



Biodiv Sci ›› 2025, Vol. 33 ›› Issue (8): 25184. DOI: 10.17520/biods.2025184 cstr: 32101.14.biods.2025184
• Special Feature: Genetic Diversity and Conservation • Previous Articles Next Articles
Xin Peng( ), Chuan Liu(
), Chuan Liu( ), Xiaolei Huang*(
), Xiaolei Huang*( )(
)( )
)
Received:2025-05-19
Accepted:2025-08-28
Online:2025-08-20
Published:2025-09-17
Contact:
*E-mail: huangxl@fafu.edu.cn
Supported by:Xin Peng, Chuan Liu, Xiaolei Huang. Spatiotemporal pattern analysis of eukaryotic genetic data based on the GenBank database[J]. Biodiv Sci, 2025, 33(8): 25184.
| 类群 Group | 总计 Total | 经纬度缺失 Missing lat. or log. (%) | 经纬度异常 Abnormal lat. or log. (%) | 高精度数据集 High-resolution dataset (%) | 采样时间异常 Abnormal sampling time (%) | 时间序列数据集 Time series dataset (%) | 全数据集 Full dataset (%) |
|---|---|---|---|---|---|---|---|
| 动物COI Animalia COI | 3,604,970 | 951,853 (26.40%) | 358,576 (9.95%) | 2,294,541 (63.65%) | 169,450 (4.70%) | 2,125,091 (58.95%) | 3,116,067 (86.44%) |
| 植物rbcL Plantae rbcL | 383,112 | 318,694 (83.19%) | 9,041 (2.36%) | 55,377 (14.45%) | 20,279 (5.29%) | 35,098 (9.16%) | 220,722 (57.61%) |
| 真菌ITS Fungi ITS | 2,073,369 | 1,908,945 (92.07%) | 14,574 (0.70%) | 149,850 (7.23%) | 78,940 (3.81%) | 70,910 (3.42%) | 1,318,800 (63.61%) |
Table 1 Statistical analysis of representative sequences across Animalia, Plantae, and Fungi
| 类群 Group | 总计 Total | 经纬度缺失 Missing lat. or log. (%) | 经纬度异常 Abnormal lat. or log. (%) | 高精度数据集 High-resolution dataset (%) | 采样时间异常 Abnormal sampling time (%) | 时间序列数据集 Time series dataset (%) | 全数据集 Full dataset (%) |
|---|---|---|---|---|---|---|---|
| 动物COI Animalia COI | 3,604,970 | 951,853 (26.40%) | 358,576 (9.95%) | 2,294,541 (63.65%) | 169,450 (4.70%) | 2,125,091 (58.95%) | 3,116,067 (86.44%) |
| 植物rbcL Plantae rbcL | 383,112 | 318,694 (83.19%) | 9,041 (2.36%) | 55,377 (14.45%) | 20,279 (5.29%) | 35,098 (9.16%) | 220,722 (57.61%) |
| 真菌ITS Fungi ITS | 2,073,369 | 1,908,945 (92.07%) | 14,574 (0.70%) | 149,850 (7.23%) | 78,940 (3.81%) | 70,910 (3.42%) | 1,318,800 (63.61%) |
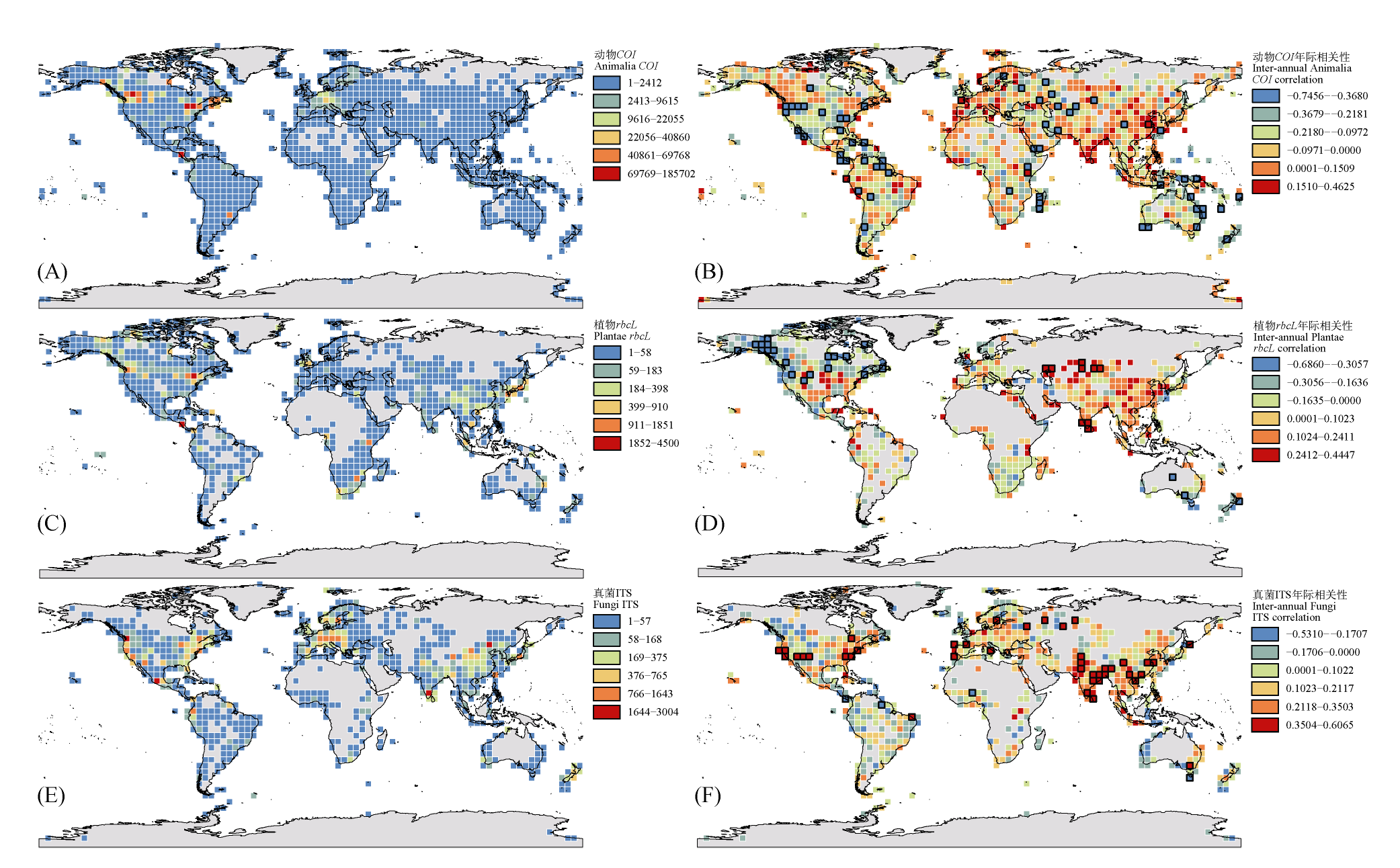
Fig. 1 Global spatiotemporal distribution patterns of representative sequences at grid level for Animalia (A, B), Plantae (C, D), and Fungi (E, F). The left panels (A, C, E) display the spatial distribution patterns of sequences from Animalia (COI), Plantae (rbcL), and Fungi (ITS), respectively, based on a 4° × 4° grid system. The color gradient represents six sequence density levels classified using the Jenks natural breaks. The right panels (B, D, F) show the interannual trends of sequences from Animalia (COI), Plantae (rbcL), and Fungi (ITS), respectively. The correlation coefficient between sequence count and year within each grid was calculated using a general linear model (positive values indicate increasing trends, and negative values indicate decreasing trends). Grids with black borders denote statistically significant correlations (P < 0.05).
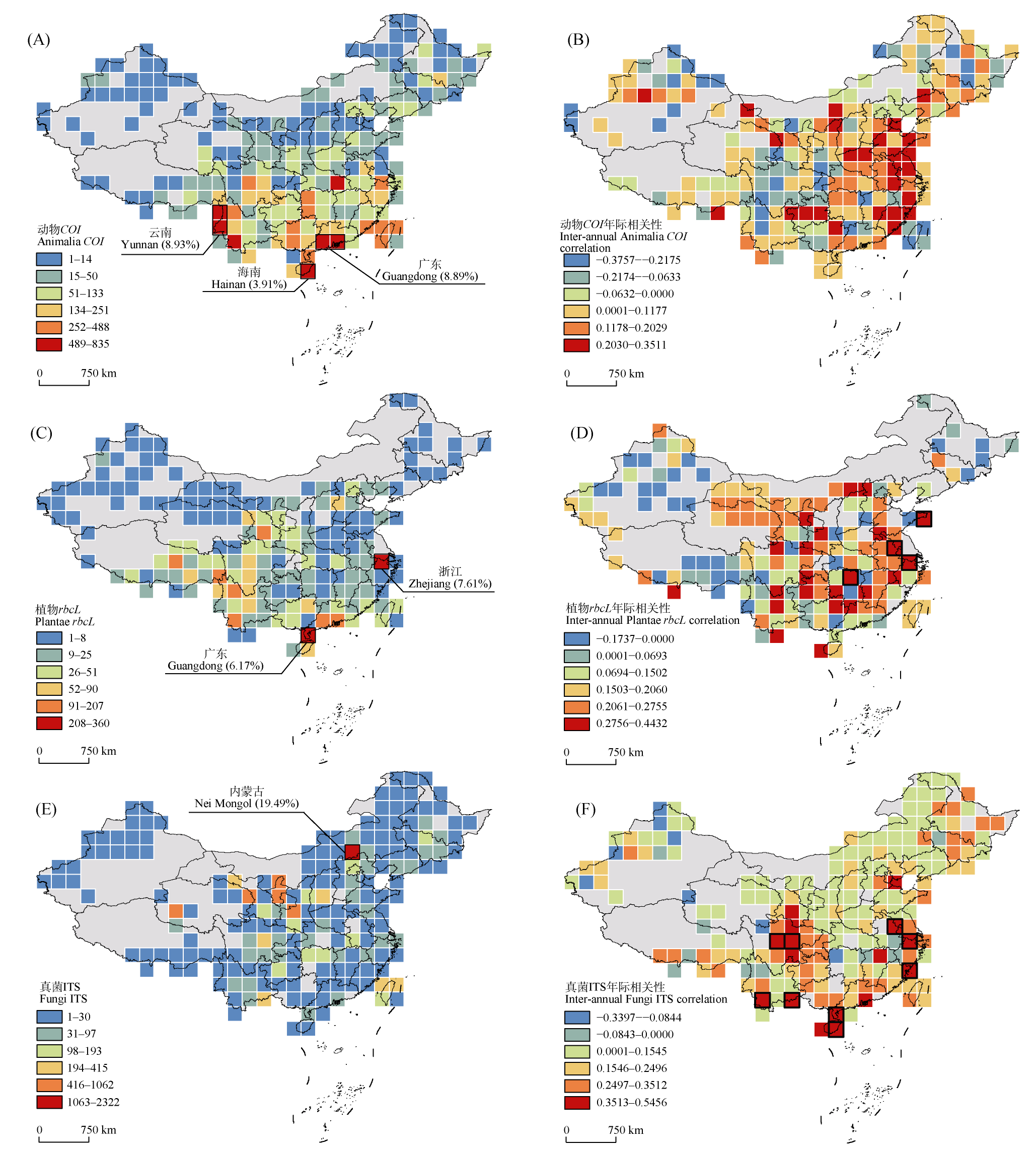
Fig. 2 China’s spatiotemporal distribution patterns of representative sequences across Animalia (A, B), Plantae (C, D), and Fungi (E, F) kingdoms. The left panels (A, C, E) display the spatial distribution patterns of sequences from Animalia (COI), Plantae (rbcL), and Fungi (ITS) within China, based on a 2° × 2° grid system. The color gradient represents six sequence density levels classified using the Jenks natural breaks. The right panels (B, D, F) show the inter-annual trends of sequences from Animalia (COI), Plantae (rbcL), and Fungi (ITS) in China. The correlation coefficient between sequence count and year within each grid was calculated using a general linear model (positive values indicate increasing trends, and negative values indicate decreasing trends). Grids with black borders denote statistically significant correlations (P < 0.05).
| [1] |
Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q (2019) Revisions to the classification, nomenclature, and diversity of eukaryotes. Journal of Eukaryotic Microbiology, 66, 4-119.
DOI PMID |
| [2] | Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW (2012) GenBank. Nucleic Acids Research, 40, D48-D53. |
| [3] |
Boettiger C (2019) Ecological metadata as linked data. Journal of Open Source Software, 4, 1276.
DOI URL |
| [4] |
Boria RA, Olson LE, Goodman SM, Anderson RP (2014) Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecological Modelling, 275, 73-77.
DOI URL |
| [5] |
Brown ED, Williams BK (2019) The potential for citizen science to produce reliable and useful information in ecology. Conservation Biology, 33, 561-569.
DOI PMID |
| [6] |
Chen ZY, Baeza JA, Chen C, Gonzalez MT, González VL, Greve C, Kocot KM, Arbizu PM, Moles J, Schell T, Schwabe E, Sun J, Wong NLWS, Yap-Chiongco M, Sigwart JD (2025) A genome-based phylogeny for Mollusca is concordant with fossils and morphology. Science, 387, 1001-1007.
DOI PMID |
| [7] | Cochrane G, Karsch-Mizrachi I, Takagi T (2016) The International Nucleotide Sequence Database Collaboration. Nucleic Acids Research, 44, D48-D50. |
| [8] | Deck J, Gaither MR, Ewing R, Bird CE, Davies N, Meyer C, Riginos C, Toonen RJ, Crandall ED (2017) The Genomic Observatories Metadatabase (GeOMe): A new repository for field and sampling event metadata associated with genetic samples. PLoS Biology, 15, e2002925. |
| [9] |
Fallon SM (2007) Genetic data and the listing of species under the U.S. Endangered Species Act. Conservation Biology, 21, 1186-1195.
PMID |
| [10] | French CM, Bertola LD, Carnaval AC, Economo EP, Kass JM, Lohman DJ, Marske KA, Meier R, Overcast I, Rominger AJ, Staniczenko PPA, Hickerson MJ (2023) Global determinants of insect mitochondrial genetic diversity. Nature Communications, 14, 5276. |
| [11] | Gaither MR, Bowen BW, Toonen RJ (2013) Population structure in the native range predicts the spread of introduced marine species. Proceedings of the Royal Society B: Biological Sciences, 280, 20130409. |
| [12] |
Gratton P, Marta S, Bocksberger G, Winter M, Trucchi E, Kühl H (2017) A world of sequences: Can we use georeferenced nucleotide databases for a robust automated phylogeography. Journal of Biogeography, 44, 475-486.
DOI URL |
| [13] |
Harris MA, Slippers B, Kemler M, Greve M (2023) Opportunities for diversified usage of metabarcoding data for fungal biogeography through increased metadata quality. Fungal Biology Reviews, 46, 100329.
DOI URL |
| [14] |
Hawksworth DL (2001) The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycological Research, 105, 1422-1432.
DOI URL |
| [15] | Hebert PDN, Cywinska A, Ball SL, Dewaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences, 270, 313-321. |
| [16] |
Hoban S, Bruford M, D’Urban Jackson J, Lopes-Fernandes M, Heuertz M, Hohenlohe PA, Paz-Vinas I, Sjögren-Gulve P, Segelbacher G, Vernesi C, Aitken S, Bertola LD, Bloomer P, Breed M, Rodríguez-Correa H, Funk WC, Grueber CE, Hunter ME, Jaffe R, Liggins L, Mergeay J, Moharrek F, O’Brien D, Ogden R, Palma-Silva C, Pierson J, Ramakrishnan U, Simo-Droissart M, Tani N, Waits L, Laikre L (2020) Genetic diversity targets and indicators in the CBD Post-2020 Global Biodiversity Framework must be improved. Biological Conservation, 248, 108654.
DOI URL |
| [17] | Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, Graham SW, James KE, Kim K, Kress WJ, Schneider H, van Alphenstahl J, Barrett SCH, van den Berg C, Bogarin D, Burgess KS, Cameron KM, Carine M, Chacón J, Clark A, Clarkson JJ, Conrad F, Devey DS, Ford CS, Hedderson TAJ, Hollingsworth ML, Husband BC, Kelly LJ, Kesanakurti PR, Kim JS, Kim Y, Lahaye R, Lee H, Long DG, Madriñán S, Maurin O, Meusnier I, Newmaster SG, Park C, Percy DM, Petersen G, Richardson JE, Salazar GA, Savolainen V, Seberg O, Wilkinson MJ, Yi D, Little DP (2009) DNA barcode for land plants. Proceedings of the National Academy of Sciences, USA, 106, 12794-12797. |
| [18] | Jenkins GB, Beckerman AP, Bellard C, Benítez López A, Ellison AM, Foote CG, Hufton AL, Lashley MA, Lortie CJ, Ma Z, Moore AJ, Narum SR, Nilsson J, O’Boyle B, Provete DB, Razgour O, Rieseberg L, Riginos C, Santini L, Sibbett B, Peres-Neto PR (2023) Reproducibility in ecology and evolution: Minimum standards for data and code. Ecology and Evolution, 13, 9961. |
| [19] | Jenks GF (1967) The data model concept in statistical mapping. International Yearbook of Cartography, 7, 186-190. |
| [20] |
Mardis ER (2008) Next-generation DNA sequencing methods. Annual Review of Genomics and Human Genetics, 9, 387-402.
DOI PMID |
| [21] |
Meineke EK, Davies TJ, Daru BH, Davis CC (2018) Biological collections for understanding biodiversity in the Anthropocene. Philosophical Transactions of the Royal Society B: Biological Sciences, 374, 20170386.
DOI URL |
| [22] |
Miraldo A, Li S, Borregaard MK, Flórez-Rodríguez A, Gopalakrishnan S, Rizvanovic M, Wang ZH, Rahbek C, Marske KA, Nogués-Bravo D (2016) An Anthropocene map of genetic diversity. Science, 353, 1532-1535.
PMID |
| [23] |
Peng X, Li Q, Cheng ZT, Huang XL (2023) The geography of genetic data: Current status and future perspectives. Frontiers in Ecology and Evolution, 11, 1112636.
DOI URL |
| [24] |
Pope LC, Liggins L, Keyse J, Carvalho SB, Riginos C (2015) Not the time or the place: The missing spatio-temporal link in publicly available genetic data. Molecular Ecology, 24, 3802-3809.
DOI PMID |
| [25] | Powers SM, Hampton SE (2019) Open science, reproducibility, and transparency in ecology. Ecological Applications, 29, e01822. |
| [26] | Ratnasingham S, Hebert PDN (2007) bold: The barcode of life data system (http://www.barcodinglife.org). Molecular Ecology Notes, 7, 355-364. |
| [27] |
Reddy S (2014) What’s missing from avian global diversification analyses? Molecular Phylogenetics and Evolution, 77, 159-165.
DOI URL |
| [28] | Rhoads A, Au KF (2015) PacBio sequencing and ITS applications. Genomics, Proteomics & Bioinformatics, 13, 278-289. |
| [29] |
Riginos C, Crandall ED, Liggins L, Gaither MR, Ewing RB, Meyer C, Andrews KR, Euclide PT, Titus BM, Therkildsen NO, Salces Castellano A, Stewart LC, Toonen RJ, Deck J (2020) Building a global genomics observatory: Using GEOME (the Genomic Observatories Metadatabase) to expedite and improve deposition and retrieval of genetic data and metadata for biodiversity research. Molecular Ecology Resources, 20, 1458-1469.
DOI PMID |
| [30] | Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Consortium FB (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences, USA, 109, 6241-6246. |
| [31] |
Tannenbaum C, Ellis RP, Eyssel F, Zou J, Schiebinger L (2019) Sex and gender analysis improves science and engineering. Nature, 575, 137-146.
DOI |
| [32] |
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology, 31, 21-32.
DOI PMID |
| [33] |
Troudet J, Grandcolas P, Blin A, Vignes-Lebbe R, Legendre F (2017) Taxonomic bias in biodiversity data and societal preferences. Scientific Reports, 7, 9132.
DOI PMID |
| [34] |
Vines TH, Andrew RL, Bock DG, Franklin MT, Gilbert KJ, Kane NC, Moore JS, Moyers BT, Renaut S, Rennison DJ, Veen T, Yeaman S (2013) Mandated data archiving greatly improves access to research data. The Federation of American Societies for Experimental Biology Journal, 27, 1304-1308.
DOI URL |
| [35] |
Warnasuriya SD, Udayanga D, Manamgoda DS, Biles C (2023) Fungi as environmental bioindicators. Science of the Total Environment, 892, 164583.
DOI URL |
| [36] |
Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J, Da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, ’t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone S, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B (2016) The FAIR guiding principles for scientific data management and stewardship. Scientific Data, 3, 160018.
DOI |
| [37] |
Winter DJ (2017) rentrez: An R package for the NCBI eUtils API. The R Journal, 9, 520.
DOI URL |
| [38] | Wood DA, Vandergast AG, Barr KR, Inman RD, Esque TC, Nussear KE, Fisher RNBM (2013) Comparative phylogeography reveals deep lineages and regional evolutionary hotspots in the Mojave and Sonoran Deserts. Diversity & Distributions, 19, 722-737. |
| [39] |
Yan DF, Mills JG, Gellie NJC, Bissett A, Lowe AJ, Breed MF (2018) High-throughput eDNA monitoring of fungi to track functional recovery in ecological restoration. Biological Conservation, 217, 113-120.
DOI URL |
| [40] |
Yao G, Zhang YQ, Barrett C, Xue B, Bellot S, Baker WJ, Ge XJ (2023) A plastid phylogenomic framework for the palm family (Arecaceae). BMC Biology, 21, 50.
DOI PMID |
| [41] |
Zizka A, Silvestro D, Andermann T, Azevedo J, Ritter CD, Edler D, Farooq H, Herdean A, Ariza M, Scharn R, Svantesson S, Wengström N, Zizka V, Antonelli A, Quental T (2019) CoordinateCleaner: Standardized cleaning of occurrence records from biological collection databases. Methods in Ecology and Evolution, 10, 744-751.
DOI URL |
| [1] | Tenzin Nyima, Wei Sun, Cong Li, Shuyi Zhang, Zhunan Zhao, Yongqiang Xu, Zhuoma Pubu, Shiqi Luo, Wa Da, Xin Zhou. The chloroplast genome dataset of flowering plants from the Jilong Valley region in Xizang [J]. Biodiv Sci, 2025, 33(9): 25270-. |
| [2] | Ruixiang Xue, Xuerong Ma, Jiongwen Wu, Aijun Liu, Xiquan Zhang, Congliang Ji, Yingshan Yin, Weijian Zhu, Qingbin Luo. Genetic diversity and genetic structure of Zhongshan partridge duck populations [J]. Biodiv Sci, 2025, 33(8): 24592-. |
| [3] | Fengying Wang, Zengyuan Wu, Han Cui, Yinlei Li, Lijuan Deng, Hong Wang, Jie Liu. Species boundaries of cannabina Clade of Urtica in the Third Pole [J]. Biodiv Sci, 2025, 33(8): 25138-. |
| [4] | Huixia Li, Yu Li, Xin Ning, Xiaochen Li, Tianrui Wang, Yigang Song, Xiling Dai, Sisi Zheng, Xin Zhong. Genetic diversity and genetic structure of Gymnospermium kiangnanense based on chloroplast genome [J]. Biodiv Sci, 2025, 33(8): 25149-. |
| [5] | Lin Zhen, Xiang Jiabao, Cai Hejiayi, Gao Bei, Yang Jintao, Li Junyi, Zhou Qingsong, Huang Xiaolei, Deng Jun. Mitochondrial genomic data for seven Hemipteran species [J]. Biodiv Sci, 2025, 33(2): 24434-. |
| [6] | Cao Dong, Li Huanlong, Peng Yang, Wei Cunzheng. Progresses in the study of the relationship between plant genome size and traits [J]. Biodiv Sci, 2025, 33(2): 24192-. |
| [7] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [8] | Xiangtan Yao, Xinyi Zhang, Yang Chen, Ye Yuan, Wangda Cheng, Tianrui Wang, Yingxiong Qiu. Genomic resequencing reveals the genetic diversity of the cultivated water caltrop, and the origin and domestication of ‘Nanhuling’ [J]. Biodiv Sci, 2024, 32(9): 24212-. |
| [9] | Xiaoyan Luo, Qiang Li, Xiaolei Huang. DNA barcode reference dataset for flower-visiting insects in Daiyun Mountain National Nature Reserve [J]. Biodiv Sci, 2023, 31(8): 23236-. |
| [10] | Zhaoyang Jing, Keguang Cheng, Heng Shu, Yongpeng Ma, Pingli Liu. Whole genome resequencing approach for conservation biology of endangered plants [J]. Biodiv Sci, 2023, 31(5): 22679-. |
| [11] | Fan Wu, Shenyun Liu, Huqiang Jiang, Qian Wang, Kaiwei Chen, Hongliang Li. Pollination difference between Apis cerana cerana and Apis mellifera ligustica during the late autumn and winter [J]. Biodiv Sci, 2023, 31(5): 22528-. |
| [12] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [13] | Wenda Cheng, Shuang Xing, Yang Liu. Wallace’s contributions and inspirations to contemporary research on the evolution of animal body color [J]. Biodiv Sci, 2023, 31(12): 23434-. |
| [14] | Jie Tao, Benqiang Li, Jinghua Cheng, Ying Shi, Peihong Liu, Guixia He, Weijie Xu, Huili Liu. Viral metagenome analysis of the viral community composition of the porcine diarrhea feaces [J]. Biodiv Sci, 2023, 31(11): 23170-. |
| [15] | Rui Luo, Ya Chen, Hanma Zhang. Research progress on whole-genome resequencing in Brassica [J]. Biodiv Sci, 2023, 31(10): 23237-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()