



Biodiv Sci ›› 2025, Vol. 33 ›› Issue (8): 25138. DOI: 10.17520/biods.2025138 cstr: 32101.14.biods.2025138
• Special Feature: Genetic Diversity and Conservation • Previous Articles Next Articles
Fengying Wang1,3( ), Zengyuan Wu2(
), Zengyuan Wu2( ), Han Cui2,3, Yinlei Li2,4, Lijuan Deng2,5, Hong Wang1(
), Han Cui2,3, Yinlei Li2,4, Lijuan Deng2,5, Hong Wang1( ), Jie Liu1,2,*(
), Jie Liu1,2,*( )(
)( )
)
Received:2025-04-15
Accepted:2025-06-11
Online:2025-08-20
Published:2025-09-17
Contact:
*E-mail: liujie@mail.kib.ac.cn
Supported by:Fengying Wang, Zengyuan Wu, Han Cui, Yinlei Li, Lijuan Deng, Hong Wang, Jie Liu. Species boundaries of cannabina Clade of Urtica in the Third Pole[J]. Biodiv Sci, 2025, 33(8): 25138.

Fig. 1 Plant morphology and habitat of six Urtica species in the Third Pole. Panels (A)-(G) represent species from the cannabina Clade, while panels (H)-(I) show outgroup species within the genus Urtica. (A) U. cannabina; (B) U. triangularis subsp. pinnatifida; (C) U. triangularis subsp. triangularis; (D) U. dioica subsp. afghanica; (E) U. dioica subsp. gansuensis; (F) U. dioica subsp. dioica; (G) U. hyperborea; (H) U. mairei; (I) Virtual specimen of U. membranifolia (from the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences).
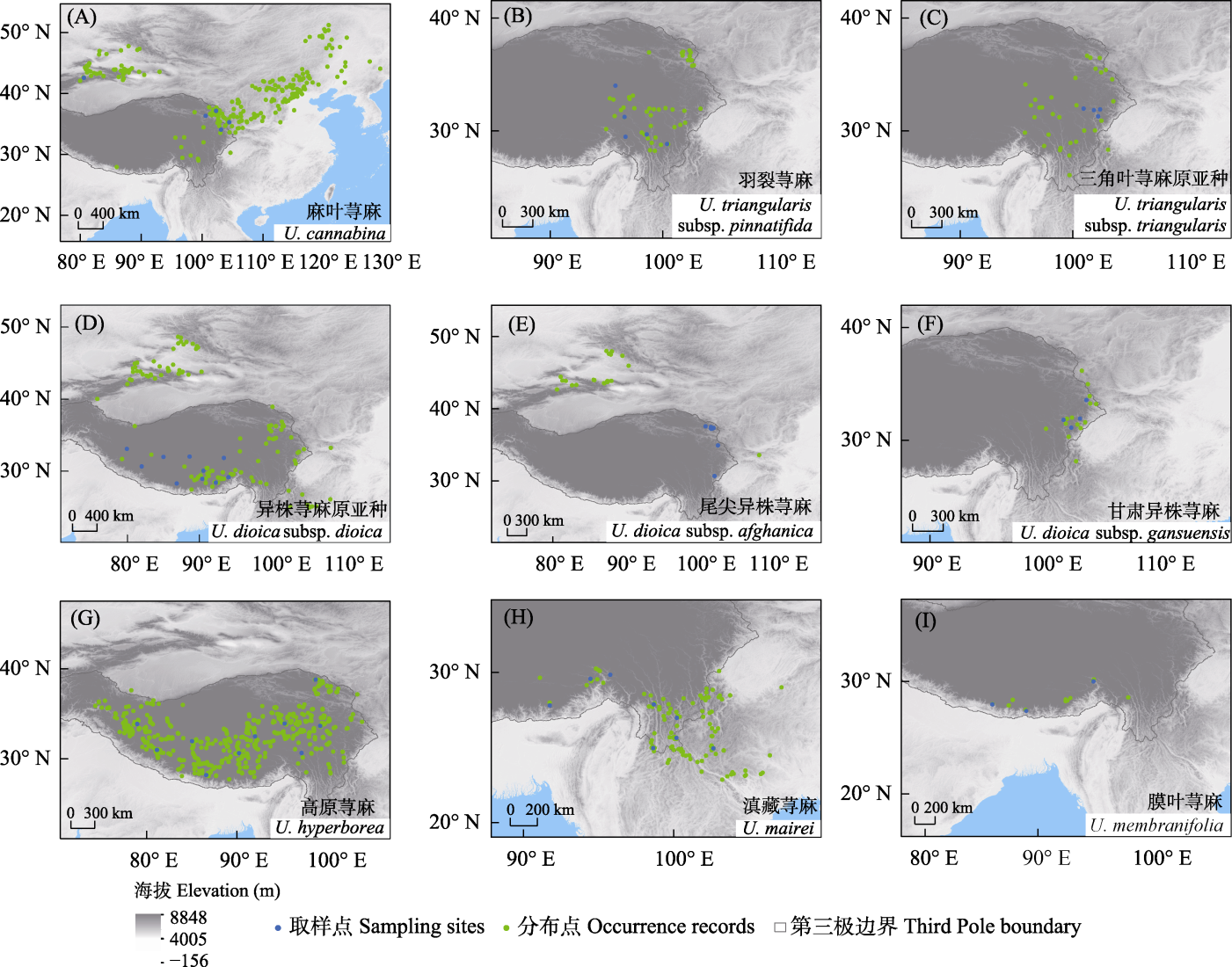
Fig. 2 Distribution of occurrence records (from GBIF (https://www.gbif.org), PPBC (https://ppbc.iplant.cn/), NPSRC (http s://www.cvh.ac.cn), iNaturalist (https://www.inaturalist.org/), field records) and sampling (collected from the wild) sites for six Urtica species in the Third Pole. The green dots represent the occurrence records, and the blue dots indicate the sampling sites in this study. Panels (A-G) represent species from the cannabina Clade, while panels (H-I) show outgroup species within the genus Urtica.
| 物种 Species | N | 质体基因组 Plastid genome | Angiosperms353核基因数据集Angiosperms353 nuclear gene set | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GL (bp) | Nh | Hd | π | NG | Nh | Hd | π | ||
| 麻叶荨麻 Urtica cannabina | 9 | 147,845-147,955 | 4 | 0.83 | 6.90 × 10-4 | 211-244 | 9 | 1.00 | 8.72 × 10-3 |
| 羽裂荨麻 U. triangularis subsp. pinnatifida | 5 | 147,933-147,948 | 5 | 1.00 | 1.50 × 10-4 | 203-224 | 5 | 1.00 | 8.96 × 10-3 |
| 三角叶荨麻原亚种 U. triangularis subsp. triangularis | 4 | 147,923-147,945 | 4 | 1.00 | 8.00 × 10-5 | 217-227 | 4 | 1.00 | 6.37 × 10-3 |
| 尾尖异株荨麻 U. dioica subsp. afghanica | 8 | 147,927-148,114 | 5 | 0.79 | 1.60 × 10-4 | 196-250 | 8 | 1.00 | 1.78 × 10-2 |
| 甘肃异株荨麻 U. dioica subsp. gansuensis | 4 | 148,021-148,164 | 4 | 1.00 | 1.00 × 10-4 | 188-199 | 4 | 1.00 | 1.15 × 10-2 |
| 异株荨麻原亚种 U. dioica subsp. dioica | 10 | 147,777-147,912 | 10 | 1.00 | 1.03 × 10-3 | 163-194 | 10 | 1.00 | 1.76 × 10-2 |
| 高原荨麻 U. hyperborea | 10 | 147,407-147,941 | 10 | 1.00 | 6.80 × 10-4 | 172-190 | 10 | 1.00 | 1.03 × 10-2 |
| 滇藏荨麻 U. mairei | 9 | 146,732-146,833 | 8 | 0.97 | 1.15 × 10-3 | 38-44 | 9 | 1.00 | 1.77 × 10-2 |
| 膜叶荨麻 U. membranifolia | 3 | 146,785-158,078 | 3 | 1.00 | 1.16 × 10-3 | 39-40 | 3 | 1.00 | 3.59 × 10-2 |
| 总计 Total | 62 | ||||||||
Table 1 Genomic characteristics and genetic diversity of six Urtica species in the Third Pole at the species level. There are four species from the cannabina Clade and two outgroup species within the genus Urtica (indicated in bold).
| 物种 Species | N | 质体基因组 Plastid genome | Angiosperms353核基因数据集Angiosperms353 nuclear gene set | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GL (bp) | Nh | Hd | π | NG | Nh | Hd | π | ||
| 麻叶荨麻 Urtica cannabina | 9 | 147,845-147,955 | 4 | 0.83 | 6.90 × 10-4 | 211-244 | 9 | 1.00 | 8.72 × 10-3 |
| 羽裂荨麻 U. triangularis subsp. pinnatifida | 5 | 147,933-147,948 | 5 | 1.00 | 1.50 × 10-4 | 203-224 | 5 | 1.00 | 8.96 × 10-3 |
| 三角叶荨麻原亚种 U. triangularis subsp. triangularis | 4 | 147,923-147,945 | 4 | 1.00 | 8.00 × 10-5 | 217-227 | 4 | 1.00 | 6.37 × 10-3 |
| 尾尖异株荨麻 U. dioica subsp. afghanica | 8 | 147,927-148,114 | 5 | 0.79 | 1.60 × 10-4 | 196-250 | 8 | 1.00 | 1.78 × 10-2 |
| 甘肃异株荨麻 U. dioica subsp. gansuensis | 4 | 148,021-148,164 | 4 | 1.00 | 1.00 × 10-4 | 188-199 | 4 | 1.00 | 1.15 × 10-2 |
| 异株荨麻原亚种 U. dioica subsp. dioica | 10 | 147,777-147,912 | 10 | 1.00 | 1.03 × 10-3 | 163-194 | 10 | 1.00 | 1.76 × 10-2 |
| 高原荨麻 U. hyperborea | 10 | 147,407-147,941 | 10 | 1.00 | 6.80 × 10-4 | 172-190 | 10 | 1.00 | 1.03 × 10-2 |
| 滇藏荨麻 U. mairei | 9 | 146,732-146,833 | 8 | 0.97 | 1.15 × 10-3 | 38-44 | 9 | 1.00 | 1.77 × 10-2 |
| 膜叶荨麻 U. membranifolia | 3 | 146,785-158,078 | 3 | 1.00 | 1.16 × 10-3 | 39-40 | 3 | 1.00 | 3.59 × 10-2 |
| 总计 Total | 62 | ||||||||
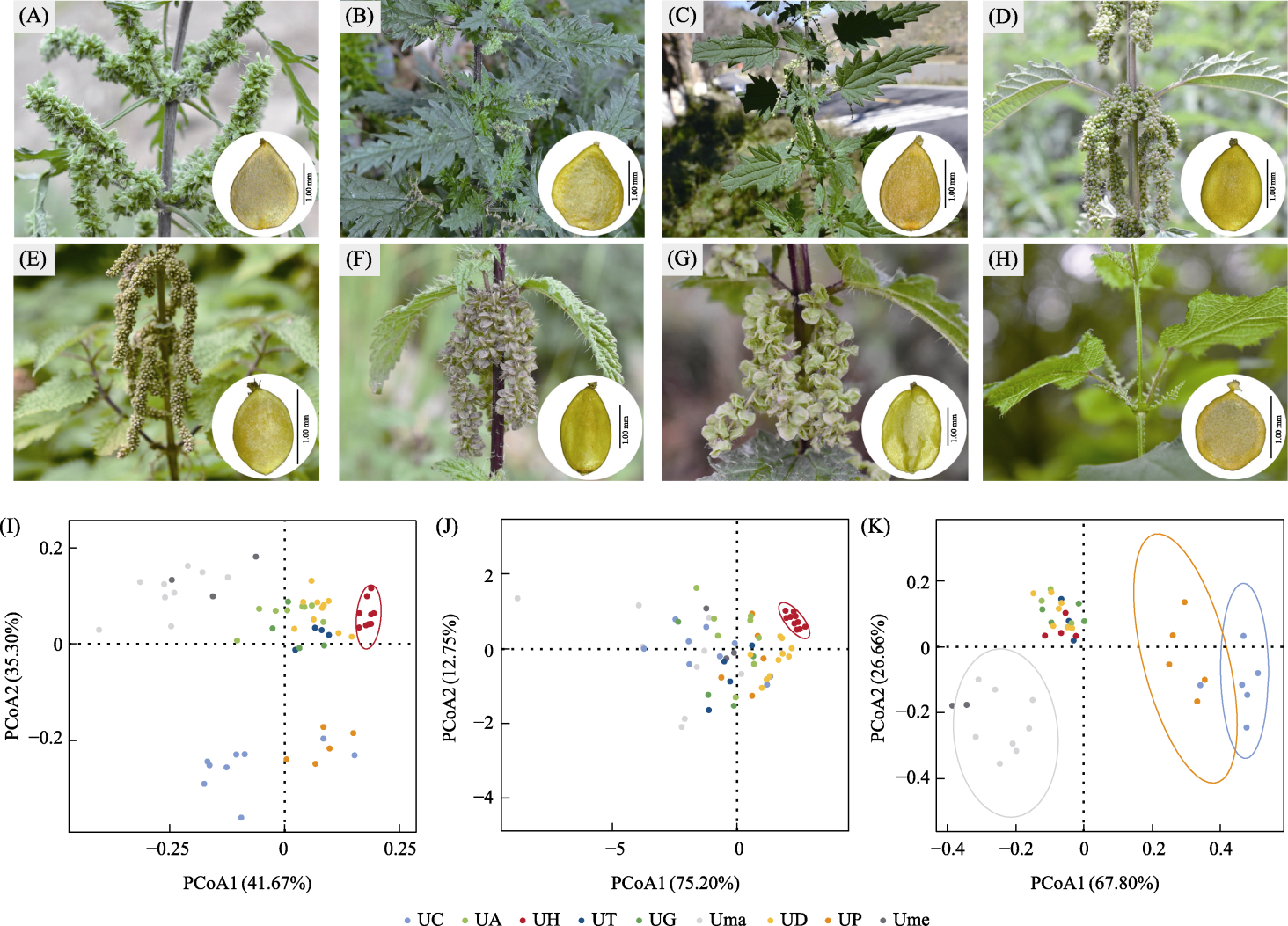
Fig. 3 Morphological traits of six Urtica species in the Third Pole. (A)-(H) Ecological photographs and achenes: (A) U. cannabina; (B) U. triangularis subsp. pinnatifida; (C) U. triangularis subsp. triangularis; (D) U. dioica subsp. afghanica; (E) U. dioica subsp. gansuensis; (F) U. dioica subsp. dioica; (G) U. hyperborea; (H) U. mairei. (I)-(K) The results of PCoA analysis: (I) All traits; (J) Quantitative traits; (K) Qualitative traits. The circular shapes represent 95% confidence intervals. UC, U. cannabina; UA, U. dioica subsp. afghanica; UH, U. hyperborea; UT, U. triangularis subsp. triangularis; UG, U. dioica subsp. gansuensis; Uma, U. mairei; UD, U. dioica subsp. dioica; UP, U. triangularis subsp. pinnatifida; Ume, U. membranifolia.
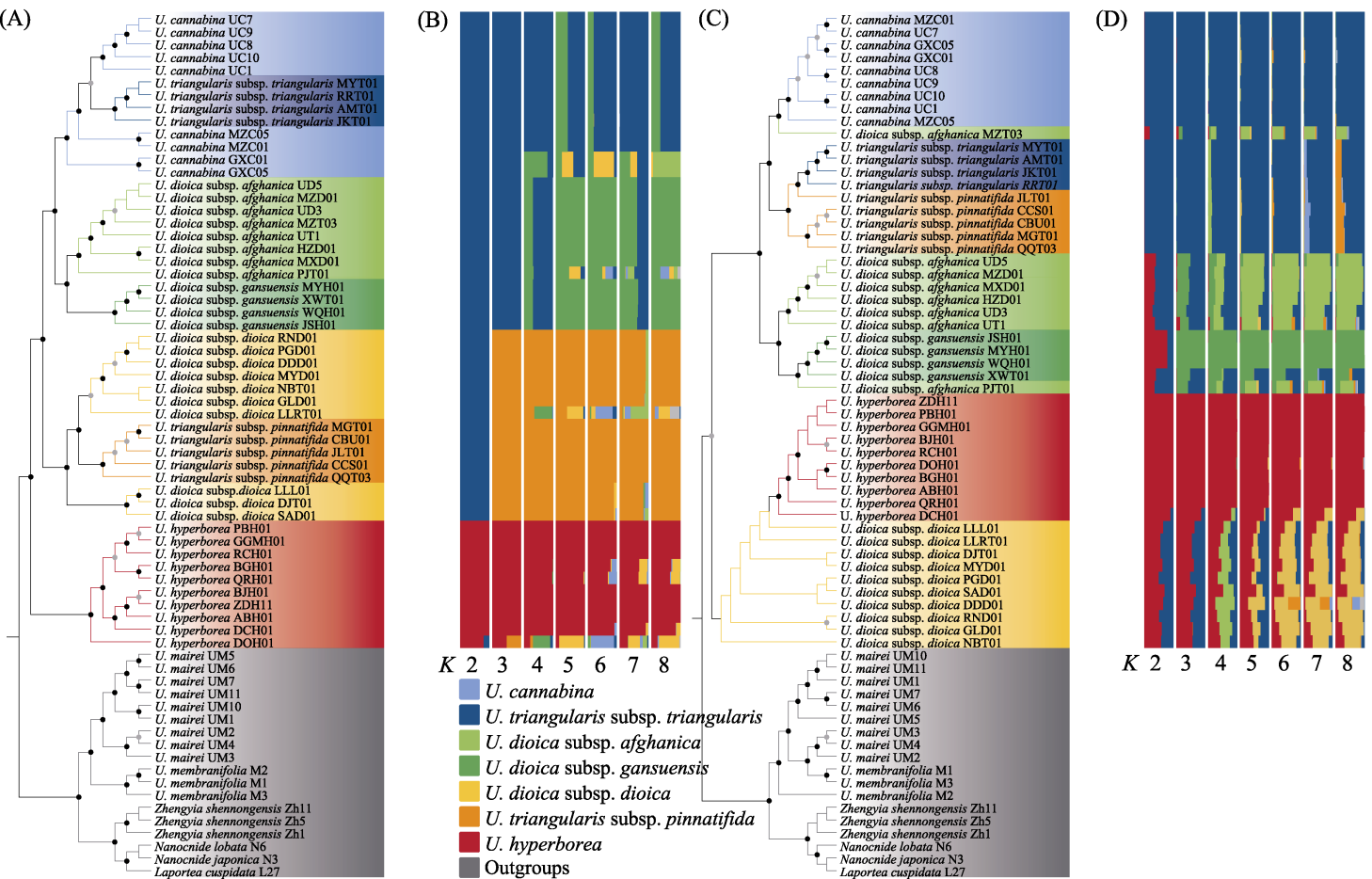
Fig. 4 Phylogenetic relationships and genetic structure based on complete plastid genome and Angiosperms353 nuclear gene set. Black dots represent the bootstrap > 90%, gray dots represent the bootstrap 70%-90%; the different species are highlighted with different colored backgrounds. (A) Phylogenetic tree of complete plastid genome based on the maximum likelihood (ML) methods; (B) Structure analysis result of complete plastid genome for four species (optimal K = 5); (C) Phylogenetic tree of Angiosperms353 nuclear gene set based on the ML methods. (D) Structure analysis result of Angiosperms353 nuclear gene set for four species (optimal K = 3).
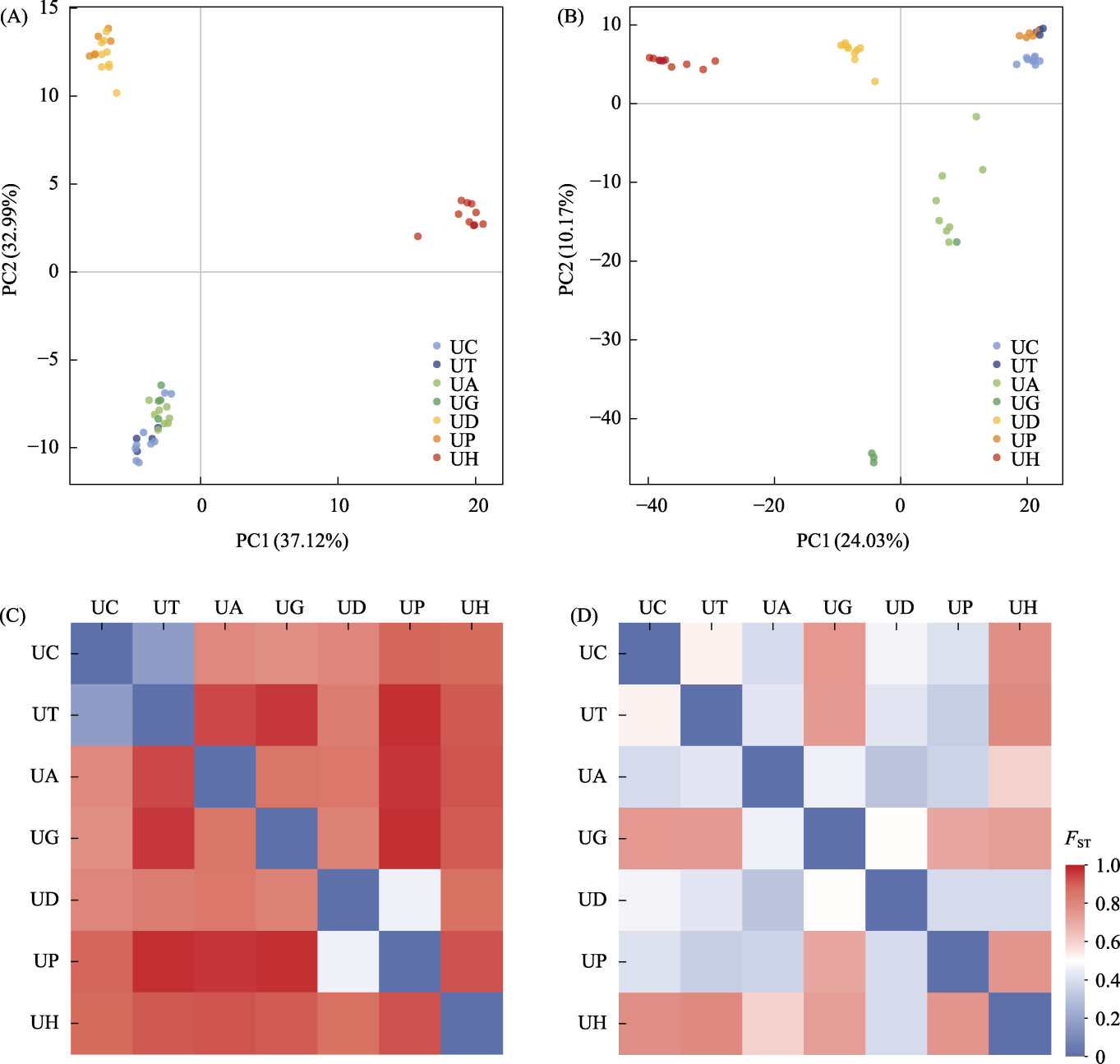
Fig. 5 Principal component analysis (PCA) and genetic differentiation heat map of four Urtica species within cannabina Clade from the Third Pole based on complete plastid genome and Angiosperms353 nuclear gene set. A-B, The result of PCA of plastid genome and concatenated Angiosperms353 nuclear gene set based on 50 individuals of four species, respectively; C-D, The genetic differentiation heat map of plastid genome and concatenated Angiosperms353 nuclear gene set based on 50 individuals of four species, respectively. U. mairei and U. membranifolia are phylogenetically distant from the four species, so they were excluded from this analysis. UC, U. cannabina; UT, U. triangularis subsp. triangularis; UA, U. dioica subsp. afghanica; UG, U. dioica subsp. gansuensis; UD, U. dioica subsp. dioica; UP, U. triangularis subsp. pinnatifida; UH, U. hyperborea. FST: Genetic differentiation index.
| [1] | Abràmoff MD, Magalhães PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics International, 11, 36-42. |
| [2] |
Ahmad M, Luo YH, Rathee S, Spicer RA, Zhang J, Wambulwa MC, Zhu GF, Cadotte MW, Wu ZY, Khan SM, Maity D, Li DZ, Liu J (2025) Multifaceted plant diversity patterns across the Himalaya: Status and outlook. Plant Diversity, 47, 529-543.
DOI |
| [3] |
Becker K, Grosse-Veldmann B, Weigend M (2017a) Weeding the nettles V: Taxonomic and phylogenetic studies of the eastern Asian species Urtica thunbergiana Sieb. & Zucc. (Urticaceae). Phytotaxa, 323, 201-216.
DOI URL |
| [4] |
Becker K, Grosse-Veldmann B, Weigend M (2017b) Weeding the nettles VI: Taxonomic and phylogenetic studies of the Southeast Asian Urtica fissa-clade (Urticaceae). Phytotaxa, 323, 217-236.
DOI URL |
| [5] | Birky CW (1995) Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proceedings of the National Academy of Sciences, USA, 92, 11331-11338. |
| [6] | Boehm MMA, Cronk QCB (2021) Dark extinction: The problem of unknown historical extinctions. Biology Letters, 17, 20210007. |
| [7] |
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 25, 1972-1973.
DOI PMID |
| [8] | Chen J, Lin Q, Friis I, Wilmot-Dear CM, Monro AK (2003) Urticaceae. In: Flora of China (eds Wu ZY, Raven PH, Hong DY), pp. 76-189. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis. |
| [9] | Chen YX, Chen YS, Shi CM, Huang ZB, Zhang Y, Li SK, Li Y, Ye J, Yu C, Li Z, Zhang XQ, Wang J, Yang HM, Fang L, Chen Q (2018) SOAPnuke: A MapReduce acceleration- supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience, 7, gix120. |
| [10] |
Coissac E, Hollingsworth PM, Lavergne S, Taberlet P (2016) From barcodes to genomes: Extending the concept of DNA barcoding. Molecular Ecology, 25, 1423-1428.
DOI PMID |
| [11] |
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772.
DOI PMID |
| [12] |
De Queiroz K (2007) Species concepts and species delimitation. Systematic Biology, 56, 879-886.
DOI PMID |
| [13] |
Deng LJ, Li YL, Wang FY, Sun XQ, Milne RI, Liu J, Wu ZY (2025) Comparative metabolomics of two nettle species unveils distinct high-altitude adaptation mechanisms on the Tibetan Plateau. BMC Plant Biology, 25, 640.
DOI |
| [14] | Dimaer DZPC (1986) Tibetan Medicine Monographs: Crystal Materia Medica. Shanghai Scientific & Technical Publishers, Shanghai. (in Chinese) |
| [帝玛尔·丹增彭措 (1986) 晶珠本草. 上海科学技术出版社, 上海.] | |
| [15] | Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19, 11-15. |
| [16] |
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI PMID |
| [17] |
Fišer C, Robinson CT, Malard F (2018) Cryptic species as a window into the paradigm shift of the species concept. Molecular Ecology, 27, 613-635.
DOI PMID |
| [18] | Friis I (1993) Urticaceae. In: Flowering Plants·Dicotyledons: Magnoliid, Hamamelid and Caryophyllid Families (eds Kubitzki K, Rohwer JG, Bittrich V), pp. 612-630. Springer-Verlag, Berlin. |
| [19] |
Fu PC, Guo QQ, Chang D, Gao QB, Sun SS (2024) Cryptic diversity and rampant hybridization in annual gentians on the Qinghai-Tibet Plateau revealed by population genomic analysis. Plant Diversity, 46, 194-205.
DOI URL |
| [20] | Greiner S, Lehwark P, Bock R (2019) OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research, 47, W59-W64. |
| [21] |
Grosse-Veldmann B, Nürk NM, Smissen R, Breitwieser I, Quandt D, Weigend M (2016) Pulling the sting out of nettle systematics—A comprehensive phylogeny of the genus Urtica L. (Urticaceae). Molecular Phylogenetics and Evolution, 102, 9-19.
DOI PMID |
| [22] |
Grosse-Veldmann B, Weigend M (2015) Weeding the nettles III: Named nonsense versus named morphotypes in European Urtica dioica L. (Urticaceae). Phytotaxa, 208, 239-260.
DOI URL |
| [23] | Guedes JJM, Moura MR, Jardim L, Diniz-Filho JAF (2025) Global patterns of taxonomic uncertainty and its impacts on biodiversity research. Systematic Biology, doi: 10.1093/sysbio/syaf045. |
| [24] |
Guo C, Luo Y, Gao LM, Yi TS, Li HT, Yang JB, Li DZ (2023) Phylogenomics and the flowering plant tree of life. Journal of Integrative Plant Biology, 65, 299-323.
DOI |
| [25] | Harris JG, Harris MW (2001) Plant Identification Terminology: An Illustrated Glossary, 2nd edn. Spring Lake Publishing, Utah. |
| [26] |
Henning T, Quandt D, Grosse-Veldmann B, Monro A, Weigend M (2014) Weeding the Nettles II: A delimitation of “Urtica dioica L.” (Urticaceae) based on morphological and molecular data, including a rehabilitation of Urtica gracilis Ait. Phytotaxa, 162, 61-83.
DOI URL |
| [27] |
Hirabayashi K, Dumigan CR, Kučka M, Percy DM, Guerriero G, Cronk Q, Deyholos MK, Todesco M (2025) A high-quality phased genome assembly of stinging nettle (Urtica dioica ssp. dioica). Plants, 14, 124.
DOI URL |
| [28] |
Hong DY (2016) Opinion of raising rationality in species delimitation. Biodiversity Science, 24, 360-361. (in Chinese)
DOI |
|
[洪德元 (2016) 关于提高物种划分合理性的意见. 生物多样性, 24, 360-361.]
DOI |
|
| [29] |
Hortal J, de Bello F, Diniz-Filho JAF, Lewinsohn TM, Lobo JM, Ladle RJ (2015) Seven shortfalls that beset large-scale knowledge of biodiversity. Annual Review of Ecology, Evolution, and Systematics, 46, 523-549.
DOI |
| [30] |
Huang XH, Deng T, Moore MJ, Wang HC, Li ZM, Lin N, Yusupov Z, Tojibaev KS, Wang YH, Sun H (2019) Tropical Asian Origin, boreotropical migration and long-distance dispersal in Nettles (Urticeae, Urticaceae). Molecular Phylogenetics and Evolution, 137, 190-199.
DOI PMID |
| [31] |
Jakobsson M, Rosenberg NA (2007) CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23, 1801-1806.
DOI PMID |
| [32] |
Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ (2020) GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology, 21, 241.
DOI |
| [33] |
Johnson MG, Pokorny L, Dodsworth S, Botigué LR, Cowan RS, Devault A, Eiserhardt WL, Epitawalage N, Forest F, Kim JT, Leebens-Mack JH, Leitch IJ, Maurin O, Soltis DE, Soltis PS, Wong GKS, Baker WJ, Wickett NJ (2019) A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Systematic Biology, 68, 594-606.
DOI PMID |
| [34] |
Jombart T (2008) Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403-1405.
DOI PMID |
| [35] |
Karbstein K, Kösters L, Hodač L, Hofmann M, Hörandl E, Tomasello S, Wagner ND, Emerson BC, Albach DC, Scheu S, Bradler S, de Vries J, Irisarri I, Li H, Soltis P, Mäder P, Wäldchen J (2024) Species delimitation 4.0: Integrative taxonomy meets artificial intelligence. Trends in Ecology & Evolution, 39, 771-784.
DOI URL |
| [36] |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772-780.
DOI PMID |
| [37] |
Kipkoech A, Li K, Milne RI, Oyebanji OO, Wambulwa MC, Fu XG, Wakhungu DA, Wu ZY, Liu J (2025) An integrative approach clarifies species delimitation and biogeographic history of Debregeasia (Urticaceae). Plant Diversity, 47, 229-243.
DOI |
| [38] | Kolbert E (2014) The Sixth Extinction:An Unnatural History. Henry Holt and Company, New York. |
| [39] | Li HT, Yi TS, Gao LM, Ma PF, Zhang T, Yang JB, Gitzendanner MA, Fritsch PW, Cai J, Luo Y, Wang H, van der Bank M, Zhang SD, Wang QF, Wang J, Zhang ZR, Fu CN, Yang J, Hollingsworth PM, Chase MW, Soltis DE, Soltis PS, Li DZ (2019) Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants, 5, 461-470. |
| [40] |
Li XW, Yang Y, Henry RJ, Rossetto M, Wang YT, Chen SL (2015) Plant DNA barcoding: From gene to genome. Biological Reviews, 90, 157-166.
DOI URL |
| [41] |
Liu BB, Ren C, Kwak M, Hodel RGJ, Xu C, He J, Zhou WB, Huang CH, Ma H, Qian GZ, Hong DY, Wen J (2022) Phylogenomic conflict analyses in the apple genus Malus s.l. reveal widespread hybridization and allopolyploidy driving diversification, with insights into the complex biogeographic history in the Northern Hemisphere. Journal of Integrative Plant Biology, 64, 1020-1043.
DOI URL |
| [42] |
Liu DH, Liu QR, Tojibaev KS, Sukhorukov AP, Wariss HM, Zhao Y, Yang L, Li WJ (2025) Phylogenomics provides new insight into the phylogeny and diversification of Asian Lappula (Boraginaceae). Molecular Phylogenetics and Evolution, 208, 108361.
DOI URL |
| [43] | Liu J, Gao LM (2011) Comparative analysis of three different methods of total DNA extraction used in Taxus. Guihaia, 31, 244-249, 159. (in Chinese with English abstract) |
| [刘杰, 高连明 (2011) 红豆杉属植物三种不同总DNA提取方法的分析比较. 广西植物, 31, 244-249, 159.] | |
| [44] |
Liu J, Luo YH, Li DZ, Gao LM (2017) Evolution and maintenance mechanisms of plant diversity in the Qing-hai-Tibet Plateau and adjacent regions: Retrospect and prospect. Biodiversity Science, 25, 163-174. (in Chinese with English abstract)
DOI |
|
[刘杰, 罗亚皇, 李德铢, 高连明 (2017) 青藏高原及毗邻区植物多样性演化与维持机制:进展及展望. 生物多样性, 25, 163-174.]
DOI |
|
| [45] | Liu J, Milne RI, Möller M, Zhu GF, Ye LJ, Luo YH, Yang JB, Wambulwa MC, Wang CN, Li DZ, Gao LM (2018) Integrating a comprehensive DNA barcode reference library with a global map of yews (Taxus L.) for forensic identification. Molecular Ecology Resources, 18, 1115-1131. |
| [46] |
Liu J, Milne RI, Zhu GF, Spicer RA, Wambulwa MC, Wu ZY, Boufford DE, Luo YH, Provan J, Yi TS, Cai J, Wang H, Gao LM, Li DZ (2022) Name and scale matter: Clarifying the geography of Tibetan Plateau and adjacent mountain regions. Global and Planetary Change, 215, 103893.
DOI URL |
| [47] |
Liu J, Möller M, Gao LM, Zhang DQ, Li DZ (2011) DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Molecular Ecology Resources, 11, 89-100.
DOI URL |
| [48] |
Liu J, Möller M, Provan J, Gao LM, Poudel RC, Li DZ (2013) Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytologist, 199, 1093-1108.
DOI PMID |
| [49] |
Liu J, Zhou SZ, Liu YL, Zhao BY, Yu DM, Zhong MC, Jiang XD, Cui WH, Zhao JX, Qiu J, Liu LM, Guo ZH, Tan DY, Hu JY, Li DZ (2024) Genomes of Meniocus linifolius and Tetracme quadricornis reveal the ancestral karyotype and features of core Brassicaceae. Plant Communications, 5, 100878.
DOI URL |
| [50] | Liu LX, Yao G, Yang J, Wang GQ (2022) One new genus, ten new species, and four new records of eriophyoid mites (Acari: Eriophyoidea) in Tibet, China. Systematic and Applied Acarology, 27, 1295-1336. |
| [51] |
Lumbsch HT, Leavitt SD (2011) Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Diversity, 50, 59-72.
DOI URL |
| [52] |
Ma PF, Zhang YX, Zeng CX, Guo ZH, Li DZ (2014) Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Systematic Biology, 63, 933-950.
DOI URL |
| [53] | Mayden RL (1997) A hierarchy of species concepts:The denouement in the saga of the species problem. In: Species: The Unit of Biodiversity (eds Claridge M, Dawah H, Wilson M), pp. 381-424. Chapman and Hall, London. |
| [54] | Meyer L, Ladle RJ, Trad R, Frazão A, Frateles L, Lessa T, Pinto RB, Freitas TMS, Tessarolo G, Guedes JJM, Castro-Souza RA, Medeiros H, Jardim L, Neves DRM, Rosado BHP, Lohmann LG, Hortal J, Diniz-Filho JAF, Stropp J (2025) Taxonomic uncertainty: Causes, consequences, and metrics. EcoEvoRxiv, doi: 10.32942/X21M04. |
| [55] |
Möller M, Gao LM, Mill RR, Liu J, Zhang DQ, Poudel RC, Li DZ (2013) A multidisciplinary approach reveals hidden taxonomic diversity in the morphologically challenging Taxus wallichiana complex. Taxon, 62, 1161-1177.
DOI URL |
| [56] |
Möller M, Liu J, Li Y, Li JH, Ye LJ, Mill R, Thomas P, Li DZ, Gao LM (2020) Repeated intercontinental migrations and recurring hybridizations characterise the evolutionary history of yew (Taxus L.). Molecular Phylogenetics and Evolution, 153, 106952.
DOI URL |
| [57] |
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature, 403, 853-858.
DOI |
| [58] |
Naciri Y, Linder HP (2015) Species delimitation and relationships: The dance of the seven veils. Taxon, 64, 3-16.
DOI URL |
| [59] | Northwest Institute of Plateau Biology, Chinese Academy of Sciences (1991) Tibetan Medicine Monographs. Qinghai People’s Publishing House, Xining. (in Chinese) |
| [中国科学院西北高原生物研究所 (1991) 藏药志. 青海人民出版社, 西宁.] | |
| [60] |
Ogoma CA, Liu J, Stull GW, Wambulwa MC, Oyebanji O, Milne RI, Monro AK, Zhao Y, Li DZ, Wu ZY (2022) Deep insights into the plastome evolution and phylogenetic relationships of the tribe Urticeae (Family Urticaceae). Frontiers in Plant Science, 13, 870949.
DOI URL |
| [61] |
Otero A, Barcenas-Peña A, Lumbsch HT, Grewe F (2023) Reference-based RADseq unravels the evolutionary history of polar species in ‘the crux lichenologorum’ genus Usnea (Parmeliaceae, Ascomycota). Journal of Fungi, 9, 99.
DOI URL |
| [62] |
Ottenlips MV, Mansfield DH, Buerki S, Feist MAE, Downie SR, Dodsworth S, Forest F, Plunkett GM, Smith JF (2021) Resolving species boundaries in a recent radiation with the Angiosperms 353 probe set: The Lomatium packardiae/L. anomalum clade of the L. triternatum (Apiaceae) complex. American Journal of Botany, 108, 1217-1233.
DOI PMID |
| [63] |
Padial JM, Miralles A, De la Riva I, Vences M (2010) The integrative future of taxonomy. Frontiers in Zoology, 7, 16-29.
DOI PMID |
| [64] |
Pérez-Escobar OA, Dodsworth S, Bogarín D, Bellot S, Balbuena JA, Schley RJ, Kikuchi IA, Morris SK, Epitawalage N, Cowan R, Maurin O, Zuntini A, Arias T, Serna-Sánchez A, Gravendeel B, Jimenez MFT, Nargar K, Chomicki G, Chase MW, Leitch IJ, Forest F, Baker WJ (2021) Hundreds of nuclear and plastid loci yield novel insights into orchid relationships. American Journal of Botany, 108, 1166-1180.
DOI PMID |
| [65] |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
DOI PMID |
| [66] |
Qin HT, Möller M, Milne R, Luo YH, Zhu GF, Li DZ, Liu J, Gao LM (2023) Multiple paternally inherited chloroplast capture events associated with Taxus speciation in the Hengduan Mountains. Molecular Phylogenetics and Evolution, 189, 107915.
DOI URL |
| [67] |
Qiu J (2008) The Third Pole. Nature, 454, 393-396.
DOI |
| [68] | R Core Team (2024) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Version R-4.4.1. https://www.R-project.org/ . (accessed on 2024-07-01) |
| [69] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34, 3299-3302.
DOI PMID |
| [70] |
Sang T (2002) Utility of low-copy nuclear gene sequences in plant phylogenetics. Critical Reviews in Biochemistry and Molecular Biology, 37, 121-147.
PMID |
| [71] |
Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH (2010) Integrative taxonomy: A multisource approach to exploring biodiversity. Annual Review of Entomology, 55, 421-438.
DOI PMID |
| [72] | Sgarlata GM, Salmona J, Le Pors B, Rasolondraibe E, Jan F, Ralantoharijaona T, Rakotonanahary A, Randriamaroson J, Marques AJ, Aleixo-Pais I, de Zoeten T, Ali Ousseni DS, Knoop SB, Teixeira H, Gabillaud V, Miller A, Ibouroi MT, Rasoloharijaona S, Zaonarivelo JR, Andriaholinirina NV, Chikhi L (2019) Genetic and morphological diversity of mouse lemurs (Microcebus spp.) in northern Madagascar: The discovery of a putative new species? American Journal of Primatology, 81, e23070. |
| [73] |
Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312-1313.
DOI PMID |
| [74] | Su RN, Luo WZ, Zhu JX, Ao WLJ, Zhong GY (2018) Research progress on medicinal plant of Urtica L. Chinese Traditional and Herbal Drugs, 49, 2722-2728. (in Chinese with English abstract) |
| [苏日娜, 罗维早, 朱继孝, 奥·乌力吉, 钟国跃 (2018) 荨麻属药用植物研究进展. 中草药, 49, 2722-2728.] | |
| [75] | Tattersall I (2007) Madagascar’s lemurs: Cryptic diversity or taxonomic inflation? Evolutionary Anthropology: Issues, News, and Reviews, 16, 12-23. |
| [76] |
Taylor K (2009) Biological flora of the British Isles: Urtica dioica L. Journal of Ecology, 97, 1436-1458.
DOI URL |
| [77] | van Elst T, Sgarlata GM, Schüßler D, Tiley GP, Poelstra JW, Scheumann M, Blanco MB, Aleixo-Pais IG, Rina Evasoa M, Ganzhorn JU, Goodman SM, Hasiniaina AF, Hending D, Hohenlohe PA, Ibouroi MT, Iribar A, Jan F, Kappeler PM, Le Pors B, Manzi S, Olivieri G, Rakotonanahary AN, Rakotondranary SJ, Rakotondravony R, Ralison JM, Ranaivoarisoa JF, Randrianambinina B, Rasoloarison RM, Rasoloharijaona S, Rasolondraibe E, Teixeira H, Zaonarivelo JR, Louis EE, Yoder AD, Chikhi L, Radespiel U, Salmona J (2025) Integrative taxonomy clarifies the evolution of a cryptic primate clade. Nature Ecology & Evolution, 9, 57-72. |
| [78] |
Viotti C, Albrecht K, Amaducci S, Bardos P, Bertheau C, Blaudez D, Bothe L, Cazaux D, Ferrarini A, Govilas J, Gusovius HJ, Jeannin T, Lühr C, Müssig J, Pilla M, Placet V, Puschenreiter M, Tognacchini A, Yung L, Chalot M (2022) Nettle, a long-known fiber plant with new perspectives. Materials, 15, 4288.
DOI URL |
| [79] |
Wang J, Ding XY, Guo CG, Zhang X, Feng HW, Yang HZ, Wang YH (2023) An ethnobotanical study of wild edible plants used by the Tibetan in the Rongjia River Valley, Tibet, China. Journal of Ethnobiology and Ethnomedicine, 19, 49.
DOI PMID |
| [80] |
Wang MZ, Zhu MM, Qian JY, Yang ZP, Shang FD, Egan AN, Li P, Liu LX (2024) Phylogenomics of mulberries (Morus, Moraceae) inferred from plastomes and single copy nuclear genes. Molecular Phylogenetics and Evolution, 197, 108093.
DOI URL |
| [81] |
Wang T, Zhang YP, Yang ZY, Liu Z, Du YY (2020) DNA barcoding reveals cryptic diversity in the underestimated genus Triplophysa (Cypriniformes: Cobitidae, Nemacheilinae) from the northeastern Qinghai-Tibet Plateau. BMC Evolutionary Biology, 20, 151.
DOI PMID |
| [82] | Wang WC, Chen JR (1995) Urticaceae. In: Flora Reipublicae Popularis Sinica, Tomus 23(2) (ed. Delecti Florae Reipublicae Popularis Sinicae Agendae Academiae Sinicae), pp. 5-27. Science Press, Beijing. (in Chinese) |
| [王文采, 陈家瑞 (1995) 荨麻科. 见: 中国植物志(第二十三卷第二分册) (中国科学院中国植物志编辑委员会), 5-27页. 科学出版社, 北京.] | |
| [83] |
Wang XY, Han XS, Csorba G, Wu Y, Chen HQ, Zhao X, Dong ZY, Yu WH, Lu Z (2025) A new species of Tube-Nosed Bat (Chiroptera: Vespertilionidae: Murina) from Qinghai-Tibet Plateau, China. Journal of Mammalogy, 106, 178-186.
DOI URL |
| [84] | Wickham H (2016) ggplot2:Elegant Graphics for Data Analysis. Springer-Verlag, New York. |
| [85] |
Wu SD, Wang Y, Wang ZF, Shrestha N, Liu JQ (2022) Species divergence with gene flow and hybrid speciation on the Qinghai-Tibet Plateau. New Phytologist, 234, 392-404.
DOI PMID |
| [86] |
Wu XT, Wang MQ, Li XY, Chen Y, Liao ZP, Zhang DL, Wen YF, Wang S (2024) Identification and characterization of a new species of Taxus—Taxus qinlingensis by multiple taxonomic methods. BMC Plant Biology, 24, 658.
DOI |
| [87] |
Wu ZY, Liu J, Provan J, Wang H, Chen CJ, Cadotte MW, Luo YH, Amorim BS, Li DZ, Milne RI (2018) Testing Darwin’s transoceanic dispersal hypothesis for the inland nettle family (Urticaceae). Ecology Letters, 21, 1515-1529.
DOI URL |
| [88] | Wu ZY, Milne RI, Chen CJ, Liu J, Wang H, Li DZ (2015) Ancestral state reconstruction reveals rampant homoplasy of diagnostic morphological characters in Urticaceae, conflicting with current classification schemes. PLoS ONE, 10, e0141821. |
| [89] |
Wu ZY, Milne RI, Liu J, Slik F, Yu Y, Luo YH, Monro AK, Wang WT, Wang H, Kessler PJA, Cadotte MW, Nathan R, Li DZ (2022) Phylogenomics and evolutionary history of Oreocnide (Urticaceae) shed light on recent geological and climatic events in SE Asia. Molecular Phylogenetics and Evolution, 175, 107555.
DOI URL |
| [90] |
Wu ZY, Monro AK, Milne RI, Wang H, Yi TS, Liu J, Li DZ (2013) Molecular phylogeny of the nettle family (Urticaceae) inferred from multiple loci of three genomes and extensive generic sampling. Molecular Phylogenetics and Evolution, 69, 814-827.
DOI URL |
| [91] | Xie JM, Chen YR, Cai GJ, Cai RL, Hu Z, Wang H (2023) Tree Visualization by One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Research, 51, W587-W592. |
| [92] |
Xu H, Zhang XQ, Yao ZY, Ali A, Li SQ (2021) Thirty-five new species of the spider genus Pimoa (Araneae, Pimoidae) from Pan-Himalaya. ZooKeys, 1029, 1-92.
DOI URL |
| [93] |
Xu KW, Yang Y, Chen H, Lin CX, Jiang L, Guo ZL, Li M, Hao MZ, Meng KK (2025) Extensive cytonuclear discordance revealed by phylogenomic analyses suggests complex evolutionary history in the holly genus Ilex (Aquifoliaceae). Molecular Phylogenetics and Evolution, 204, 108255.
DOI URL |
| [94] | Yan LJ, Fan PZ, Wambulwa MC, Qi HL, Chen Y, Wu ZY, Milne RI, Khan R, Luo YH, Gao LM, Shen SK, Rashid I, Khan SM, Maity D, Li DZ, Liu J (2024) Human-associated genetic landscape of walnuts in the Himalaya: Implications for conservation and utilization. Diversity and Distributions, 30, e13809. |
| [95] |
Yang FS, Peng D, Ma XT, Ban Q, Du YF, Yu SX, Gao TG, Wang JX, Liu B, Lu LM, Chen ZD, Hong DY, Wang Q (2025) A review of plant diversity surveys and monographic studies of the Pan-Himalaya region. Journal of Systematics and Evolution, 63, 5-11.
DOI |
| [96] |
Zhang C, Rabiee M, Sayyari E, Mirarab S (2018) ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics, 19, 153.
DOI PMID |
| [97] |
Zhang GJ, Ma H (2024) Nuclear phylogenomics of angiosperms and insights into their relationships and evolution. Journal of Integrative Plant Biology, 66, 546-578.
DOI |
| [98] |
Zhang KL, Leng YN, Hao RR, Zhang WY, Li HF, Chen MX, Zhu FY (2024) Adaptation of high-altitude plants to harsh environments: Application of phenotypic-variation-related methods and multi-omics techniques. International Journal of Molecular Sciences, 25, 12666.
DOI URL |
| [99] |
Zhang X, Kuang TH, Dong WL, Qian ZH, Zhang HJ, Landis JB, Feng T, Li LJ, Sun YX, Huang JL, Deng T, Wang HC, Sun H (2023) Genomic convergence underlying high-altitude adaptation in alpine plants. Journal of Integrative Plant Biology, 65, 1620-1635.
DOI |
| [100] | Zhang Z, Xie PL, Guo YL, Zhou WB, Liu EY, Yu Y (2022) Easy353: A tool to get Angiosperms353 genes for phylogenomic research. Molecular Biology and Evolution, 39, msac261. |
| [101] |
Zhou AT, Ge BRX, Chen S, Kang DX, Wu JR, Zheng YL, Ma HC (2024) Leaf ecological stoichiometry and anatomical structural adaptation mechanisms of Quercus sect. Heterobalanus in southeastern Qinghai-Tibet Plateau. BMC Plant Biology, 24, 325.
DOI |
| [102] |
Zhou WW, Wen Y, Fu JZ, Xu YB, Jin JQ, Ding L, Min MS, Che J, Zhang YP (2012) Speciation in the Rana chensinensis species complex and its relationship to the uplift of the Qinghai-Tibetan Plateau. Molecular Ecology, 21, 960-973.
DOI URL |
| [103] |
Zuntini AR, Carruthers T, Maurin O, Bailey PC, Leempoel K, Brewer GE, Epitawalage N, Francoso E, Gallego-Paramo B, McGinnie C, …… Bellot S, Crayn DM, Grace OM, Kersey PJ, Leitch IJ, Sauquet H, Smith SA, Eiserhardt WL, Forest F, Baker WJ (2024) Phylogenomics and the rise of the angiosperms. Nature, 629, 843-850.
DOI |
| [1] | Hong Du. “Species” versus “individuals”: Which is the right target for biodiversity conservation? [J]. Biodiv Sci, 2023, 31(8): 23140-. |
| [2] | Xinyu Du, Jinmei Lu, Dezhu Li. Advances in the evolution of plastid genome structure in lycophytes and ferns [J]. Biodiv Sci, 2019, 27(11): 1172-1183. |
| [3] | Zhigang Jiang,Lili Li,Yiming Hu,Huijian Hu,Chunwang Li,Xiaoge Ping,Zhenhua Luo. Diversity and endemism of ungulates on the Qinghai-Tibetan Plateau: Evolution and conservation [J]. Biodiv Sci, 2018, 26(2): 158-170. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()