 348-384.
DOI URL |
| [9] |
Kazutaka K, Kazuharu M, Kei-Ichi K, Takashi M (2002) MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30, 3059-3066.
DOI URL |
| [10] |
Kearse M, Moir R, Wilson A, Stoneshavas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647-1649.
DOI URL |
| [11] | Kim KJ, Lee HL (2005) Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Research, 11, 247-261. |
| [12] | Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology & Evolution, 33, 1870-1874. |
| [13] |
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research, 41, W575.
DOI URL |
| [14] |
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Qi P, Liu Y (2012) SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience, 1, 18.
DOI URL |
| [15] |
McCauley DE, Stevens JE, Peroni PA, Raveill JA (1996) The spatial distribution of chloroplast DNA and allozyme polymorphisms within a population of Silene alba (Caryophyllaceae). American Journal of Botany, 83, 727-731.
DOI URL |
| [16] | Peden JF (1999) CodonW. PhD Dissertation, University of Nottingham, Nottinghamshire, UK. |
| [17] |
Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H (2006) Complete plastid genome sequence of Daucus carota: Implications for biotechnology and phylogeny of angiosperms. BMC Genomics, 7, 222-235.
DOI URL |
| [18] | Shan RH, She ML (1979) Flora Reipublicae Popularis Sinicae, Tomus 14, pp. 12-67. Science Press, Beijing. (in Chinese) |
| [ 单人骅, 佘孟兰 (1979) 中国植物志, 第十四卷, 12-67页. 科学出版社, 北京.] | |
| [19] | Sichuan Provincial Health Department (1979) Sichuan Chinese Herbal Medicine Standard (Trial Draft). Sichuan Provincial Health Department, Chengdu. (in Chinese) |
| [ 四川省卫生局 (1979) 四川省中草药标准(试行稿). 四川省卫生局, 成都.] | |
| [20] |
Small RL, Cronn RC, Wendel JF (2004) Use of nuclear genes for phylogeny reconstruction in plants. Australian Systematic Botany, 17, 145-170.
DOI URL |
| [21] |
Soltis PS, Soltis DE (2000) The role of genetic and genomic attributes in the success of polyploids. Proceedings of the National Academy of Sciences, USA, 97, 7051-7057.
DOI URL |
| [22] | Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theoretical & Applied Genetics, 106, 411-422. |
| [23] | Wang L, Dong WP, Zhou SL (2012) Structural mutations and reorganizations in chloroplast genomes of flowering plants. Acta Botanica Boreali-Occidentalia Sinica. 32, 1282-1288. (in Chinese with English abstract) |
| [ 王玲, 董文攀, 周世良 (2012) 被子植物叶绿体基因组的结构变异研究进展. 西北植物学报, 32, 1282-1288.] | |
| [24] | Xie ZW (1996) The Compilation of Chinese Herbal Medicine. People’s Medical Publishing House, Beijing. (in Chinese) |
| [ 谢宗万 (1996) 全国中草药汇编. 人民卫生出版社, 北京.] | |
| [25] | Xing SC, Clarke JL (2008) Process in chloroplast genome analysis. Progress in Biochemistry and Biophysics, 35, 21-28. (in Chinese with English abstract) |
| [ 邢少辰 , Clarke JL (2008) 叶绿体基因组研究进展. 生物化学与生物物理进展, 35, 21-28.] | |
| [26] | Zhang HY (1994) Annals of Chinese Traditional Medicine Resources. Science Press, Beijing. (in Chinese) |
| [ 张惠源 (1994) 中国中药资源志要. 科学出版社, 北京.] | |
| [27] |
Zhang T, Fang Y, Wang X, Deng X, Zhang X, Hu S, Yu J (2012) The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: Insights into the evolution of plant organellar genomes. PLoS ONE, 7, e30531.
DOI URL |
| [28] | Zhao YB, Yin JL, Guo HY, Zhang YY, Xiao W, Sun C, Wu JY, Qu XB, Yu J, Wang XM, Xiao JF (2015) The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Frontiers in Plant Science, 5, 696-709. |
| [1] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [2] | Zhengming Luo, Jinxian Liu, Bianhua Zhang, Yanying Zhou, Aihua Hao, Kai Yang, Baofeng Chai. Diversity characteristics and driving factors of soil protist communities in subalpine meadow at different degradation stages [J]. Biodiv Sci, 2023, 31(8): 23136-. |
| [3] | Yinger Mao, Xiumei Zhou, Nan Wang, Xiuxiu Li, Yuke You, Shangbin Bai. Impact of Phyllostachys edulis expansion to Chinese fir forest on the soil bacterial community [J]. Biodiv Sci, 2023, 31(6): 22659-. |
| [4] | Wen Zhao, Dandan Wang, Mumin Reyila, Kaichuan Huang, Shun Liu, Baokai Cui. Soil microbial community structure of Larix gmelinii forest in the Aershan area [J]. Biodiv Sci, 2023, 31(2): 22258-. |
| [5] | Fan Xia, Jing Yang, Jian Li, Yang Shi, Lixin Gai, Wenhua Huang, Jingwei Zhang, Nan Yang, Fuli Gao, Yingying Han, Weidong Bao. Gut bacterial composition of four leopard cat subpopulations in Beijing [J]. Biodiv Sci, 2022, 30(9): 22103-. |
| [6] | Qi Zhao, Jibao Jiang, Zenglu Zhang, Qing Jin, Jiali Li, Jiangping Qiu. Species composition and phylogenetic analysis of earthworms on Hainan Island [J]. Biodiv Sci, 2022, 30(12): 22224-. |
| [7] | Yixin Sun, Yingbin Li, Yuhui Li, Bing Li, Xiaofang Du, Qi Li. Application of high-throughput sequencing technique in the study of nematode diversity [J]. Biodiv Sci, 2022, 30(12): 22266-. |
| [8] | Cheng Gao, Liang-Dong Guo. Progress on microbial species diversity, community assembly and functional traits [J]. Biodiv Sci, 2022, 30(10): 22429-. |
| [9] | Dongmei Li, Weihong Yang, Qingduo Li, Xi Han, Xiuping Song, Hong Pan, Yun Feng. High prevalence and genetic variation of Bartonella species inhabiting the bats in southwestern Yunnan [J]. Biodiv Sci, 2021, 29(9): 1245-1255. |
| [10] | Qifeng Lu, Zhihuan Huang, Wenhua Luo. Characterization of complete chloroplast genome in Firmiana kwangsiensis and F. danxiaensis with extremely small populations [J]. Biodiv Sci, 2021, 29(5): 586-595. |
| [11] | Nan Wang, Jinghua Huang, Na Huo, Panpan Yang, Xinyue Zhang, Shiwei Zhao. Characteristics of soil nematode community under different vegetation restoration approaches in the mountainous region of southern Ningxia: A comparative study based on morphological identification and high-throughput sequencing methods [J]. Biodiv Sci, 2021, 29(11): 1513-1529. |
| [12] | Benfeng Han, Xin Zhou, Xue Zhang. Verification of virus identity and host association using genomics technology [J]. Biodiv Sci, 2020, 28(5): 587-595. |
| [13] | Quanjian Zhang, Biao Yang, Qiang Fu, Lei Wang, Xu Gong, Yuanbin Zhang. The winter diet of sambar (Rusa unicolor) in the Qionglai Mountains [J]. Biodiv Sci, 2020, 28(10): 1192-1201. |
| [14] | Qi Lu,Qiang Hu,Xiaogang Shi,Senlong Jin,Sheng Li,Meng Yao. Metabarcoding diet analysis of snow leopards (Panthera uncia) in Wolong National Nature Reserve, Sichuan Province [J]. Biodiv Sci, 2019, 27(9): 960-969. |
| [15] | Zhang Xue, Li Xing’an, Su Qinzhi, Cao Qina, Li Chenyi, Niu Qingsheng, Zheng Hao. A curated 16S rRNA reference database for the classification of honeybee and bumblebee gut microbiota [J]. Biodiv Sci, 2019, 27(5): 557-566. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()