



Biodiv Sci ›› 2025, Vol. 33 ›› Issue (9): 25270. DOI: 10.17520/biods.2025270 cstr: 32101.14.biods.2025270
• Data Paper • Previous Articles Next Articles
Tenzin Nyima1,#( ), Wei Sun2,#(
), Wei Sun2,#( ), Cong Li2(
), Cong Li2( ), Shuyi Zhang2(
), Shuyi Zhang2( ), Zhunan Zhao2(
), Zhunan Zhao2( ), Yongqiang Xu1(
), Yongqiang Xu1( ), Zhuoma Pubu1(
), Zhuoma Pubu1( ), Shiqi Luo2(
), Shiqi Luo2( ), Wa Da1,*(
), Wa Da1,*( )(
)( ), Xin Zhou2,*(
), Xin Zhou2,*( )(
)( )
)
Received:2025-07-14
Accepted:2025-08-20
Online:2025-09-20
Published:2025-10-31
Contact:
*E-mail: tsea2@163.com; xinzhoucaddis@icloud.com
About author:#Co-first authors
Supported by:Tenzin Nyima, Wei Sun, Cong Li, Shuyi Zhang, Zhunan Zhao, Yongqiang Xu, Zhuoma Pubu, Shiqi Luo, Wa Da, Xin Zhou. The chloroplast genome dataset of flowering plants from the Jilong Valley region in Xizang[J]. Biodiv Sci, 2025, 33(9): 25270.
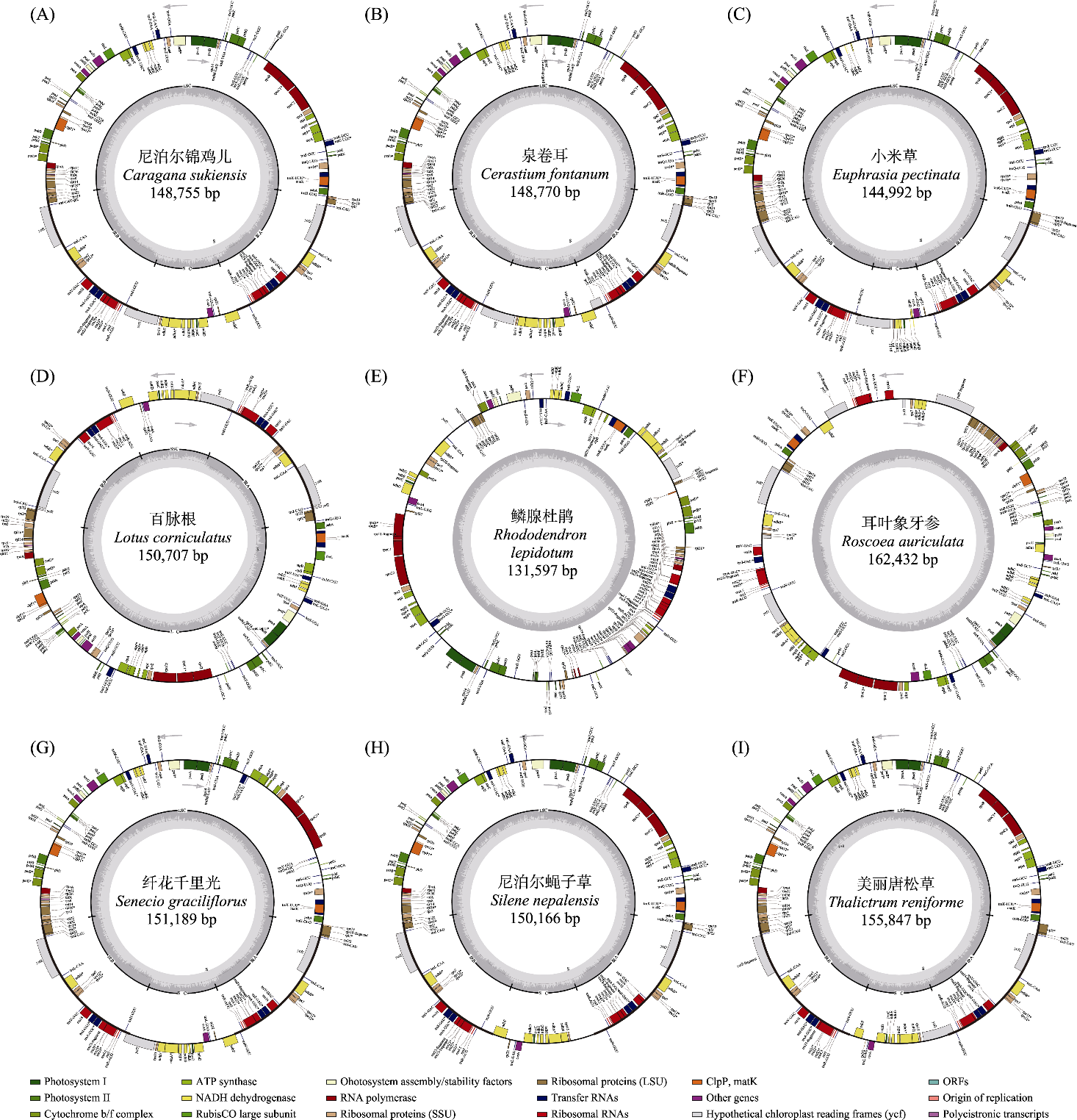
Fig. 2 Chloroplast genome circular maps of the Caragana sukiensis (A), Cerastium fontanum (B), Euphrasia pectinate (C), Lotus corniculatus (D), Rhododendron lepidotum (E), Roscoea auriculata (F), Senecio graciliflorus (G), Silene nepalensis (H) and Thalictrum reniforme (I). The outer circle of the chloroplast genome circular diagram displays the genes within the chloroplast genome along with their arrangement and orientation; the inner circle shows the CG content of different genes and regulatory regions.
| 序号 ID | 基因 Gene | 序号 ID | 基因 Gene | 序号 ID | 基因 Gene | 序号 ID | 基因 Gene |
|---|---|---|---|---|---|---|---|
| 1 | atpA | 17 | petL | 33 | psbL | 49 | rpoC2 |
| 2 | atpB | 18 | petN | 34 | psbM | 50 | rps11 |
| 3 | atpE | 19 | psaA | 35 | psbN | 51 | rps12 |
| 4 | atpF | 20 | psaB | 36 | psbT | 52 | rps14 |
| 5 | atpI | 21 | psaC | 37 | psbZ | 53 | rps15 |
| 6 | ccsA | 22 | psaI | 38 | rbcL | 54 | rps16 |
| 7 | cemA | 23 | psaJ | 39 | rpl14 | 55 | rps18 |
| 8 | clpP | 24 | psbA | 40 | rpl16 | 56 | rps19 |
| 9 | matK | 25 | psbB | 41 | rpl20 | 57 | rps2 |
| 10 | ndhD | 26 | psbC | 42 | rpl22 | 58 | rps3 |
| 11 | ndhF | 27 | psbD | 43 | rpl23 | 59 | rps4 |
| 12 | ndhJ | 28 | psbE | 44 | rpl32 | 60 | rps7 |
| 13 | ndhK | 29 | psbF | 45 | rpl36 | 61 | rps8 |
| 14 | petA | 30 | psbI | 46 | rpoA | 62 | ycf1 |
| 15 | petB | 31 | psbJ | 47 | rpoB | 63 | ycf2 |
| 16 | petD | 32 | psbK | 48 | rpoC1 | 64 | ycf3 |
Table 1 List of chloroplast protein coding genes used for database construction
| 序号 ID | 基因 Gene | 序号 ID | 基因 Gene | 序号 ID | 基因 Gene | 序号 ID | 基因 Gene |
|---|---|---|---|---|---|---|---|
| 1 | atpA | 17 | petL | 33 | psbL | 49 | rpoC2 |
| 2 | atpB | 18 | petN | 34 | psbM | 50 | rps11 |
| 3 | atpE | 19 | psaA | 35 | psbN | 51 | rps12 |
| 4 | atpF | 20 | psaB | 36 | psbT | 52 | rps14 |
| 5 | atpI | 21 | psaC | 37 | psbZ | 53 | rps15 |
| 6 | ccsA | 22 | psaI | 38 | rbcL | 54 | rps16 |
| 7 | cemA | 23 | psaJ | 39 | rpl14 | 55 | rps18 |
| 8 | clpP | 24 | psbA | 40 | rpl16 | 56 | rps19 |
| 9 | matK | 25 | psbB | 41 | rpl20 | 57 | rps2 |
| 10 | ndhD | 26 | psbC | 42 | rpl22 | 58 | rps3 |
| 11 | ndhF | 27 | psbD | 43 | rpl23 | 59 | rps4 |
| 12 | ndhJ | 28 | psbE | 44 | rpl32 | 60 | rps7 |
| 13 | ndhK | 29 | psbF | 45 | rpl36 | 61 | rps8 |
| 14 | petA | 30 | psbI | 46 | rpoA | 62 | ycf1 |
| 15 | petB | 31 | psbJ | 47 | rpoB | 63 | ycf2 |
| 16 | petD | 32 | psbK | 48 | rpoC1 | 64 | ycf3 |
| [1] |
Bagella S, Satta A, Floris I, Caria MC, Rossetti I, Podani J (2013) Effects of plant community composition and flowering phenology on honeybee foraging in Mediterranean sylvo-pastoral systems. Applied Vegetation Science, 16, 689-697.
DOI URL |
| [2] |
Bell KL, de Vere N, Keller A, Richardson RT, Gous A, Burgess KS, Brosi BJ (2016) Pollen DNA barcoding: Current applications and future prospects. Genome, 59, 629-640.
DOI PMID |
| [3] |
Bell KL, Petit RA III, Cutler A, Dobbs EK, MacPherson JM, Read TD, Burgess KS, Brosi BJ (2021) Comparing whole-genome shotgun sequencing and DNA metabarcoding approaches for species identification and quantification of pollen species mixtures. Ecology and Evolution, 11, 16082-16098.
DOI PMID |
| [4] |
Chen SF, Zhou YQ, Chen YR, Gu J (2018) fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34, 884-890.
DOI PMID |
| [5] |
Corso G, Torres Cruz CP, Pinto MP, de Almeida AM, Lewinsohn TM (2015) Binary versus weighted interaction networks. Ecological Complexity, 23, 68-72.
DOI URL |
| [6] |
Crampton-Platt A, Yu DW, Zhou X, Vogler AP (2016) Mitochondrial metagenomics: Letting the genes out of the bottle. GigaScience, 5, 15.
DOI PMID |
| [7] | Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Research, 45, e18. |
| [8] |
Fang Q, Huang SQ (2012) Progress in pollination networks: Network structure and dynamics. Biodiversity Science, 20, 300-307. (in Chinese with English abstract)
DOI |
|
[方强, 黄双全 (2012) 传粉网络的研究进展: 网络的结构和动态. 生物多样性, 20, 300-307.]
DOI |
|
| [9] | Greiner S, Lehwark P, Bock R (2019) OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research, 47, W59-W64. |
| [10] |
Hua ZY, Tian DM, Jiang C, Song SH, Chen ZY, Zhao YY, Jin Y, Huang LQ, Zhang Z, Yuan Y (2022) Towards comprehensive integration and curation of chloroplast genomes. Plant Biotechnology Journal, 20, 2239-2241.
DOI PMID |
| [11] |
Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ (2020) GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology, 21, 241.
DOI |
| [12] | Kaiser-Bunbury CN, Mougal J, Whittington AE, Valentin T, Gabriel R, Olesen JM, Blüthgen N (2017) Ecosystem restoration strengthens pollination network resilience and function. Nature, 542, 223-227. |
| [13] |
Khansari E, Zarre S, Alizadeh K, Attar F, Aghabeigi F, Salmaki Y (2012) Pollen morphology of Campanula (Campanulaceae) and allied genera in Iran with special focus on its systematic implication. Flora, 207, 203-211.
DOI URL |
| [14] |
Lamb PD, Hunter E, Pinnegar JK, Creer S, Davies RG, Taylor MI (2019) How quantitative is metabarcoding: A meta-analytical approach. Molecular Ecology, 28, 420-430.
DOI PMID |
| [15] |
Lang DD, Tang M, Hu JH, Zhou X (2019) Genome-skimming provides accurate quantification for pollen mixtures. Molecular Ecology Resources, 19, 1433-1446.
DOI PMID |
| [16] |
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754-1760.
DOI PMID |
| [17] |
Pornon A, Escaravage N, Burrus M, Holota H, Khimoun A, Mariette J, Pellizzari C, Iribar A, Etienne R, Taberlet P, Vidal M, Winterton P, Zinger L, Andalo C (2016) Using metabarcoding to reveal and quantify plant-pollinator interactions. Scientific Reports, 6, 27282.
DOI PMID |
| [18] |
Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, Kunin WE (2010) Global pollinator declines: Trends, impacts and drivers. Trends in Ecology & Evolution, 25, 345-353.
DOI URL |
| [19] |
Potts SG, Imperatriz-Fonseca V, Ngo HT, Aizen MA, Biesmeijer JC, Breeze TD, Dicks LV, Garibaldi LA, Hill R, Settele J, Vanbergen AJ (2016) Safeguarding pollinators and their values to human well-being. Nature, 540, 220-229.
DOI |
| [20] |
Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E (2012) Towards next-generation biodiversity assessment using DNA metabarcoding. Molecular Ecology, 21, 2045-2050.
DOI PMID |
| [21] | Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S (2017) GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Research, 45, W6-W11. |
| [1] | Huixia Li, Yu Li, Xin Ning, Xiaochen Li, Tianrui Wang, Yigang Song, Xiling Dai, Sisi Zheng, Xin Zhong. Genetic diversity and genetic structure of Gymnospermium kiangnanense based on chloroplast genome [J]. Biodiv Sci, 2025, 33(8): 25149-. |
| [2] | Tingting Sun, Yanwen Zhang. Research progress on pollen dimorphism [J]. Biodiv Sci, 2025, 33(7): 25031-. |
| [3] | Liao Yaqing, Huang Zefeng, Wang Xiaoyun, Zhang Libiao, Wu Yi, Yu Wenhua. An updated checklist of Chiroptera in Guangdong Province and a molecular barcode database [J]. Biodiv Sci, 2025, 33(4): 24584-. |
| [4] | Suyan Ba, Chunyan Zhao, Yuan Liu, Qiang Fang. Constructing a pollination network by identifying pollen on insect bodies: Consistency between human recognition and an AI model [J]. Biodiv Sci, 2024, 32(6): 24088-. |
| [5] | Kaiying He, Xinhui Xu, Chengyun Zhang, Zezhou Hao, Zhishu Xiao, Yingying Guo. Bioacoustics data archives management standards and management technology progress [J]. Biodiv Sci, 2024, 32(10): 24266-. |
| [6] | Xiaoqin Lü, Yang Li, Shunyu Wang, Renxiu Yao, Xiaoyue Wang. No significant differences found in chemical traits of pollen and nectar located in different positions across Aconitum piepunense racemes [J]. Biodiv Sci, 2024, 32(1): 23371-. |
| [7] | Feifei Zhang, Tianfeng Yang, Lirong Chen, Dongmei Liu, Liuyuan Yang, Duyu Yang, Peng Ju, Lu Lu. Review of pollen color diversity in Angiosperms [J]. Biodiv Sci, 2024, 32(1): 23346-. |
| [8] | Chao Xing, Yi Lin, Zhiqiang Zhou, Lianjun Zhao, Shiwei Jiang, Zhenzhen Lin, Jiliang Xu, Xiangjiang Zhan. The establishment of terrestrial vertebrate genetic resource bank and species identification based on DNA barcoding in Wanglang National Nature Reserve [J]. Biodiv Sci, 2023, 31(7): 22661-. |
| [9] | Fan Wu, Shenyun Liu, Huqiang Jiang, Qian Wang, Kaiwei Chen, Hongliang Li. Pollination difference between Apis cerana cerana and Apis mellifera ligustica during the late autumn and winter [J]. Biodiv Sci, 2023, 31(5): 22528-. |
| [10] | Demei Hu, Renxiu Yao, Yan Chen, Xiansong You, Shunyu Wang, Xiaoxin Tang, Xiaoyue Wang. Tirpitzia sinensis improves pollination accuracy by promoting the compatible pollen growth [J]. Biodiv Sci, 2021, 29(7): 887-896. |
| [11] | Qifeng Lu, Zhihuan Huang, Wenhua Luo. Characterization of complete chloroplast genome in Firmiana kwangsiensis and F. danxiaensis with extremely small populations [J]. Biodiv Sci, 2021, 29(5): 586-595. |
| [12] | Yichao Li, Yongsheng Chen, Denis Sandanov, Ao Luo, Tong Lü, Xiangyan Su, Yunpeng Liu, Qinggang Wang, Viktor Chepinoga, Sergey Dudov, Wei Wang, Zhiheng Wang. Patterns and environmental drivers of Ranunculaceae species richness and phylogenetic diversity across eastern Eurasia [J]. Biodiv Sci, 2021, 29(5): 561-574. |
| [13] | Yeqin Du, Di Zhang, Sai Wang, Lei Wang, Xingfu Yan, Zhanhui Tang. Sexual system characteristics of Lilium concolor var. megalanthum in peatland [J]. Biodiv Sci, 2021, 29(10): 1321-1335. |
| [14] | Xuehua Liu, Yuke Zhang, Xiangyu Zhao, Xiangbo He, Qiong Cai, Yun Zhu, Baisuo He, Qiang Jiu. Introduction to the wildlife camera-trapping database of the middle Qinling Mountains [J]. Biodiv Sci, 2020, 28(9): 1075-1080. |
| [15] | Yaqiong Wan, Jiaqi Li, Xingwen Yang, Sheng Li, Haigen Xu. Progress of the China mammal diversity observation network (China BON-Mammal) based on camera-trapping [J]. Biodiv Sci, 2020, 28(9): 1115-1124. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()